Abstract
The complete capsule (cap) loci from three Haemophilus influenzae strains, one serotype b (Hib) and two nonencapsulated b capsule-negative variants, were sequenced. Two new open reading frames, hcsA and hcsB, were identified in region III and thought to be involved in postpolymerization modification of the capsule. The location of the cap locus in the Haemophilus influenzae chromosome was identified within section 97 of the Rd genome (chromosomal coordinates 1074542 to 1086327) and found to be the same for the Hib and two Hib− strains as well as some other encapsulated division I H. influenzae strains.
Haemophilus influenzae is a pathogenic gram-negative bacterium responsible for a wide variety of infections in both children and adults, ranging from bronchitis to meningitis. H. influenzae is classified into two main groups: typeable (encapsulated) and nontypeable (nonencapsulated) strains. Six capsule serotypes have been described (a to f) (30). Prior to the introduction of H. influenzae serotype b (Hib) conjugate vaccines into routine use in infants in 1991, Hib caused more than 95% of all invasive H. influenzae infections in children in the United States (1, 31, 36). Nontypeable H. influenzae is most often responsible for localized respiratory tract infections (32).
Based on genetic analysis, two major phylogenetic divisions of H. influenzae, divisions I and II, have been described (27, 28). Division I consists of the majority of serotype a and b strains and all of serotype c, d, and e strains. Division II includes all of serotype f strains and some serotype a and b strains. All encapsulated H. influenzae strains, whether division I or II, contain common genes for the production of their respective polysaccharide capsules (cap genes) found within the cap locus (21). However, in division I strains, the cap locus is flanked by direct repeats of insertion element IS1016 and is frequently amplified (5, 13, 17). Although often present elsewhere in the chromosome, IS1016 does not appear to be physically associated with the cap genes in division II strains (17).
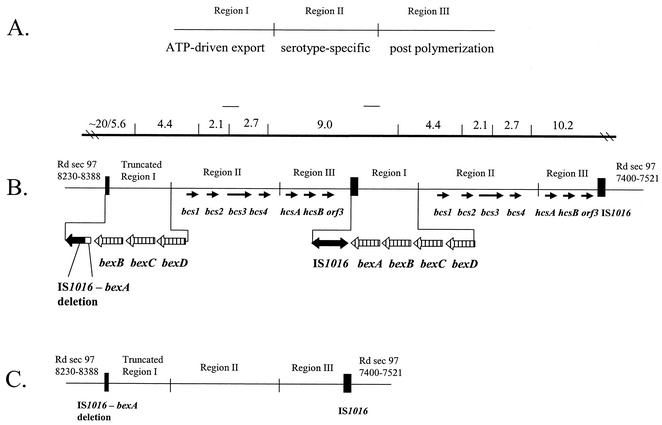
The cap loci for all serotypes consist of functionally unique regions I, II, and III (Fig. 1A) (17, 21). Regions I and III are common to all six capsular types and contain genes necessary for the processing and exportation of the capsular material. Homologs for the genes in these regions can be found in a number of other gram-negative bacteria including Escherichia coli, Neisseria meningitidis, Actinobacillus pleuropneumoniae, Pasteurella multocida, and Mannheimia haemolytica (24). Region I genes (bexDCBA) code for an ATP-driven capsule export apparatus (16). Region II contains serotype-specific biosynthesis genes that appear to be unique to each of the six capsule types (34). Region III genes appear to be involved in capsule postpolymerization steps (10, 24).
FIG. 1.
(A) Common organization of H. influenzae cap locus. Regions I and III are shared among all encapsulated H. influenzae strains. Region II contains serotype-specific genes. (B) Partially duplicated cap locus of Hib Hi 1007 showing the truncated region I with the 1.2-kb deletion between IS1016 and bexA. The top line represents an EcoRI restriction map with fragment sizes (in kilobases) shown; the line segments above the restriction map indicate the locations of probes used in Southern blot hybridization analysis to distinguish a duplicated Hib cap locus from a Hib− variant. (C) Organization of the cap locus in Hib− variants GA834 and Hi 373. Rd section 97 bp 7400 to 7521 and 8230 to 8388 correspond to Rd chromosomal coordinates 1081723 to 1081844 and 1082530 to 1082711, respectively.
The genetic elements of H. influenzae capsule production have been characterized most extensively in Hib (12, 13, 18). The majority of cap loci in Hib contain a partial duplication that results in two complete copies of regions II and III, one complete copy of region I, and a truncated copy of region I with a 1.2-kb deletion within the bexA gene and IS1016 (Fig. 1B) (18, 19). The presence of an incomplete IS1016-bexA on the end of the duplicated Hib cap locus is thought to stabilize capsule production by reducing chances of recombination events that could result in loss of cap genes. However, if during a recombination event, a complete copy of the cap locus were lost, a truncated copy would remain and would contain a partially deleted bexA gene, the ATP-binding component which is essential for the exportation of the capsule material to the surface of the bacteria. The truncated copy results in a capsule-negative phenotype known as Hib− or b− (Fig. 1C) (12, 15).
Two Hib− strains were identified, one from a collection of clinical isolates from invasive H. influenzae disease (strain GA834) (Active Bacterial Core Surveillance of the Georgia Emerging Infections Program) (M. M. Farley, unpublished data) and the other from a survey of nasopharyngeal carriage (strain Hi 373) (C. Whitney, J. Elliott, and Y.-H. Yang, unpublished data). Both isolates were found by Southern blot hybridization analysis to have 2.1- and 2.7-kb EcoRI fragments when probed with a region II capsule b-specific probe and to lack a 9-kb EcoRI fragment (normally present in the wild-type Hib cap locus) when probed with a region I (bexA-specific) probe (Fig. 1B; data not shown) (5, 15).
We sequenced the entire cap locus from clinical Hib isolate Hi 1007 (7, 25) and the Hib− variants GA834 and Hi 373, described above. Hi 1007 was isolated from cerebrospinal fluid of a child with bacterial meningitis and was confirmed as serotype b by both serologic and molecular methods in our laboratory. PCR and primers based on the known sequence of the Hib cap locus were used to generate overlapping PCR products ranging from 500 bp to 6 kb to cover the entire cap locus. Primers and the region of the cap locus amplified are shown in Table 1. Each amplicon was subcloned into pCR4-TOPO (Invitrogen, Carlsbad, Calif.), and both strands of the insert from each plasmid were sequenced by the Atlanta Veterans Affairs Medical Center/Emory University School of Medicine DNA core facility with an ABI 377 version 3.0 with d-rhodamine chemistry. The nucleotide sequences were analyzed with LaserGene, version 5.03, software programs SeqMan II, MapDraw II, and MegAlign II (DNASTAR, Inc., Madison, Wis.). Nucleotide and amino acid sequence homology comparisons were carried out with GenBank DNA and protein sequence databases by using the National Center for Biotechnology Information BLAST network server (3, 4).
TABLE 1.
PCR primers and products used for sequencing
| Primer seta | Region amplified | Template(s) | Size (bp) |
|---|---|---|---|
| Rd97(8388)-bexBb | 3′ end junction-bexB | Hi 373, GA834, Hi 1007 | 621 |
| ISLOUTb-bexBb | IS1016-bexB | Hi 373, GA834, Hi 1007 | 345 |
| bexBreva-bexD1 | bexB-bexD | Hi 373, GA834, Hi 1007 | 3,113 |
| bexC2-b5 | bexC-region II orf3 | Hi 373, GA834, Hi 1007 | 7,487 |
| b3c-orf6rev | Region II orf3-region III orf2 | Hi 373, GA834, Hi 1007 | 5,985 |
| ISLOUTb-bexBb | IS1016-bexB (bridge region) | Hi 1007 | 1,541 |
| orf6b-capHIc | Region III-bexA | Hi 1007 | 3,381 |
| orf6b-Rd97(7400) | Region III orf2-5′ end junction | Hi 373, GA834, Hi 1007 | 2,943 |
| endregIII-Rd97(7400) | End region III-5′ end junction | Hi 373, GA834, Hi 1007 | 906 |
Sequences of primers not described elsewhere are as follows: Rd97(8388), 5′-TTCCTAGTTTCCTACGTCAG-3′ (8388 corresponds to Rd chromosomal coordinate 1082711); bexBrev, 5′-CGTAAGTAACCACTGTATCGCC-3′; bexD1, 5′-CGCATAGAGGTGTGGTGGATTG-3′; bexC2, 5′-GTTGATAATCCGCAGTTTGTGTCG-3′; b5, 5′-CGTTTTTCAGCGGCGATCGC-3′; orf6rev, 5′-ACGATCACGCAAGTAATAAC-3′; endregIII, 5′-GGCCCTGTCTGCTTAATATC-3′; Rd97(7400), 5′-GCTTGGGTTCCTGTCTTGAG-3′ (7400 corresponds to Rd chromosomal coordinate 1081723).
Previously described by Leaves et al. (23).
Previously described by Falla et al. (6).
Hib cap locus of Hi 1007.
Although there is some nucleotide sequence from the Hib cap locus (accession no. X54987, X78559, and S62752), the sequence for the entire partially duplicated Hib cap locus, from end to end, particularly region III and the end junctions within the chromosome, has not been previously described. Hi 1007 contains a complete copy of region I with four genes, bexDCBA, that are 99% identical to the previously described Hib region I genes (Fig. 1B) (16) and a second, truncated copy of region I with bexDCB and bexA with the deletion previously described (19). IS1016-V5 (accession no. X58177), a truncated copy of IS1016, was found at the end adjacent to the partially deleted bexA. IS1016-V5 is one of six IS1016 variants (V1 to V6) previously described by Kroll et al. (17). Table 2 shows a comparison of Hi 1007 proteins from region I with other capsular polysaccharide (CPS) biosynthesis proteins.
TABLE 2.
Comparison of the proteins from the cap locus of H. influenzae to CPS proteins from other organisms
| H. influenzae Hi 1007 region, protein | Similar protein (source) | Accession no. | % Identity | % Similarity |
|---|---|---|---|---|
| I, BexA | CpxA (A. pleuropneumoniae) | AAB64445 | 83 | 91 |
| Wzt (M. haemolytica) | AAF08240 | 81 | 92 | |
| CtrD (N. meningitidis) | P32016 | 82 | 91 | |
| KpsT (E. coli K5) | P24586 | 43 | 66 | |
| I, BexB | CpxB (A. pleuropneumoniae) | AAB64444 | 73 | 84 |
| Wzm (M. haemolytica) | AAF08241 | 74 | 86 | |
| CtrC (N. meningitidis) | P32015 | 65 | 79 | |
| KpsM (E. coli K5) | P24584 | 25 | 43 | |
| I, BexC | CpxC (A. pleuropneumoniae) | AAB64443 | 67 | 77 |
| Wzf (M. haemolytica) | AAF08242 | 70 | 80 | |
| CtrB (N. meningitidis) | P32014 | 55 | 72 | |
| KpsE (E. coli K5) | CAA52655 | 27 | 47 | |
| I, BexD | CpxD (A. pleuropneumoniae) | AAB64442 | 74 | 86 |
| Wza (M. haemolytica) | AAF08243 | 74 | 87 | |
| CtrA (N. meningitidis) | P32013 | 55 | 72 | |
| Wza (E. coli K-12) | NP_416566 | 26 | 42 | |
| II, Orf1 (Bcs1) | Acs1 (Hia RM107) | CAA85750 | 95 | 96 |
| II, Orf2 (Bcs2) | Asc2 (Hia RM107) | CAA85751 | 71 | 67 |
| II, Orf3 (Bcs3) | Cps19bR (S. pneumoniae) | AAB66523 | 19 | 39 |
| Cj1432c (C. jejuni) | NP_282573 | 17 | 38 | |
| II, Orf4 (Bcs4) | Cps14K (S. pneumoniae) | CAA59783 | 19 | 40 |
| III, HcsA | PhyA (P. multocida) | AF067175 | 54 | 70 |
| WbrA (M. haemolytica) | AAF08250 | 56 | 69 | |
| LipA (N. meningitidis) | Q05013 | 57 | 71 | |
| KpsC (E. coli K5) | P42217 | 46 | 61 | |
| III, HcsB | PhyB (P. multocida) | AF067175 | 65 | 80 |
| WbrB (M. haemolytica) | AAF08251 | 60 | 73 | |
| LipB (N. meningitidis) | Q05014 | 55 | 68 | |
| KpsS (E. coli K5) | P42218 | 39 | 58 |
Immediately adjacent to both copies of region I is region II, containing four serotype b-associated open reading frames (ORFs) previously identified by van Eldere et al. (34) (Fig. 1B). The nucleotide sequences for region II of Hib strain RM135 (accession no. X78559) and H. influenzae type a (Hia) strain Hia RM107 (accession no. Z37516) have been reported. Only the protein products from Hib RM135 orf1 and Hia acs1 have been assigned a function, that of bifunctional ribulose 5-phosphate reductase/CDP-ribitol pyrophoshorylase (9, 34). The predicted protein encoded by Hi 1007 region II orf1 was 99% similar to the previously described Hib region II orf1 protein and 96% similar to Acs1 (Table 2). The predicted protein encoded by Hi 1007 region II orf2 is 67% similar to Acs2 from Hia. We therefore suggest that Hib region II orf1 and orf2 be named bcs1 and bcs2, for b capsule specific, based on the nomenclature proposed for the serotype a capsule-specific genes acs1 and acs2 (9).
Hi 1007 region II orf3 is 100% identical to the previously sequenced Hib region II orf3 but showed no nucleotide similarity to acs3, from region II of Hia, and their products showed no amino acid similarity. There was low-level similarity between the predicted protein of Hi 1007 orf3 and a probable sugar transferase from Campylobacter jejuni and a protein of unknown function from the cap locus of Streptococcus pneumoniae type 19B (Table 2). Hi 1007 region II orf4 is 100% identical to the previously sequenced Hib region II orf4 but showed no nucleotide similarity to acs4, from region II of Hia, and their products showed no amino acid similarity. There was low-level similarity between the predicted protein of Hi 1007 orf4 and Cps14K, a CPS synthesis protein from Streptococcus pneumoniae type 14 (Table 2). We propose that capsule-specific region II ORFs orf3 and orf4 be named bcs3 and bcs4.
Although region III of Hib is common to all serotypes, little is known about it. No sequence data from any H. influenzae serotype had been published for region III. We have identified two ORFs in region III of Hi 1007. Table 2 shows the strong similarity of both genes to cap genes involved in postpolymerization of capsules from E. coli, N. meningitidis, A. pleuropneumoniae, P. multocida, and M. haemolytica. The function of these genes in H. influenzae has yet to be determined. It has been proposed that LipA and LipB from N. meningitidis are involved in phospholipid substitution, necessary for translocation of the capsule (10). The strong consensus among the various genes and proteins involved in CPS biosynthesis suggests a shared function. In many cases the genes and proteins from region III, as well as those from region I, have been shown to be functionally interchangeable with other region III and region I genes, respectively, by complementation studies (10, 24). Furthermore, evidence strongly suggests that a phospholipid moiety is covalently associated with the Hib CPS (22). We propose naming orf1 and orf2 from region III hcsA and hcsB for Haemophilus capsule synthesis.
Hib− GA834 and Hi 373 cap loci.
The two Hib− strains identified did not contain a central 9-kb EcoRI fragment, suggesting that there was only one copy of regions I, II, and III and that the region I present was the site of the previously described IS1016-bexA deletion (19). We confirmed this finding and sequenced a single truncated copy of region I and only one copy of region II and III in both strains as described above. The sequence and arrangement of region I from GA834 and Hi 373 were found to be >99% identical to the those of the truncated copy of region I in Hi 1007. Furthermore, regions II and III from GA834 and Hi 373 were nearly identical to regions II and III from Hi 1007 with one exception. Hi 373 contained a 24-bp direct repeat found at the 3′ end of orf4 (now bcs4), an in-frame insertion that results in a slightly larger protein.
Location of the cap locus within the H. influenzae chromosome.
The cap locus from division I H. influenzae strains is flanked by direct repeats of IS1016 (17). However, it has not been previously determined whether there is a common location for the cap locus within the chromosome. The nonencapsulated H. influenzae Rd strain, for which the first completed H. influenzae genomic sequence is available, contains copies of IS1016 at three different locations within its chromosome. HI1018, a complete, putative copy of IS1016, is found in section 97 at chromosomal coordinates 1082514 to 1081941 on the negative strand of the Rd genome (8) (accession no. NC_000907). Two incomplete copies of IS1016, HI1329 and HI1577 (chromosomal coordinates 1407406 to 1407095, negative strand, and 1646128 to 1646346, respectively), are found in sections 127 and 143 of the Rd genome (8). Rd, a division I H. influenzae strain, was formerly an encapsulated serotype d strain. Therefore, if Rd lost its capsule in a recombination event between the IS1016 repeats flanking the ends of the cap locus, the predicted result would be a complete copy of IS1016 remaining, as found in section 97 (HI1018; 1082514 to 1081941 on the negative strand of the complete Rd genome). In addition, Herbert et al. suggest that the site of insertion of the cap locus in an Rd b+ transformant was at HI1018 (11).
Primers based on the region of DNA just outside of IS1016 from section 97 in Rd (Table 1) were generated and used in combination with region I (bexB) or region III (endregIII) primers to amplify DNA at the ends of the cap locus from Hi 1007, GA834, and Hi 373, as well as American Type Culture Collection (Manassas, Va.) strains ATCC 9006 (9), ATCC 9007 (14), ATCC 9008, ATCC 8142, and ATCC 700222 (33) (serotypes a, c, d, e, and f, respectively). All PCRs were performed by using an annealing temperature 5°C less than the melting temperature of the primer as calculated by the manufacturer (Sigma-Genosys, The Woodlands, Tex.).
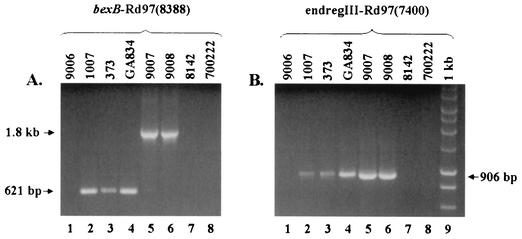
PCR amplification of the end junctions adjacent to region I [primers bexB and Rd 97(8388)] gave a 621-bp product for Hi 1007, GA834, and Hi 373, consistent with a partial deletion of IS1016-bexA at the junction in section 97 (Fig. 2A). When the same primers were used with ATCC 9007 and ATCC 9008 (serotypes c and d), a product of 1.8 kb was seen (Fig. 2A). The increase of 1.2 kb indicates the presence of complete copies of IS1016 and bexA at the left junction in these strains. Each PCR product was sequenced, confirming the presence of the truncated IS1016-V5 and partially deleted bexA in Hi 1007, GA834, and Hi 373 and a complete IS1016 (IS1016-V2, accession no. X58174) and a complete bexA in the serotype c and d strains at Rd section 97 bp 8230. PCR analysis of the end adjacent to region III [primers endregIII and Rd97(7400)] demonstrated a 906-bp product for Hi 1007, GA834, Hi 373, and the serotype c and d strains (Fig. 2B). These products were sequenced, and region III was found to contain a complete copy of IS1016-V2 adjacent to the sequence found at Rd section 97 bp 7521. We conclude that the cap loci from serotypes b, c, and d (all division I H. influenzae) appear to be located within the same chromosomal site between bp 7520 and 8230 of Rd in section 97 (Fig. 1). The only variation noted was the IS1016-bexA deletion at the region I junction in duplicated Hib strains and Hib− variants compared with a complete IS1016 and bexA forming the region I junction in serotypes c and d.
FIG. 2.
PCR analysis of the end junctions of the H. influenzae cap locus. Lanes 1, H. influenzae serotype a strain ATCC 9006; lanes 2, serotype b strain Hi 1007; lanes 3, Hib− variant Hi 373; lanes 4, Hib− variant GA834; lanes 5, serotype c strain ATCC 9007; lanes 6, serotype d strain ATCC 9008; lanes 7, serotype e strain ATCC 8142; lanes 8, serotype f strain ATCC 700222; lane 9, 1-kb standard marker (Promega Corp., Madison, Wis.). (A) Samples were amplified with primer set bexB-Rd97(8388) (Table 1). The truncated end junctions of Hib (lane 2) and the Hib− variants (lanes 3 and 4) yielded a product of 621 bp due to the 1.2-kb deletion within the IS1016-bexA region. Strains ATCC 9007 (lane 5) and ATCC 9008 (lane 6), serotypes c and d, respectively, yielded a product of 1.8 kb, consistent with an intact IS1016 and bexA. (B) Samples were amplified with primer set endregIII-Rd97(7400) (Table 1). The end junctions adjacent to region III for Hi 1007, Hi 373, GA834, ATCC 9007, and ATCC 9008 are the same size (906 bp) (lanes 2 to 6). No product is seen for serotype a (lane 1), e (lane 7), or f (lane 8).
No PCR product was amplified with either primer pair for serotypes a, e, and f (Fig. 2), suggesting the possibility of an alternative location for the capsule genes in some H. influenzae strains. Capsule serotype f strains and some serotype a strains are classified as division II, distinguished by the lack of an IS1016 element associated with the cap locus (17). Division II cap loci may therefore not be linked to the IS1016 site located in section 97 of Rd. Although serotype e strains are division I, the failure to amplify cap-specific material from the serotype e strain tested suggests that it may also contain capsule genes in a different location. However, given the greater genetic distance between serotype e and other capsule serotypes within division I (27), failure to amplify a PCR product may reflect sequence variation rather than a different cap location. Further studies will be necessary to document cap locus sequence variation among non-b serotypes and to identify alternative chromosomal sites for capsule genes.
The possession of a polysaccharide capsule greatly enhances the ability of H. influenzae to cause invasive disease (26). The Hib conjugate vaccine has resulted in the near elimination of invasive serotype b disease (1). However, the success of the Hib conjugate vaccines should not bring about complacency. The occurrence of invasive Haemophilus disease due to other capsular serotypes as well as nontypeable H. influenzae during the postvaccine era (2, 29, 33, 35) suggests that greater understanding of non-b capsules and events leading to acquisition and/or loss of capsule genes may be relevant in the future. The IS1016-bexA deletion has been demonstrated in some serotype a strains, and it potentially stabilizes encapsulation and enhances virulence in these strains (2, 20). The natural transformability of H. influenzae and the evidence that exchange of cap genes has occurred between strains in the past (20) suggest that further investigation of the cap genes from all six capsular serotypes is warranted.
The nucleotide sequences reported in this paper have been deposited in GenBank under the accession numbers AF549213 (Hi 1007), AF549210 (GA834), and AF549212 (Hi 373).
Acknowledgments
We thank Yong-Hong Yang, Beijing Children's Hospital, Beijing, People's Republic of China, and John Elliott and Cynthia Whitney of the Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention (CDC), for providing the nasopharyngeal carriage isolate; the Active Bacterial Core surveillance (ABCs) of the Emerging Infections Program Network of CDC and Wendy Baughman of the Georgia Emerging Infections Program for surveillance support; Timothy Read for critical review of the manuscript; Julie Turner Collins for technical support and help with preparing the manuscript; and the hospitals and laboratories participating in ABCs surveillance for their contributions to this project.
Editor: J. N. Weiser
REFERENCES
- 1.Adams, W. G., K. A. Deaver, S. L. Cochi, B. D. Plikaytis, E. R. Zell, C. V. Broome, and J. D. Wenger. 1993. Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA 269:221-226. [PubMed] [Google Scholar]
- 2.Adderson, E. E., C. L. Byington, L. Spencer, A. Kimball, M. Hindiyeh, K. Carroll, S. Mottice, E. K. Korgenski, J. C. Christenson, and A. T. Pavia. 2001. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era? Pediatrics 108:E18.. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corn, P. G., J. Anders, A. K. Takala, H. Kayhty, and S. K. Hoiseth. 1993. Genes involved in Haemophilus influenzae type b capsule expression are frequently amplified. J. Infect. Dis. 167:356-364. [DOI] [PubMed] [Google Scholar]
- 6.Falla, T. J., D. W. M. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farley, M. M., D. S. Stephens, S. L. Kaplan, and E. O. Mason, Jr. 1990. Pilus- and non-pilus-mediated interactions of Haemophilus influenzae type b with human erythrocytes and human nasopharyngeal mucosa. J. Infect. Dis. 161:274-280. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 9.Follens, A., M. Veiga-da-Cunha, R. Merckx, E. van Schaftingen, and J. van Eldere. 1999. acs1 of Haemophilus influenzae type a capsulation locus region II encodes a bifunctional ribulose 5-phosphate reductase-CDP-ribitol pyrophosphorylase. J. Bacteriol. 181:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frosch, M., and A. Muller. 1993. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol. Microbiol. 8:483-493. [DOI] [PubMed] [Google Scholar]
- 11.Herbert, M. A., S. Hayes, M. E. Deadman, C. M. Tang, D. W. Hood, and E. R. Moxon. 2002. Signature tagged mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb. Pathog. 33:211-223. [DOI] [PubMed] [Google Scholar]
- 12.Hoiseth, S. K., C. J. Connelly, and E. R. Moxon. 1985. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect. Immun. 49:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., E. R. Moxon, and R. P. Silver. 1986. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc. Natl. Acad. Sci. USA 83:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krieg, N. R., and J. G. Holt (ed.). 1984. Bergey's manual of systematic bacteriology, vol. 1. Lippincott, Williams & Wilkins, Baltimore, Md.
- 15.Kroll, J. S., I. Hopkins, and E. R. Moxon. 1988. Capsule loss in H. influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell 53:347-356. [DOI] [PubMed] [Google Scholar]
- 16.Kroll, J. S., B. Loynds, L. N. Brophy, and E. R. Moxon. 1990. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol. Microbiol. 4:1853-1862. [DOI] [PubMed] [Google Scholar]
- 17.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 18.Kroll, J. S., and E. R. Moxon. 1988. Capsulation and gene copy number at the cap locus of Haemophilus influenzae type b. J. Bacteriol. 170:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroll, J. S., E. R. Moxon, and B. M. Loynds. 1993. An ancestral mutation enhancing the fitness and increasing the virulence of Haemophilus influenzae type b. J. Infect. Dis. 168:172-176. [DOI] [PubMed] [Google Scholar]
- 20.Kroll, J. S., E. R. Moxon, and B. M. Loynds. 1994. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J. Infect. Dis. 169:676-679. [DOI] [PubMed] [Google Scholar]
- 21.Kroll, J. S., S. Zamze, B. Loynds, and E. R. Moxon. 1989. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J. Bacteriol. 171:3343-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo, J. S., V. W. Doelling, J. F. Graveline, and D. W. McCoy. 1985. Evidence for covalent attachment of phospholipid to the capsular polysaccharide of Haemophilus influenzae type b. J. Bacteriol. 163:769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leaves, N. I., T. J. Falla, and D. W. M. Crook. 1995. The elucidation of novel capsular genotypes of Haemophilus influenzae type b with the polymerase chain reaction. J. Med. Microbiol. 43:120-124. [DOI] [PubMed] [Google Scholar]
- 24.Lo, R. Y., L. J. McKerral, T. L. Hills, and M. Kostrzynska. 2001. Analysis of the capsule biosynthetic locus of Mannheimia (Pasteurella) haemolytica A1 and proposal of a nomenclature system. Infect. Immun. 69:4458-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason, E. O., Jr., S. L. Kaplan, B. L. Wiedermann, E. P. Norrod, and W. A. Stenback. 1985. Frequency and properties of naturally occurring adherent piliated strains of Haemophilus influenzae type b. Infect. Immun. 49:98-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moxon, E. R., and J. S. Kroll. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 150:65-85. [DOI] [PubMed] [Google Scholar]
- 27.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings., W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 28.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 85:7758-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nitta, D. M., M. A. Jackson, V. F. Burry, and L. C. Olson. 1995. Invasive Haemophilus influenzae type f disease. Pediatr. Infect. Dis. J. 14:157-160. [PubMed] [Google Scholar]
- 30.Pittman, M. 1931. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 53:471-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro, E. D., and J. I. Ward. 1991. The epidemiology and prevention of disease caused by Haemophilus influenzae type b. Epidemiol. Rev. 13:113-142. [DOI] [PubMed] [Google Scholar]
- 32.Turk, D. C. 1982. Clinical importance of Haemophilus influenzae, p. 3-9. In S. H. Sell and P. F. Wright (ed.), Haemophilus influenzae: epidemiology, immunology and prevention of disease. Elsevier/North-Holland Publishing Co., New York, N.Y.
- 33.Urwin, G., J. A. Krohn, K. Deaver-Robinson, J. D. Wenger, and M. M. Farley. 1996. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiologic characteristics in the H. influenzae serotype b vaccine era. The Haemophilus influenzae Study Group. Clin. Infect. Dis. 22:1069-1076. [DOI] [PubMed] [Google Scholar]
- 34.van Eldere, J., L. Brophy, B. Loynds, P. Celis, I. Hancock, S. Carman, J. S. Kroll, and E. R. Moxon. 1995. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107-118. [DOI] [PubMed] [Google Scholar]
- 35.Waggoner-Fountain, L. A., J. O. Hendley, E. J. Cody, V. A. Perriello, and L. G. Donowitz. 1995. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin. Infect. Dis. 21:1322-1324. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg, G. A., and D. M. Granoff. 1988. Polysaccharide-protein conjugate vaccines for the prevention of Haemophilus influenzae type b disease. J. Pediatr. 113:621-631. [DOI] [PubMed] [Google Scholar]