Abstract
Telomerase is an RNA-dependent reverse transcriptase that maintains telomeric DNA at a species-specific equilibrium length. To determine an upper limit for the number of telomerase molecules in a Saccharomyces cerevisiae cell, we have established real-time RT-PCR assays to quantify the noncoding telomerase RNA, TLC1. We find that the number of TLC1 molecules in a haploid yeast cell is ∼29, less than the number of chromosome ends (64) in late S-phase. Wild-type diploid cells contain ∼37 telomerase RNAs, while diploids heterozygous for a null tlc1 allele have half the wild-type amount, ∼19 TLC1 molecules. For comparison, there are ∼480 molecules of the U2 snRNA per haploid cell. We show that a biological consequence of this low level of telomerase is haploinsufficiency: A TLC1/tlc1Δ heterozygote maintains shorter telomeres. A dominant-negative telomerase RNA, with a deletion of the template for telomeric DNA synthesis, further demonstrates that yeast telomere length is sensitive to telomerase dosage. Sixfold overexpression of tlc1Δtemplate establishes a new telomere length set point, ∼160 bp shorter than wild type. Removing telomerase protein-interaction sites from the tlc1Δtemplate RNA mitigates the dominant-negative effect, suggesting that the tlc1Δtemplate RNA competes with wild-type TLC1 for a limited supply of telomerase proteins or for telomeres. Because yeast telomerase is tethered at chromosome ends, the finding that it may be outnumbered by its telomeric DNA substrates provides a new perspective for interpreting the results of telomere maintenance studies.
Keywords: TLC1, telomerase, real-time PCR, telomere length regulation, haploinsufficiency
INTRODUCTION
Telomeres are nucleoprotein structures that protect the ends of linear chromosomes from deleterious degradation and chromosomal fusions. Conventional DNA polymerases are unable to completely replicate telomeric DNA (de Lange et al. 2006), a dilemma that leads to progressive telomere shortening with every cell cycle. To combat this end-replication problem, most eukaryotic cells employ a reverse transcriptase called telomerase (Greider and Blackburn 1985, 1989). Telomerase is a ribonucleoprotein that minimally requires a catalytic protein subunit, called TERT (Telomerase Reverse Transcriptase) or Est2p (Ever Shorter Telomeres 2) in budding yeast (Lendvay et al. 1996; Lingner et al. 1997b), and a template-containing RNA subunit, called TERC or hTR in humans, mTR in mice, or TLC1 in budding yeast (Singer and Gottschling 1994). Telomerase works to maintain telomeric DNA at an equilibrium length that is species- and even strain-specific (Greider 1996; Smogorzewska and de Lange 2004; Shippen 2006). Recent studies in humans, mice, and budding yeast demonstrate that a threshold amount of telomerase is required to preserve telomeres at the species-specific length.
Telomerase deficiency in humans causes dyskeratosis congenita (DC), an inherited disease characterized by abnormal skin pigmentation, nail dystrophy, and bone marrow failure (Marrone et al. 2005). The autosomal dominant form of DC is linked to mutations in one copy of the gene encoding the TERC telomerase RNA (Mitchell et al. 1999; Vulliamy et al. 2001; Chen and Greider 2004). These heterozygous TERC mutations reduce the catalytic activity of telomerase via haploinsufficiency (Fu and Collins 2003; Marrone et al. 2004; Ly et al. 2005), resulting in short telomeres (Vulliamy et al. 2001). Telomeres become shorter with successive generations in DC families, concomitant with earlier and more severe disease onset (Vulliamy et al. 2004; Goldman et al. 2005). This phenomenon is called genetic anticipation.
Successive generations of mice heterozygous for the null allele of telomerase RNA (mTR+/−) also display genetic anticipation; progressive telomere shortening leads to more pronounced telomere dysfunction phenotypes in later mTR+/− generations (Hao et al. 2005). mTR+/− mice have half as much telomerase RNA as their wild-type counterparts (Hathcock et al. 2002), suggesting that mTR haploinsufficiency underlies the inability of mTR+/− mice to maintain telomeres at the equilibrium length observed in isogenic littermates (Hathcock et al. 2002; Hao et al. 2005).
In budding yeast, telomerase RNA haploinsufficiency has not been documented (Harrington 2005); however, strains heterozygous for null alleles of two telomere components show additive haploinsufficiency for telomere length maintenance (Lendvay et al. 1996; Lingner et al. 1997a). For example, a doubly heterozygous TLC1/tlc1Δ EST1/est1Δ strain has shorter telomeres than either single heterozygote (Lendvay et al. 1996).
The sensitivity of telomere length homeostasis to telomerase abundance suggests that the equilibrium length is maintained by a limiting amount of telomerase. To quantify the telomerase RNA, TLC1, in yeast, we have developed standard-based, real-time RT-PCR assays. The number of TLC1 molecules defines the maximum number of telomerase holoenzyme complexes because (1) all active telomerase complexes contain TLC1 (Hughes et al. 2000) and (2) TLC1 is a noncoding RNA, so the number of TLC1 molecules is the biologically relevant number (as opposed to mRNAs, whose messages are amplified). Furthermore, if telomerase is a dimer when active at chromosome ends (Prescott and Blackburn 1997), then the number of functional telomerase complexes could be no greater than half the number of TLC1 molecules.
We find that the yeast telomerase RNA is less abundant than its telomeric DNA substrates, i.e., newly replicated chromosome ends. A biological consequence of limiting telomerase RNA is that telomere length homeostasis is exquisitely sensitive to reductions in telomerase dosage. We show that diploid cells establish shorter telomere length set points in response to TLC1 haploinsufficiency and to dominant-negative interference by the tlc1Δtemplate RNA. Several tlc1Δtemplate RNAs that lack one or more telomerase protein-interaction sites have a diminished telomere-shortening effect, suggesting that tlc1Δtemplate RNA competes with wild-type TLC1 for limiting telomerase proteins or for telomere substrates. This study suggests a mechanism for telomerase RNA haploinsufficiency and may explain why only a fraction of yeast telomeres is extended during each cell cycle (Teixeira et al. 2004).
RESULTS
Standard-based RNA quantification assays
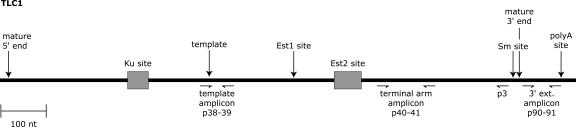
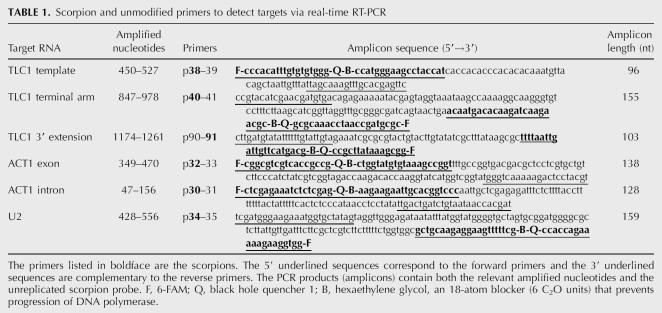
We sought to determine the number of molecules of TLC1 per yeast cell by using standard-based, real-time RT-PCR quantification. We also wanted to assess the relative abundances of two in vivo forms of TLC1—the predominant mature form (∼1.157 kb long) and a less-abundant, polyadenylated, 3′-extended form (Fig. 1 and references therein). To determine the total number (all forms) of TLC1 molecules in a cell, we selected two regions of TLC1 for quantification by real-time RT-PCR: the template amplicon and the terminal arm amplicon (Fig. 1). A third target region, the 3′-extension amplicon (Fig. 1), was chosen to detect the 3′-extended form. To monitor PCR amplification, we used unimolecular scorpion probe-primers (Whitcombe et al. 1999). The scorpion and unmodified primer sequences are listed in Table 1.
FIGURE 1.
Diagram of TLC1 RNA. The RNA shown is 1301 nt in length, based on the endpoints identified by overlapping TLC1 cDNAs (Singer and Gottschling 1994). Arrows indicate sequences <16 nt, while boxes represent sequences ≥48 nt. The mature 5′ end, at nt 11, was mapped by primer extension (Dandjinou et al. 2004). The Ku site (nt 288–335) is a stem–loop that interacts with the DNA end-binding protein Ku (Peterson et al. 2001; Stellwagen et al. 2003). The Est1 site (nt 660–664) is a bulged stem that interacts with the telomerase regulatory subunit, Est1p (Seto et al. 2002). The Est2 site (nt 751–812) is a stem–loop required for interaction with the telomerase catalytic subunit, Est2p (Chappell and Lundblad 2004). p3 marks the position (nt 1114–1143) of a TLC1-specific RT primer used in some real-time PCR assays described in this paper. The Sm site (nt 1153–1160) interacts with Sm proteins, an association critical for accumulation of the mature TLC1 RNA (Seto et al. 1999). Mature TLC1 terminates within a run of uridine residues downstream from the Sm site, around nt 1167 (Bosoy et al. 2003; Dandjinou et al. 2004). The polyA site (nt 1261) is the major 3′ end that is polyadenylated (Chapon et al. 1997).
TABLE 1.
Scorpion and unmodified primers to detect targets via real-time RT-PCR
Because there were no previously published estimates of TLC1 abundance, we also selected two control RNAs (ACT1 mRNA and U2 snRNA) that had been quantified by methods other than real-time RT-PCR. This allowed us to check whether our PCR-based quantification was consistent with independent measures. Two amplicons were chosen within the ACT1 cDNA, one within the second exon and the other within the intron (Table 1). A single amplicon was targeted in the U2 snRNA (Table 1).
In order to count molecules of TLC1, ACT1, or U2 RNA in a total RNA sample, we generated external standards, i.e., in vitro transcribed RNAs that were identical to or closely matched the in vivo targets. The four RNA standards were: TLC1(1167) to quantify total TLC1, TLC1(1261A55) to quantify 3′-extended TLC1, ACT1, and U2 (see Materials and Methods for details on the T7 transcription vectors). All purified standard RNAs were quantified by A260 and by a fluorescence-based RiboGreen assay.
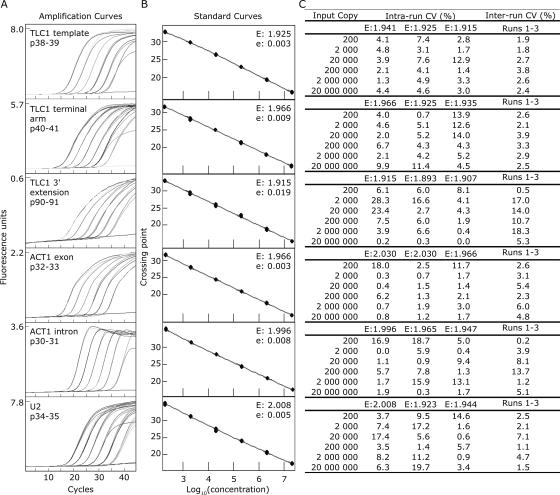
Tenfold serial dilutions of each standard RNA (from 2 × 107 to 2 × 102 molecules) were reverse-transcribed and then amplified by real-time PCR. The amplification curves were evenly spaced (Fig. 2A) and produced linear standard curves spanning 5 orders of magnitude (Fig. 2B). The slopes of the standard curves were used to calculate the amplification efficiency (E) of each assay. An ideal E value of 2 would indicate that the PCR product precisely doubled during each cycle and that the amplification efficiency was 100% ((E−1) × 100%). The observed amplification efficiencies fell between 90% and 100% (Fig. 2B). To test the reproducibility of the assays, we calculated coefficients of variation (CV%) for replicates in the same run (intrarun CV) and in three runs performed months apart with different batches of RT and PCR reagents (inter-run CV) (Fig. 2C). The inter-run variation reflected the cumulative differences in standard dilutions, reverse-transcription reactions, PCR amplifications, and reagent batches. The assays for TLC1 template, TLC1 terminal arm, ACT1 exon, ACT1 intron, and U2 were highly reproducible, with an average inter-run CV of 3.6%. The TLC1 3′-extension assay was less reproducible but still acceptable, with <20% inter-run variation. A common way to reduce assay variability would be to select different primers; however, the small size (96 nt) of the 3′ extension constrained primer and probe design. The efficiencies (E) varied slightly between different runs of the same assay (Fig. 2C); this was not unexpected because different batches of RT and PCR master mixes can perform differently. To control for these differences, we included a standard curve, amplified with the same master mixes as the unknowns, in every run.
FIGURE 2.
Real-time RT-PCR assays for TLC1, ACT1, and U2 RNAs are sensitive and reproducible. (A) Amplification curves of 10-fold serial dilutions of in vitro transcribed standard RNAs, ranging from 2 × 107 molecules (the leftmost curve) to 2 × 102 molecules (the rightmost curve). The flat line in each graph corresponds to the no-template control. Primer pairs (such as p38–39) are defined in Table 1. The standard RNA was TLC1(1261A55) for the three TLC1 assays, ACT1 for the ACT1 assays, and U2 for the U2 assay. Each PCR was run for 45 cycles. (B) Standard curves of the assays shown in A. “E” denotes the efficiency of the reaction, calculated as E = 10−(1/slope). The error associated with the fit of the line is denoted “e.” (C) Intra- and inter-run coefficients of variation (CV [%]) for the calculated number of molecules for each standard point run in triplicate. The mean and the standard deviation of the replicates were calculated and the coefficient of variation determined by dividing the standard deviation by the mean and multiplying by 100%. Runs 1–3 were performed months apart with different batches of RT and PCR reagents.
To determine the fewest number of target molecules each assay could reproducibly detect, we examined six replicates of standard dilutions containing 200, 20, or two RNA molecules. The lower detection limit, defined as the highest dilution that fell on the linear standard curve and that showed <20% variation between replicates, was 200 copies for all the assays.
Real-time RT-PCR quantification of TLC1
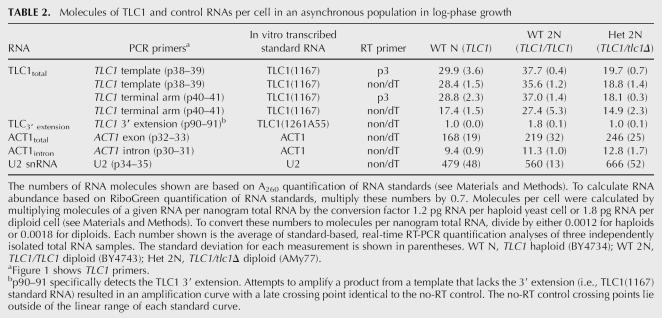
Before applying the real-time assays to yeast total RNA, we verified that (1) the primers amplified a single product of predicted size from yeast total cDNA, (2) the assays worked as efficiently with total RNA as with purified standard RNA, and (3) genomic DNA contamination did not appreciably contribute to the amplification signal (see Materials and Methods for details). We then used the standard-based assays (Fig. 2) to determine the number of telomerase RNA molecules in various haploid and diploid strains (Table 2). Two assays (TLC1 template and terminal arm) combined with the TLC1(1167) standard curve and the TLC1-specific RT primer, p3, counted the same number of TLC1 molecules per wild-type haploid yeast cell: ∼29 TLC1 RNAs. These assays also counted ∼37 TLC1 RNAs in a diploid yeast cell, and half that amount, ∼19, in a TLC1/tlc1Δ heterozygous diploid. These estimates suggest that there are fewer TLC1 RNAs than newly replicated chromosome ends per cell (haploids in late S-phase contain 64 chromosome ends and diploids contain 128 ends). Because the number of telomerase holoenzymes cannot exceed the number of TLC1 molecules, we conclude that chromosome ends outnumber telomerase in yeast.
TABLE 2.
Molecules of TLC1 and control RNAs per cell in an asynchronous population in log-phase growth
To evaluate whether our PCR quantification was consistent with other RNA quantification methods, we compared our ACT1 mRNA and U2 snRNA quantification with published estimates. Using the ACT1 exon assay, we counted ∼170 molecules per haploid cell (Table 2). Previous estimates of ACT1 mRNA abundance were: 60 copies per cell by serial analysis of gene expression (SAGE) (Velculescu et al. 1997); 40 copies per cell by kinetic PCR (Kang et al. 2000); and 20 copies per cell by high-density oligonucleotide arrays (HDAs) (Holstege et al. 1998). So our ACT1 estimate is nearly threefold higher than the highest published estimate. We estimated 480 U2 snRNAs per haploid cell, which falls within the 200–500 molecule-per-cell range determined by Northern analysis (Riedel et al. 1986).
We took advantage of these published estimates to quantify TLC1 (target) relative to U2 snRNA (Riedel et al. 1986), using a relative quantification approach that assumes perfect efficiency (E = 2) for both the target and reference assays. This ΔΔCT method does not depend on the accuracy of our standard curves. To determine the concentration ratio of TLC1 to U2 in a total RNA sample, we used a reverse transcription mix of random nonamers and oligo(dT) primers (non/dT, Qiagen) to generate an inclusive cDNA pool. The TLC1 template assay counted the same number of TLC1 molecules in cDNA populations primed either with TLC1 sequence-specific primers or with random primers (Table 2). In contrast, the TLC1 terminal arm assay detected ∼40% less TLC1 in a random-primed cDNA pool than in a TLC1 sequence specific-primed population (Table 2). This discrepancy could be explained by less frequent random priming 3′ of the terminal arm amplicon due to the close proximity (<200 nt) of the terminal arm amplicon to the mature 3′ end (for reference, the template amplicon is ∼640 nt from the end). We based the TLC1/U2 concentration ratio on the TLC1 template assay because it was consistent between RT primers. The ACT1 and U2 amplicons, each located more than 600 nt from its encoded 3′ end, were comparably detected in both random-primed and gene-specific-primed cDNA populations (data not shown). The ΔΔCT concentration ratio of total TLC1 to U2 snRNA was 0.05. Assuming the published maximum amount of U2 snRNA per cell (500 copies) (Riedel et al. 1986), the concentration ratio predicts ∼25 copies of TLC1 per cell. Both relative and standard-based quantification indicate that telomerase RNA is less abundant than chromosome ends.
Northern blot quantification of TLC1
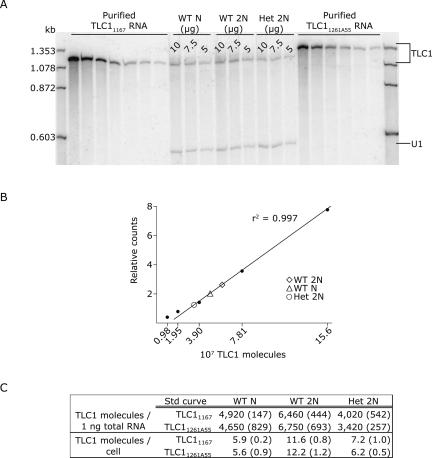
Real-time RT-PCR indicated an unexpectedly low number of telomerase RNA molecules per yeast cell. To assess TLC1 abundance by a different method, we performed quantitative Northern blot analysis using the same TLC1 RNA standards used in the real-time assays (Fig. 3A). We first experimentally determined the linear concentration range for PhosphorImager detection of TLC1 RNA. Unlike the real-time PCR assays that had a linear range spanning 5 orders of magnitude, Northern analysis of TLC1 was linear over two logs, from ∼107 to ∼109 copies; 5000-fold more total RNA was also required to detect TLC1 with the Northern. We generated a standard curve by plotting the number of TLC1 molecules against the relative PhosphorImager counts in each TLC1 RNA peak (Fig. 3B). The numbers of TLC1 molecules in the total RNA samples were interpolated from the standard curve (Fig. 3C). Northern analysis estimated the number of TLC1 molecules per cell to be about threefold lower than the real-time estimates. Quantitative Northern was also not as reproducible as real-time. A repeat Northern loaded with the identical samples shown in Figure 3A gave a twofold lower estimate of TLC1 copy number (data not shown). The following explanations may contribute to the difference between real-time and Northern estimates: (1) TLC1 RNAs greater or less than 1.157 kb (degraded, incompletely synthesized, or precursor forms) would not be counted in the Northern analysis but would be counted by real-time, providing the target amplicon was intact; (2) vagaries in band shape and size influenced the ImageQuant analysis; and (3) the signal-to-noise ratio confounded quantification by northern more than by real-time. Despite the different absolute values for TLC1 obtained by Northern and real-time, both measures led to the same biological conclusion: Yeast telomerase is not more abundant than chromosome ends.
FIGURE 3.
Quantitative Northern blot analysis of TLC1 molecules in total RNA. (A) The leftmost and rightmost sets of lanes, respectively, show twofold serial dilutions of in vitro transcribed TLC11167 RNA (beginning with 6.25 × 108 molecules) and TLC11261A55 RNA (beginning with 3.12 × 108 molecules). The center lanes show 10 μg, 7.5 μg, and 5 μg total RNA isolated from wild-type haploid cells (WT N), wild-type diploid cells (WT 2N), and TLC1/tlc1Δ heterozygous diploid cells (Het 2N). RNAs were hybridized with TLC1 and U1 snRNA probes. (B) A standard curve for TLC1 quantification was made by plotting the counts per peak area against the known amount of each standard in the TLC11167 RNA dilution series. Linear regression analysis of the seven-point curve yielded an r2 value of 0.997, indicating that the Northern assay is linear from 9.76 × 106 to 6.25 × 108 TLC1 molecules; only the five lower points are shown in the graph because the interpolated TLC1 copy numbers in total RNA fall within this lower range. The TLC1 signal in each 10 μg total RNA sample is plotted on the line. (C) The number of TLC1 molecules in each total RNA sample was interpolated from either the TLC11167 standard curve or the TLC11261A55 standard curve (r2 = 0.998). TLC1 molecules per nanogram total RNA were calculated by dividing the interpolated number by the amount of RNA loaded, i.e., either 10 μg, 7.5 μg, or 5 μg. To express these numbers relative to the A260 quantification of the RNA standards, we then multiplied the RiboGreen-based values by the concentration ratio of A260/RiboGreen (see Materials and Methods). TLC1 molecules per cell were calculated by multiplying TLC1 molecules per nanogram RNA by 0.0012 ng RNA per haploid cell or 0.0018 ng RNA per diploid cell. Each number shown is the average of the 10 μg, 7.5 μg, and 5 μg samples, with the standard deviations shown in parentheses.
TLC1 is cell density– but not cell cycle–regulated
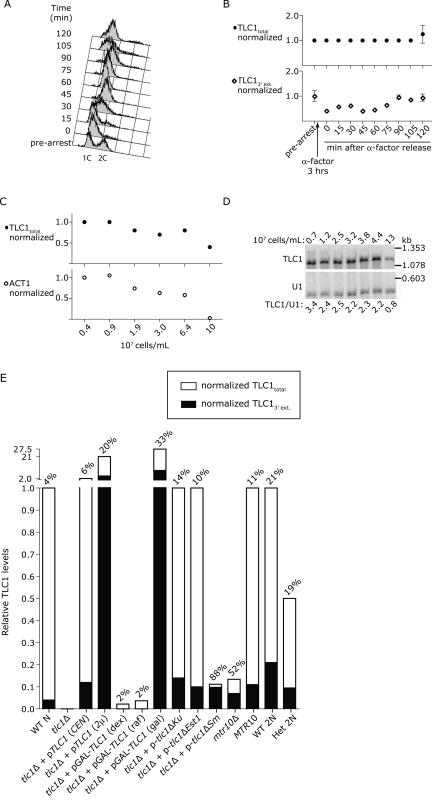
TLC1 RNA was detected at an average of 29 molecules/cell in an asynchronously growing population. To determine whether TLC1 abundance fluctuated during the cell cycle, we arrested log-phase cells in G1 with α-factor, then washed the cells to release them synchronously into the cell cycle (Fig. 4A). Total TLC1 RNA levels normalized to U2 RNA remained constant throughout the ∼1 hr cell cycle (Fig. 4B). The amount of TLC1 3′ extension decreased following α-factor arrest and recovered to prearrest levels by 90 min (Fig. 4B), suggesting that α-factor may have depressed new TLC1 transcription. The finding that total TLC1 abundance is not cell cycle–regulated corroborates previous data based on end-point RT-PCR analysis (Fisher et al. 2004) and Northern analysis (L. Harrington, pers. comm.).
FIGURE 4.
TLC1 levels are constant throughout the cell cycle, depressed at saturating cell density, and variable depending on tlc1 allele and plasmid type. (A) FACS analysis of wild-type haploid cells following release from a 3-h arrest in 5 μM α-factor. “1C” indicates haploid DNA content. At each time point shown, cells were removed from culture and either fixed for FACS anlaysis of DNA content or flash-frozen for RNA isolation. The RNA samples were reverse-transcribed using random nonamers and oligo(dT). The resulting cDNA was amplified with three PCR primer sets: p38–39 to quantify TLC1 total, p90–91 to quantify TLC1 3′ extension, and p34–35 to quantify U2. The concentration ratio of TLC1/U2 at each time point was then divided by the concentration ratio of the prearrest sample, i.e., (TLC1/U2)x/(TLC1/U2)pre-arrest, where x is each time point. (B) Graphical representation of TLC1total RNA levels (top) and TLC13′ extension RNA levels (bottom) at the time points shown in A. Each point on the graphs is the mean of two RT-PCR reactions, and the error bars show standard deviation. At each density point, RNA was isolated and reverse-transcribed using random nonamers and oligo(dT). The cDNAs were amplified with the following PCR primer sets: p38–39 to quantify TLC1 total, p32–33 to quantify ACT1 exon, and p34–35 to quantify U2. (C) Graphical representation of normalized TLC1total RNA levels (top) and ACT1 mRNA levels (bottom) at the cell density points shown below the graph. (D) Northern blot analysis of TLC1 RNA over a cell density course. RNA was isolated from the same strain as in C but from an independent cell density course. Note that U2 is used for normalization in real-time RT-PCR experiments because the U2 RNA (1.175 kb) is similar in size to TLC1 (1.167 kb), while U1 is used for normalization in Northern blots because its smaller size (0.568 kb) permits adequate separation from TLC1. (E) Relative TLC1 RNA levels in strains with different genetic contexts or different tlc1 alleles. WT N indicates wild-type haploid; (CEN), low-copy CEN plasmid; (2μ), high-copy 2-micron plasmid; (dex), dextrose media; (raf), raffinose media; (gal), galactose media; WT 2N, wild-type diploid; and Het 2N, TLC1/tlc1Δ heterozygous diploid. Total TLC1 was normalized to U2 as described in Materials and Methods, then the TLC1/U2 concentration ratio of each sample was divided by the concentration ratio of the WT N sample, i.e., (TLC1tot/U2)x/(TLC1tot/U2)WT N, where x is each strain. The TLC13′ extension was normalized to TLC1total, and the resulting percentage of total TLC1 that contains the 3′ extension is shown above each bar. Note that the Y axis is split; the bottom bar extends from 0 to 1, while the upper bar extends from 1 to 27.5.
To examine whether TLC1 abundance changed depending on cell density, we harvested RNA from cells approximately every doubling beginning with 3.5 × 106 cells/mL and ending at saturating cell density (108 cells/mL in synthetic complete media). The TLC1 levels at each density point were normalized against U2, and the concentration ratio of each sample was divided by the concentration ratio of the first density point sample, i.e., (TLC1/U2)x / (TLC1/U2)0 where x is each cell density point and 0 is the 3.5 × 106 cells/mL point. TLC1 abundance decreased by 2.5-fold at saturating cell density (Fig. 4C). These samples were also analyzed by Northern blot. The same 2.5-fold reduction in TLC1 signal was observed when TLC1 was normalized against U1. ACT1 mRNA levels decreased by 33-fold (Fig. 4C). We conducted another density course, with cells grown in YPD to allow higher saturating cell density. Northern analysis showed that TLC1 abundance decreased fourfold (Fig. 4D). Because growth phase clearly impacted TLC1 abundance, we harvested RNA only from exponentially growing cells for all the other experiments described here.
TLC1 abundance is affected by genetic context
TLC1 abundance was also affected by plasmid copy number, tlc1 allele, and genetic background (Fig. 4E). TLC1 RNA abundance in each strain tested was expressed relative to TLC1 abundance in cells where TLC1 is expressed from its endogenous locus. TLC1 levels increased twofold when expressed from a low-copy CEN plasmid, 22-fold when expressed from a high-copy 2-μ plasmid, and 27-fold when expressed from a GAL promoter in the presence of galactose for 3 h. TLC1 RNA was almost undetectable when expressed from a GAL promoter in the presence of dextrose or raffinose, and completely undetectable in tlc1::kanr cells. Two mutant RNAs, tlc1ΔKu (Peterson et al. 2001) and tlc1ΔEst1 (Seto et al. 2002), accumulated to wild-type levels, while the tlc1ΔSm RNA (Seto et al. 1999) was ∼90% less abundant than TLC1 RNA. The majority of the tlc1ΔSm RNA contained the 3′ extension, as was previously shown by Northern (Seto et al. 1999). mtr10Δ cells also showed decreased total TLC1, with an increase in the percentage of unprocessed TLC1, as was previously shown by Northern (Ferrezuelo et al. 2002). Finally, a TLC1/tlc1Δ heterozygous diploid had precisely half as much TLC1 as the wild-type diploid. The real-time assays revealed changes in TLC1 abundance that replicated published results obtained by Northern analyses, confirming that real-time PCR distinguished between biologically relevant changes in TLC1 abundance and processing.
A TLC1 heterozygote maintained short telomeres
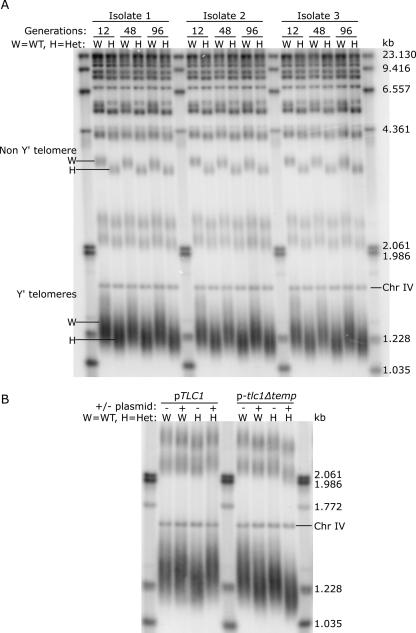
Diploid yeast cells maintained Y′-containing telomeres at an average length that varied by <10 bp among samples (Fig. 5A, W lanes). To test whether this telomere length set point was sensitive to telomerase abundance, we analyzed telomere length in TLC1/tlc1Δ heterozygotes that contained ∼19 telomerase RNAs, half the ∼37 molecules in a wild-type diploid. TLC1 heterozygosity resulted in a 40 ± 10 bp reduction in the average length of Y′-containing telomeres (Fig. 5A, H lanes). Telomeres that lacked Y′ elements were also visibly shorter (Fig. 5A, Non Y′ telomere). To determine whether the shortened telomeres reflected a new, stable set point or whether telomeres would progressively shorten with increasing generation time, we surveyed telomere lengths from cells grown for 12, 48, and 96 generations. The telomeres remained stable at the haploinsufficient set point (Fig. 5A, H lanes, 12, 48, and 96 generations).
FIGURE 5.
TLC1 haploinsufficiency leads to stably shortened telomeres, a phenotype that can be rescued by ectopic expression of wild-type TLC1. (A) To assess effects of TLC1 haploinsufficiency on telomere length, three independent isolates of TLC1/tlc1Δ heterozygous diploids (H) and an isogenic wild-type diploid (W) were each grown in liquid culture from a density of 4.88 × 104 cells/mL to 2 × 108 cells/mL, every 24 h for eight days. Genomic DNA was harvested from cells isolated on day one (12 generations), day four (48 generations), and day eight (96 generations). The Southern blot shows XhoI telomeric fragments from these samples hybridized with a telomeric probe. Chr IV is a 1.621-kb fragment of chromosome IV used as an internal mobility marker. (B) Wild-type diploid cells (W) and TLC1/tlc1Δ heterozygous diploid cells (H) were transformed with either pRS416-empty (− lanes), pRS416-TLC1 (pTLC1, + lanes) or pRS416-tlc1Δtemplate (p-tlc1Δtemp, + lanes). Genomic DNA was prepared from these strains and analyzed via Southern blot as described in A.
Three independently isolated heterozygotes had the same telomere-length phenotype, minimizing the possibility that the shortened telomeres resulted from a random, second-site mutation incurred during yeast transformation. A second indication that telomere shortening was caused by TLC1 haploinsufficiency was that exogenous TLC1 expression reversed the phenotype. Telomeres returned to wild-type lengths in TLC1/tlc1Δ cells expressing TLC1 from a CEN plasmid (Fig. 5B, pTLC1 lanes). The observed rescue depended on the functional competence of TLC1; tlc1 RNA lacking the 16-nt template (Livengood et al. 2002) failed to restore telomeres to wild-type length (Fig. 5B, p-tlc1Δtemp lanes). In fact, tlc1Δtemplate RNA had a dominant-negative effect on telomere length in both wild-type diploids and TLC1 heterozygotes.
tlc1Δtemplate RNA caused dominant-negative telomere shortening
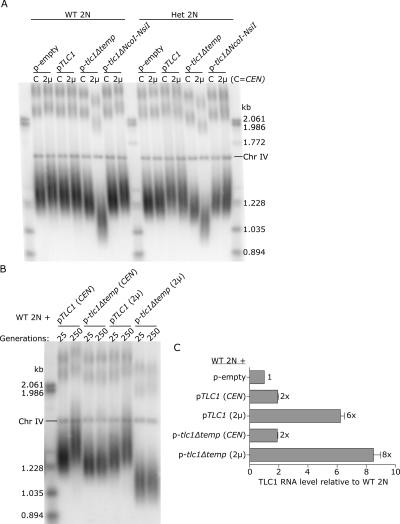
The tlc1Δtemplate RNA caused a subtle telomere-shortening defect (Fig. 5B, p-tlc1Δtemp lanes). We hypothesized that the tlc1Δtemplate RNA affected telomere length by sequestering some limiting protein subunits, thereby decreasing the number of catalytically active telomerase molecules per cell. One prediction of this hypothesis was that increasing the ratio of nonfunctional to functional telomerase RNA would result in even shorter telomeres. To test this, we expressed tlc1Δtemplate from a high-copy 2-μ plasmid in wild-type diploid cells, and analyzed telomere length (Fig. 6A). Telomeres shortened by ∼40 bp in wild-type diploids expressing a twofold excess of tlc1Δtemplate RNA and by ∼160 bp in cells expressing a sixfold excess of tlc1Δtemplate RNA. The same trends were observed in a TLC1 heterozygote (Fig. 6A, Het 2N lanes). For comparison, telomeres shortened by ∼250 bp in tlc1Δ haploid cells prior to senescence (data not shown). Telomeres did not shorten in the presence of overexpressed TLC1 or tlc1ΔNcoI-NsiI (Fig. 6A), suggesting that telomere length is not negatively affected by 2-μ expression of just any tlc1 allele.
FIGURE 6.
Telomeres become stably shorter with increasing tlc1Δtemplate overexpression. (A) Southern blot analysis of telomere lengths in wild-type diploid cells (WT 2N) and TLC1/tlc1Δ heterozygous diploid cells (Het 2N) expressing tlc1 alleles from either low-copy CEN vectors (C) or high-copy 2-μ vectors (2μ). (B) Wild-type diploid cells (WT 2N) expressing tlc1 alleles from either low-copy (CEN) or high-copy (2μ) vectors were successively streaked onto plates 10 times, for an approximate 250 generations. Genomic DNA was prepared from cells at 25 generations and at 250 generations and analyzed by Southern blot. (C) Northern blot and ImageQuant TL analysis of TLC1 RNA levels in the four strains shown in B relative to a wild-type diploid strain containing an empty plasmid. Relative RNA levels were calculated by dividing the ratio (TLC1/U1)sample by the ratio (TLC1/U1)wild-type control. Note that Northern blot analysis and real-time RT-PCR showed that 2-μ overexpression in a diploid strain is approximately threefold less than in a haploid strain. This difference may be explained by the observation that some plasmids are maintained at a lower copy number in diploids (Gullov and Friis 1985).
We next tested whether telomeres became progressively shorter with continued overexpression of tlc1Δtemplate, or whether the shortened telomeres reflected a new, stable set point. After growth on plates for 250 successive generations, cells overexpressing tlc1Δtemplate showed no signs of senescence; the control tlc1Δ haploid cells senesced by generation 100 (data not shown). The telomeres in cells expressing tlc1Δtemplate remained a constant length over the 250 generations (Fig. 6B, p-tlc1Δtemp lanes). Interestingly, exogenous TLC1 expression for 250 generations caused telomeres to lengthen; telomeres lengthened by ∼90 bp in cells expressing TLC1 from a CEN plasmid and ∼70 bp in cells expressing TLC1 from a 2-μ plasmid (Fig. 6B, pTLC1 lanes; see Fig. 6C for the relevant TLC1 RNA levels). Combined, these results suggested that yeast cells responded to the decreased relative abundance of catalytically active telomerase RNA by decreasing the telomere length set point, and responded to increased TLC1 abundance by increasing the telomere length set point. Furthermore, the telomere length regulation system is highly responsive: Even a twofold increase or decrease in TLC1 RNA levels gives a telomere-length phenotype.
The dominant-negative effect of tlc1Δtemplate is mitigated by deleting telomerase protein-interaction sites
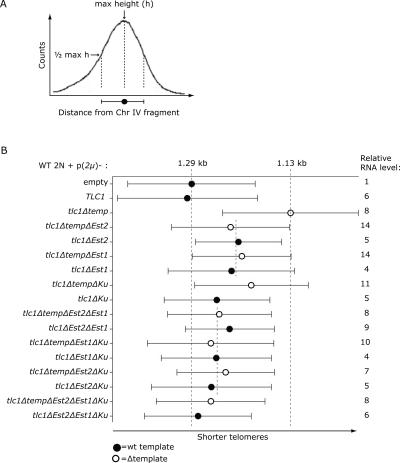
As little as a twofold excess of tlc1Δtemplate RNA over endogenous TLC1 RNA perturbed telomere length homeostasis (Fig. 6). We postulated that the dominant-negative character of the tlc1Δtemplate RNA was a consequence of its ability to bind one or more of the telomerase-associated proteins: Est2p, Est1p, and Ku. To test this, we removed the Est2p-, Est1p-, and/or Ku-binding sites (Fig. 1 and references therein) from both the tlc1Δtemplate and wild-type TLC1 RNAs. We then performed Southern blot analysis of telomere lengths in cells expressing each of the constructs shown in Figure 7. The Y′-telomere fragment lengths and length distributions were determined as shown in Figure 7A.
FIGURE 7.
The extent of telomere shortening caused by overexpressed tlc1Δtemplate RNA is proportional to the number of intact telomerase-protein binding sites. (A) Graph illustrating how telomere lengths were quantified and presented in B. An XhoI digest of yeast genomic DNA generated heterogeneously sized telomeric fragments that appeared as a broad distribution on a telomere Southern blot. The maximum height of the curve corresponds to the most highly represented telomere fragment size. The max height is indicated by a circle. The bars to either side represent the width of the distribution at half-maximum. (B) Telomere lengths in cells overexpressing tlc1 RNAs that contain targeted deletions. Wild-type diploid cells (WT 2N) were transformed with a high-copy plasmid (2 μ) containing one of the tlc1 alleles shown on the left. Telomere lengths in each strain were quantified and depicted as described in A. The RNA levels relative to the WT 2N strain expressing an empty plasmid are shown to the right. The processed form of each RNA was quantified by Northern blot and normalized against the U1 snRNA internal loading control.
The most common telomere fragment length in wild-type cells was 1.29 kb, indicated by the first long dashed line (Fig. 7B, p(2μ)-empty). The most common telomere length in cells containing the dominant-negative tlc1Δtemplate RNA was 1.13 kb, indicated by the second long dashed line (Fig. 7B, p(2μ)-tlc1Δtemp). The 160-bp difference in peak telomere length between these two samples defined the maximum telomere shortening observed. Despite the expectation that TLC1 RNAs lacking either template or the Est2p-binding site would be equally catalytically impaired, telomeres are shorter in cells containing the tlc1Δtemplate RNA (Fig. 7B, cf. third and fifth bars). An obvious difference between these mutant RNAs was that the tlc1Δtemplate RNA possessed the Est2p-binding site. When the Est2p-binding site was deleted from tlc1Δtemplate RNA, telomeres were similar in length to those observed in cells expressing the tlc1Δest2 RNA (Fig. 7B, fourth and fifth bars). These results supported the idea that the tlc1Δtemplate RNA acted as a dominant-negative either by reducing the pool of telomerase proteins available to assemble with wild-type TLC1 or by occupying telomerase-recruitment sites at telomeres.
This interpretation was upheld by the following general trend: The extent of telomere shortening was correlated with the number of protein-interaction sites present in the tlc1Δtemplate RNA. The topmost short dashed line (Fig. 7B) shows that single deletions of either the Est2p- or the Est1p-interaction site from tlc1Δtemplate RNA had a comparable effect on telomere length. The bottommost short dashed line shows that simultaneously deleting any two protein-interaction sites from the tlc1Δtemplate RNA further attenuated the telomere-shortening defect. The differences in telomere length were not an artifact of reduced RNA abundance; all the mutant RNAs accumulated stably at the predicted sizes as observed by Northern blot (data not shown) and were similarly overexpressed relative to endogenous TLC1 (Fig. 7B, relative RNA level).
DISCUSSION
We have developed standard-based real-time RT-PCR assays to count the number of the noncoding telomerase RNA (TLC1) molecules per yeast cell. This number, ∼29 TLC1 molecules in a haploid cell, defines an upper limit for the number of telomerase holoenzymes available to maintain the ends of a yeast cell's 16 linear chromosomes. A biological consequence of this low abundance of telomerase is haploinsufficiency: Diploid cells lacking one copy of TLC1 contain half as much TLC1 RNA as wild-type diploids and consequently maintain shorter telomeres. Telomere length also decreases in response to a dominant-negative tlc1Δtemplate RNA that perturbs length maintenance either by sequestering limiting telomerase proteins or by competing for telomere substrates.
Precision and accuracy of standard-based real-time RT-PCR quantification of TLC1
The measurements presented here, like all biochemical measurements, have limits on their precision and accuracy. The low intra- and inter-run coefficients of variation (Fig. 2; mostly <10%) and the low standard deviations between multiple yeast total RNA samples (Table 2) demonstrate high precision of our assays and technique. The accuracy of our measurements largely depends on two factors: how accurately we quantified the standard RNAs and how efficiently we extracted RNA from yeast cells.
We quantified the standards both by A260 and by RiboGreen fluorescence calibrated against an rRNA standard of known concentration provided by Molecular Probes (see Materials and Methods). The concentrations determined by A260 measurements were 35%–45% higher than those determined by RiboGreen. The accuracy of A260 depended on the sample's extinction coefficient, which we corrected for hypochromicity due to base-stacking, but the measurements could still be affected by free nucleotides from any RNA degradation. While RiboGreen measurements are unaffected by single nucleotides or short (<30 nt) oligos, their accuracy ultimately depends on how accurately the rRNA standards were quantified by A260. We report our TLC1 measurements based on A260 because we prefer to provide an upper limit for the number of telomerase RNPs per cell.
We isolated RNA from yeast cells by a method likely to give optimal recovery. First, we compared two methods of yeast cell lysis (see Materials and Methods) and chose the one that gave the higher and more consistent yield. Our RNA yields were extremely consistent from prep to prep (60 ± 4 μg RNA per 5 × 107 haploid cells), and we recovered 1.5-fold more RNA from equivalent numbers of diploids than haploids, consistent with the roughly 1.5-fold larger diploid cell volume. Furthermore, the resulting value of 1.2 pg RNA per haploid yeast cell agrees with the independent measurement of another group (Kang et al. 2000). Still, our RNA recovery was probably <100%, leading to some underestimation of cellular RNA and underestimation of TLC1 molecules per cell. Even an improbable 50% loss of RNA would not invalidate our conclusion that telomeric DNA substrates are more abundant than telomerase (58 molecules of TLC1 per haploid cell instead of 29 would still be <64 ends in a cell containing a replicated genome).
Finally, our quantitative Northern blot analysis (Fig. 3) suggested that the real-time analysis may overestimate TLC1 abundance by more than threefold. This discrepancy can be partially explained by the fact that the real-time analysis counts all forms of TLC1, including precursor, partially synthesized, and partially degraded molecules, which are unlikely to be assembled into active telomerase RNPs, while the Northern only counts the mature, presumably active form. While this study provides an upper limit for the number of telomerase complexes in a yeast cell, an exact number awaits a microscopic approach capable of visualizing single telomerases in a cell.
Telomerase RNA is less abundant than its telomeric DNA substrates in yeast
Both real-time RT-PCR and quantitative Northern blot analyses indicated that there are fewer TLC1 molecules per cell than newly replicated chromosome ends. This finding that telomerase is limiting with respect to its substrate suggests that telomerase recruitment and/or activation mechanisms select which telomeres to elongate. Indeed, an elegant assay designed to examine the frequency of telomerase-mediated telomere elongation events during a single cell cycle revealed that telomerase preferentially elongates the shortest telomeres, leading to a model where telomerase recruitment/activation is influenced by the “extendibility” of the telomere (Teixeira et al. 2004). Under conditions where telomere lengths are wild-type, <10% of telomeres were extended in a single cell cycle (Teixeira et al. 2004). Because telomerase is bound to telomeres by Est1p–Cdc13p interaction (Evans and Lundblad 1999), limiting telomerase may also contribute to an explanation for why so few telomeres are extended during a single cell cycle. The diploid cells analyzed in the Teixeira et al. (2004) study were TLC1/tlc1Δ. Our quantification indicates that these diploid cells would have at most ∼19 molecules of TLC1 RNA, or ∼15% as much telomerase as the 128 chromosome ends in late S-phase (when telomerase acts [Marcand et al. 2000]). Similarly low telomere elongation frequencies were observed in EST1/est1Δ diploids (Teixeira et al. 2004), and, although EST1 haploinsufficiency has not been reported, it would be interesting to know whether telomeres are elongated more frequently in cells containing the full diploid complement of telomerase components. Clearly though, telomerase abundance is not the only determinant of telomere elongation frequency. Elongation frequency increases to nearly 50% for very short telomeres and for telomeres lacking the bound length regulators Rif1p and Rif2p (Teixeira et al. 2004), suggesting that limiting telomerase RNPs can elongate more than one chromosome end in a single S-phase.
The low number of TLC1 molecules per cell also could account for the shortened-telomere phenotype observed in cells expressing mini-T forms of TLC1 (Zappulla et al. 2005). Various mini-Ts that support stably short telomeres are present at only ∼7%–23% of wild-type TLC1 levels (Zappulla et al. 2005) or, given our estimates, two to seven molecules per cell. If yeast telomerase acts as a dimer (Prescott and Blackburn 1997), then the number of enzyme complexes per cell could not exceed half the number of RNA molecules—one enzyme per cell in the case of TLC1 RNAs expressed at 7% of the wild type level.
TLC1 is not the only telomerase component expressed at low levels in yeast. Visualization of the telomerase proteins Est2p and Est1p by fluorescence microscopy required their galactose-induced overexpression from a GAL1 promoter (Teixeira et al. 2002). Additionally, the telomerase proteins Est1p, Est2p, and Est3p were among the ∼27% of the complete set of ORFs in the yeast genome that failed to be detected by immunopurification (Ghaemmaghami et al. 2003), suggesting that these protein levels are below the detection threshold for that approach. Why do yeast cells maintain telomerase components at such low levels? Would more telomerase be deleterious? GAL1-overexpression of the TLC1 RNA leads to shorter but stable telomeres (Singer and Gottschling 1994), but whether these cells also have fitness defects is unknown. An intriguing possibility is that excess telomerase could lead to more frequent healing of chromosome double-strand breaks by de novo telomere addition, a healing pathway that can result in genome instability (Pennaneach et al. 2006). Low telomerase abundance may be another safeguard against telomere addition at nontelomeric sites, along with the Pif1p helicase that promotes genome stability by inhibiting telomerase activity at broken chromosomes (Mangahas et al. 2001).
In vertebrates, harmful effects of increased telomerase components have been documented. For example, in mice, inappropriate expression of mTERT in keratinocytes resulted in the increased development of skin tumors upon carcinogen exposure (Gonzalez-Suarez et al. 2001). Similarly, activation of telomerase in human somatic cells, which normally are telomerase-negative, is a hallmark of human cancers (Kim et al. 1994). The missing or limiting telomerase component in normal human somatic cells is probably TERT, because the telomerase RNA, hTR, is present in all cell types (Feng et al. 1995). Another indication that hTR is not normally the limiting component for telomerase activity in human cells is the sheer abundance of hTR in human cell lines, as determined by competitive RT-PCR: Telomerase-negative cells contain ∼11,000–13,500 hTR molecules, and cancer cells contain ∼15,000 molecules (Yi et al. 2001). The quantitative relationships between human telomerase components in single cells isolated directly from the body remain to be elucidated, though. Clearly, hTR is not present in such vast excess that the telomerase maintenance system is buffered from twofold changes in its abundance: hTR haploinsufficiency results in the devastating bone-marrow failure disease, dyskeratosis congenita (for review, see Marrone et al. 2005). Furthermore, concomitant overexpression of both TERT and hTR was necessary to substantially increase telomerase activity in telomerase-positive cell lines (Cristofari and Lingner 2006).
Telomere length set points respond to twofold changes in telomerase abundance
This study demonstrated that diploid yeast cells heterozygous for a null mutation in TLC1 maintain shorter telomeres than wild-type diploids. A similar inability to maintain wild-type telomere lengths is observed in mTR+/− mice: mTR haploinsufficiency precludes the elongation of short telomeres to the new length set point normally attained in crosses between mice with long telomeres and mice with short telomeres (Hathcock et al. 2002). The mTR haploinsufficiency phenotypes are more severe in mice with a short-telomere genetic background; progressive telomere shortening in these mTR+/− mice correlates with limited renewal capacity of bone marrow, intestines, and testes (Hao et al. 2005).
Why is telomere length homeostasis sensitive to a twofold reduction in telomerase RNA? After all, haploinsufficiency is relatively rare. In budding yeast, only ∼3% of protein-encoding genes display haploinsufficient fitness defects in rich growth media (Deutschbauer et al. 2005). TLC1 was a priori excluded from this haploinsufficient gene set, because the yeast heterozygous deletion collection that was screened contained only open reading frames and no noncoding RNAs. Because we have not looked for a fitness defect of the TLC1/tlc1Δ strain in a competitive growth assay, we cannot predict whether TLC1 would be picked up in a 20-generation competitive fitness screen like the one described in Deutschbauer et al. (2005). It is likely that >3% of the yeast genome would be found to be haploinsufficient if sensitive assays for specific phenotypes, like telomere length, were employed.
The abundance of telomerase RNA is only one of many factors influencing telomere length: More than 150 genes have been shown to affect telomere length (Askree et al. 2004). Despite the complexity of telomere length regulation, the abundance of telomerase RNA influences telomere length in a very straightforward way: As we have shown here, the yeast telomere-maintenance machinery responds to as little as a twofold reduction or a twofold increase in the number of TLC1 molecules by establishing a telomere length set point respectively shorter or longer than wild type. These observations are compatible with a recently expanded model of telomere length regulation (Cristofari and Lingner 2006), which proposes that telomerase abundance affects the equilibrium between telomerase-associated and nonassociated states of extendible telomeres and, thereby, affects elongation frequency.
Tlc1Δtemplate RNA has a dominant-negative effect on telomere length
We demonstrated that the tlc1Δtemplate RNA caused dominant-negative telomere shortening in an abundance-dependent manner; i.e., the more tlc1Δtemplate RNA present, the shorter the telomeres. Deleting sites required for interactions with Est2p, Est1p, or Ku reduced the dominant-negative effect of the tlc1Δtemplate RNA. Two possible models to explain this observation are the following: (1) Deleting sites required for interaction with telomerase proteins reduces the likelihood that the tlc1Δtemplate RNA will interact with a telomere, and (2) deleting sites required for interaction with telomerase proteins frees up limiting telomerase proteins to associate with wild-type TLC1.
The first model posits two pools of assembled telomerase RNPs, one that incorporated wild-type TLC1 and is active, and the other that incorporated tlc1Δtemplate and is inactive. If only a limited number of telomeres recruit telomerase during each cell cycle (Teixeira et al. 2004), then the chances of recruiting an active complex from a mixed population are reduced. Preventing the association of the tlc1Δtemplate RNA with telomerase proteins involved in telomere recruitment would then reduce its competitiveness for telomere substrates. Support for this telomere occupancy model comes from the observation that tethering catalytically inactive Est2p to telomeres resulted in shortened telomeres (Downey et al. 2006). Competition for telomeres may be affected by association of telomerase with the nuclear envelope (NE); a recently proposed model reasons that a telomerase subcomponent may be sequestered at the NE, thus enhancing interactions with the NE-anchored telomeres (Hediger et al. 2006). This is an intriguing model that could explain how yeast cells promote efficient interaction between telomeres and limiting telomerase.
The second model assumes that one or more of the telomerase proteins are present in limiting amounts. While it is not clear whether the yeast telomerase proteins are limiting under the tlc1Δtemplate overexpression conditions, our observation that 2-μ overexpression of TLC1 results in telomere lengthening rather than shortening suggests either that the telomerase proteins are not limiting with respect to a sixfold increase in TLC1 RNA abundance or that a hierarchical assembly pathway controls telomerase RNP assembly, as observed in Tetrahymena (O'Connor and Collins 2006). On the other hand, two observations suggest that telomerase proteins are not present in vast excess over telomerase RNA. First, diploids heterozygous for null alleles of two telomere components show additive haploinsufficiency for telomere length maintenance (Lendvay et al. 1996; Lingner et al. 1997a). Second, mice heterozygous for a null mutation in TERT establish new, shorter telomere-length set points (Erdmann et al. 2004), demonstrating that telomerase protein components can also be limiting for telomere-length maintenance. Precise quantification of the telomerase proteins in yeast would shed light on the cellular stoichiometry of telomerase components.
The following findings implicate the abundance of the yeast telomerase RNA, TLC1, in contributing to telomere length regulation: (1) There are less telomerase RNAs per cell than chromosome ends at the end of S-phase; (2) half the normal complement of the TLC1 RNA is insufficient to maintain telomeres at the wild-type length; and (3) twice the wild-type amount of TLC1 RNA leads to an increase in telomere length over many generations. Understanding how TLC1 abundance affects the abundance and/or activities of other telomere-length regulators will ultimately increase our understanding of telomere length maintenance.
MATERIALS AND METHODS
Yeast strains and plasmids
All yeast strains were S288C descendants and were grown at 30°C in 2% dextrose, unless otherwise noted. The wild-type haploid strain (BY4734) and diploid strain (BY4743) (Brachmann et al. 1998) were purchased from ATCC (American Type Culture Collection, 200,896 and 201,390, respectively). To generate a full TLC1 knockout, the first 1262 nt of one copy of TLC1 in the diploid BY4743 were replaced with a kanMX6 module according to the PCR-mediated gene deletion protocol outlined in Longtine et al. (1998). The kanr gene was amplified from the plasmid pFA6a-kanMX6 (Longtine et al. 1998) with primers p97 (5′-GACCTTCTTTGTAGCTTTTAGTGTGATTTTTCTGGTTTGAGcggatccccgggttaattaa) and p98 (5′-TATAAATATTAAGAGGCATACCTCCGCCTATCCGCCTATCCgaattcgagctcgtttaaac) (TLC1 sequence is shown in uppercase and kanMX6 sequence in lowercase). tlc1::kanr integrants were identified by G418 resistance and verified by PCR using primer p99 (5′-CGAGCATCAAATGAAACTGC) that annealed within the kanMX6 module and primer p100 (5′-CTTCCTCTTTAGCAATGGTGAC) that annealed to the TLC1 locus 23 nt upstream of the TLC1 sequence used for homologous targeting of the kanMX6 module. Upon sporulation of the TLC1/tlc1::kanr (TLC1/tlc1Δ) strains (AMy77, AMy78, and AMy79), G418 resistance segregated 2:2 in 12 tetrads, suggesting that the kanMX6 module integrated exclusively at the TLC1 locus. One of the tlc1::kanr haploid strains, AMy87, was used as a negative control for TLC1 RNA detection in real-time RT-PCR experiments. The MTR10/mtr10Δ strain (Winzeler et al. 1999) was purchased from Open Biosystems (clone ID 22,416) and sporulated to generate an mtr10Δ haploid strain. We monitored culture growth, i.e., cell density, by counting cells with a hemacytometer.
Precise deletions in the pRS426-TLC1 and pRS426-tlc1Δtemplate vectors were made according to the QuikChange II XL site-directed mutagenesis kit protocol (Stratagene). The full TLC1 coding region was sequenced in every plasmid.
Yeast total RNA isolation
Yeast total RNA was prepared from ∼5 × 107 yeast cells harvested from cultures grown to densities between 9 × 106 and 2 × 107 cells/mL (unless otherwise stated). The cells were lysed either by spheroplasting in 50 units of zymolyase (Zymo Research), followed by addition of lysis buffer RLT (Qiagen), or by glass-bead lysis in a FastPrep machine (QBiogene). The lysates were processed according to the RNeasy Mini Kit protocol (Qiagen), with an on-column DNase treatment (Qiagen).
To determine empirically the amount of total RNA in a yeast cell, we isolated RNA from eight identical samples of 5 × 107 cells treated with zymolyase. The RNA yield from each replicate was calculated based on the A260 and total elution volume. The average yield was 60 μg ± 4 μg RNA per 5 × 107 cells, or 1.2 ± 0.08 pg RNA per haploid cell grown in YPD. The average RNA yield from a diploid cell grown in YPD was 1.8 ± 0.05 pg. The RNeasy column excludes RNAs <200 nt, leading to the systematic overestimation of the number of large RNAs per picogram of total RNA; however, this should have no effect on the calculated per-cell number of RNA molecules >200 nt. In our hands, RNA content measurements from glass-bead-treated cells were artificially low due to incomplete lysis (as determined by microscopic examination).
Preparation of RNA standards
The following DNA templates for in vitro transcription were PCR-amplified and cloned behind the T7 promoter into the EcoRI-BamHI sites of the pUC19 vector: TLC1(1167), TLC1(1261A55), ACT1, and LSR1 (U2 snRNA) (primer sequences available upon request). The TLC1(1167) and TLC1(1261A55) templates begin at TLC1 nt 11 (Fig. 1) to match the predominant 5′ end observed in vivo (Dandjinou et al. 2004). A tail of 55 adenines was incorporated into the TLC1(1261A55) construct immediately following nt 1261 to approximate the in vivo unprocessed, polyadenylated form of TLC1 (Chapon et al. 1997). The ACT1 sequence corresponds to the genomic sequence and contains an intron. The LSR1 (U2 snRNA) sequence published in the Saccharomyces Genome Database differs at 10 positions from the LSR1 sequence amplified from the S288C yeast background. The real-time PCR primers that detect U2 cDNA were designed against the S288C sequence.
Each plasmid was linearized so that runoff transcription would generate full-length transcripts with the correct 3′ end. Following in vitro transcription, the TLC1 RNAs were either gel-purified or column-purified. Prior to gel purification, each transcription reaction was phenol/chloroform-extracted, ethanol-precipitated, resuspended in water, and incubated with RQ1 DNase (Promega) for 15 min at 37°C. The RNA was then phenol/chloroform-extracted, ethanol-precipitated, and gel-purified. For the column purification, the in vitro transcription reactions were purified with the RNeasy Mini Kit (Qiagen) with an on-column DNase digestion step according to Qiagen's protocols. Gel analysis showed that the gel-purified batch was homogeneously full-length, while the column-purified RNAs showed slight heterogeneity, with faint but discrete bands in the 300- to 900-bp size range (data not shown). To test whether the two TLC1(1167) RNA preparations behaved differently in real-time RT-PCR, 10-fold serial dilutions of each were amplified with the same RT and PCR mixes. The gel-purified standard gave a 25% higher TLC1 estimate (per nanogram total RNA) than the column-purified RNA. We report the number of TLC1 molecules interpolated from the gel-purified standard (Table 2), both because the more homogeneous, gel-purified standard was used for the Northern analysis (Fig. 3) and because we preferred to err on the side of over- rather than underestimation in determining an upper limit for TLC1 abundance. The ACT1 and U2 RNAs were purified on Qiagen columns. Gel analysis verified the integrity and size of each purified RNA.
RNA quantification
The in vitro transcribed standard RNAs were quantified with the RiboGreen RNA Quantitation Kit (Molecular Probes) essentially according to the pack insert, with the following modifications to monitor fluorescence using smaller assay volumes in a LightCycler 2.0 (Roche). First, seven dilutions of the provided rRNA standard (Molecular Probes) ranging from 1000 pg/μL to 20 pg/μL in a final volume of 100 μL were prepared. Then 20 μL of each dilution were added to a 20-μL glass capillary tube (Roche). Fluorescence was monitored using Roche's LightCycler 4.0 Nucleic Acid Quantification program, which generates a standard curve of known standard concentration versus fluorescence from which the concentration of unknown samples can be calculated. Five dilutions of each “unknown” in vitro transcribed RNA were assayed, and the concentration of each was calculated from the standard curve. The lower limit of detection using a 20-μL assay volume in a LightCycler is 20 pg/μL RNA.
The concentration of each transcribed RNA was converted into number of molecules using the following equation: (6.0 × 1023 molecules/mol) × (concentration (g/μL)) / (molecular weight (g/mol)) = number of molecules/μL. The molecular weight of each RNA was based on its actual nucleotide composition. Each standard RNA was then diluted to a concentration of 1010 molecules/4 μL in water containing 10 ng/μL MS2 carrier RNA (Roche). Portions (20 μL) of this stock were flash-frozen in liquid nitrogen and stored at −80°C. Fresh dilution series were made for every PCR run.
Standard RNAs were also assayed for absorbance at 260 nm (A260). We calculated a conversion factor for the TLC1 RNAs based on nucleotide composition and the following equation: μg/mL = 1000(329.2A + 345.2G + 306.2U + 305.2C) (1 + H/100)/(15,300A + 11,800G + 10,100U + 7,400C), where 329.2 is the molecular weight of AMP within a polynucleotide, A is the number of adenines in the RNA, and 15,300 is the molar extinction coefficient of ATP at 260 nm. H is the percent hyperchromicity determined by hydrolysis of each RNA to single nucleotides or short oligonucleotides, as follows: First, we incubated ∼3 μg standard RNA with 100 units of RNase I, which cleaves after all four bases of single-stranded RNA (Ambion), and 0.2 units of RNase V1, which cleaves base-paired nucleotides (Ambion), in 15 μL total volume at 37°C for 30 min. We then measured the A260 of the undigested TLC1(1167) RNA (A260 = 0.232) and its digested counterpart (A260 = 0.301) against matched blanks, and calculated H = ((A260 digested/A260 undigested) − 1) × 100, from which we calculated ∼30% hyperchromicity for the TLC1 RNA standards. The conversion factors corrected for hypochromicity of the intact RNAs were 1 A260 unit = 36.2 μg/mL for TLC1(1167) and 35.9 μg/mL for TLC1(1261A55). The hyperchromicity for total yeast RNA was 32%, which brings the conversion factor for a mixture of free bases calculated on the basis of their individual extinction coefficients to 1 A260 unit = 39.6 μg/mL RNA, close to the standard conversion factor of 1 A260 unit = 40 μg/mL RNA. We used the standard conversion factor to calculate the concentrations of total yeast RNA and the ACT1 and U2 standard RNAs, although this would lead to an overestimation of ACT1 and U2 if the actual ACT1 and U2 conversion factors (not determined) are <40 μg/mL.
A260 measurements consistently estimated RNA concentration to be 35%–45% greater than the RiboGreen measurements. We chose to report numbers (Table 2) based on the A260 quantification of the RNA standards because we wanted to err on the side of overestimating TLC1 to see whether our conclusion that telomerase is less abundant than chromosome ends was valid for the maximum amount of TLC1 measured. Although numbers based on the A260 measurement of RNA standards are shown in Table 2, we measured each standard by both A260 and RiboGreen so that we could use the relevant concentration ratios as multiplication factors to express the numbers relative to either A260- or RiboGreen-quantified standards.
Reverse-transcription and PCR reactions
All RT and PCR reactions were set up in a Dead Air Box (AirClean Systems). To prevent PCR contamination, the 254-nm UV bulbs were turned on for 15 min prior to setting up every RT or PCR run. PCR-grade water (Roche) was used for all dilutions and reactions. Because precision in real-time RT-PCR is mostly affected by volume handling, we minimized pipetting errors by (1) pre-wetting each pipette tip before dispensing reagent, i.e., drawing liquid up, expelling, and drawing up again, (2) pre-warming the water to room temperature before pipetting to ensure accurate measurement of volumes from a room-temperature pipette tip, (3) selecting 4 μL as the minimum volume we could reproducibly pipette, (4) diluting RNAs in stepwise dilutions of ≤10-fold, and (5) double-checking the dispensed volume of water for all dilutions by drawing it back up to see whether the liquid fills the tip.
cDNAs were synthesized in 20 μL reactions using the QuantiTect Reverse Transcription kit (Qiagen), according to the protocol in the handbook (Qiagen's reverse transcriptase is neither the MMLV nor AMV reverse transcriptase but, rather, a blend of two proprietary recombinant heterodimeric proteins expressed in Escherichia coli). To synthesize cDNA from the standard RNAs, 4 μL of each dilution (equivalent to 108…103 molecules) were added to the QuantiTect mix. To synthesize cDNAs from yeast total RNA, the RNAs were serially diluted to 1.25 ng/μL into water containing 10 ng/μL MS2 carrier RNA (Roche); 4 μL (equivalent to 5 ng total RNA) were added to the QuantiTect mix. The RNAs were primed with either the random nonamer/oligo(dT) mix provided by Qiagen, or 300 nM sequence-specific RT primers (the TLC1-specific p3 sequence is: 5′-GCATCGAAGGCATTAGGAGAAGTAGCTGTG). Following reverse transcription, the cDNAs were mixed by brief vortexing, spun down in a minifuge, and placed on ice. All cDNAs were used in PCR reactions the same day they were made.
The LightCycler FastStart DNA Master-PLUS HybProbe PCR master mix (Roche) was used for all real-time PCR reactions according to Roche's protocol. Scorpion primer/probes were designed by DxS Limited and synthesized by Biosearch Technologies. The scorpions and primers were stored at a concentration of 10 μM in single-use, 34 μL aliquots at −80°C. Scorpions were thawed on ice in the dark and used within several hours of thawing. The final scorpion and primer concentration in each reaction was 500 nM each. As PCR template, 4 μL of an RT reaction were added to 16 μL of master mix directly in a glass capillary. Note that 4 μL is one-fifth of the RT reaction, so, assuming a 1:1 RNA:cDNA ratio, the 108 molecule standard becomes the 2 × 107 molecule standard; only 1 ng of the 5 ng cDNA made from yeast total RNA was PCR-amplified. The reactions were mixed by centrifugation in the LightCycler Carousel Centrifuge 2.0 and placed in the LightCycler 2.0 (Roche). The cycling parameters for all assays except the TLC1 3′-extension assay were 95°C denaturation for 10 min, followed by 45 cycles of 95°C for 10 sec, 57°C for 5 sec, and 72°C for 10 sec, followed by a 42°C cooling period for 30 sec. The cycling parameters for the TLC1 3′-extension assay were identical except the annealing temperature was 55°C.
To ascertain primer specificity, we analyzed PCR products on an agarose gel. Each primer set amplified a product of the predicted size; however, the products all appeared as closely spaced pairs of bands. These doublets most likely reflected single products with different secondary structures, owing to the covalently attached, single-stranded scorpion probe. For example, the probe could be folded in a stem–loop or could be hybridized to its target sequence within the PCR product. Removing the scorpion tail with a restriction enzyme from one of the products resolved the doublet into a single band of expected size (data not shown). Further confirmation of primer specificity came from analyses of tlc1::kanr strains. The TLC1 assays failed to amplify any product from tlc1::kanr total RNA (data not shown).
No-RT controls were included in every PCR run to assess whether genomic DNA contributed to target RNA quantification. In some cases, the no-RT controls were indistinguishable from the no-template controls; i.e., no amplification curves were produced. In cases where the no-RT control yielded an amplification curve, the crossing point was beyond the linear range of the standard curve, and the calculated copy number showed that genomic DNA contributed <1% to the target RNA quantification (data not shown).
Standard-based and relative quantification
To prepare a standard curve, an aliquot of in vitro transcribed RNA was thawed and 4 μL (containing 1010 molecules) were diluted 100-fold into 396 μL water containing 10 ng/μL MS2 carrier RNA. Then 10-fold serial dilutions were made by diluting 10 μL RNA solution into 90 μL MS2-containing water, from 108 to 10 molecules/4 μL. A portion (4 μL) of each dilution was used as template in a 20 μL RT reaction, then one-fifth of the RT reaction was added to a real-time PCR reaction. The absolute quantification module of the LightCycler 4.0 software calculated crossing points based on the first maximum of the second derivative of the amplification curve of each standard dilution. The software then plotted the crossing points against the log10 of the corresponding empirically determined concentrations and fitted a line to the data using linear regression with nonlinear polynomial fit functionality. The slope of the line is related to the amplification efficiency (E) of the reaction by the following equation: E = 10−(1/slope). The mean squared error of the fit of each data point to the regression line is denoted “e.” For absolute, e.g., standard-based, quantification, the software calculates the number of target molecules in a total RNA sample by plugging the crossing point of the unknown sample into the equation of the homologous standard curve.
Roche's relative quantification monocolor software was used to determine the concentration ratio of target to reference RNA. This software accounted for efficiency differences between the PCR assays.
Northern and Southern blot analyses
Northern and Southern blots were performed as described (Zappulla et al. 2005); the resulting PhosphorImager files were analyzed with ImageQuant (either 5.2 or TL). The molecular weight calibration feature of ImageQuant TL determined band sizes on Southern blots fairly accurately; the software measured the chromosome IV fragment internal size control (Friedman and Cech 1999) within 10 bp of its actual size.
ACKNOWLEDGMENTS
We thank Duane Marks (Roche Applied Science) and Rudy Spangler for valuable technical advice on real-time RT-PCR; David Whitcombe for scorpion design; and Alex Stemm-Wolf, Shelly Jones, and Mark Winey for help with FACS analysis. We also thank Jeffrey Pleiss, Joachim Lingner, and Lea Harrington for their helpful comments on this article. This research was supported by the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.134706.
REFERENCES
- Askree S.H., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosoy D., Peng Y., Mian I.S., Lue N.F. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chapon C., Cech T.R., Zaug A.J. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- Chappell A.S., Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Greider C.W. Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandjinou A.T., Levesque N., Larose S., Lucier J.F., Abou Elela S., Wellinger R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- de Lange T., Lundblad V., Blackburn E.H., editors. Telomeres. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- Deutschbauer A.M., Jaramillo D.F., Proctor M., Kumm J., Hillenmeyer M.E., Davis R.W., Nislow C., Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M., Houlsworth R., Maringele L., Rollie A., Brehme M., Galicia S., Guillard S., Partington M., Zubko M.K., Krogan N.J., et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Erdmann N., Liu Y., Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl. Acad. Sci. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Feng J., Funk W.D., Wang S.S., Weinrich S.L., Avilion A.A., Chiu C.P., Adams R.R., Chang E., Allsopp R.C., Yu J., et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Ferrezuelo F., Steiner B., Aldea M., Futcher B. Biogenesis of yeast telomerase depends on the importin mtr10. Mol. Cell. Biol. 2002;22:6046–6055. doi: 10.1128/MCB.22.17.6046-6055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher T.S., Taggart A.K., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Cech T.R. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes & Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goldman F., Bouarich R., Kulkarni S., Freeman S., Du H.Y., Harrington L., Mason P.J., Londono-Vallejo A., Bessler M. The effect of TERC haploinsufficiency on the inheritance of telomere length. Proc. Natl. Acad. Sci. 2005;102:17119–17124. doi: 10.1073/pnas.0505318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E., Samper E., Ramirez A., Flores J.M., Martin-Caballero J., Jorcano J.L., Blasco M.A. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Gullov K., Friis J. Maintenance and copy number control of ARS1 plasmids in Saccharomyces cerevisiae. Evidence of a mating type effect. Curr. Genet. 1985;10:21–27. doi: 10.1007/BF00418489. [DOI] [PubMed] [Google Scholar]
- Hao L.Y., Armanios M., Strong M.A., Karim B., Feldser D.M., Huso D., Greider C.W. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Harrington L. Making the most of a little: Dosage effects in eukaryotic telomere length maintenance. Chromosome Res. 2005;13:493–504. doi: 10.1007/s10577-005-0994-5. [DOI] [PubMed] [Google Scholar]
- Hathcock K.S., Hemann M.T., Opperman K.K., Strong M.A., Greider C.W., Hodes R.J. Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Berthiau A.S., van Houwe G., Gilson E., Gasser S.M. Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J. 2006;25:857–867. doi: 10.1038/sj.emboj.7600976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Evans S.K., Weilbaecher R.G., Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- Kang J.J., Watson R.M., Fisher M.E., Higuchi R., Gelfand D.H., Holland M.J. Transcript quantitation in total yeast cellular RNA using kinetic PCR. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Lendvay T.S., Morris D.K., Sah J., Balasubramanian B., Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Cech T.R., Hughes T.R., Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Livengood A.J., Zaug A.J., Cech T.R. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ly H., Schertzer M., Jastaniah W., Davis J., Yong S.L., Ouyang Q., Blackburn E.H., Parslow T.G., Lansdorp P.M. Identification and functional characterization of 2 variant alleles of the telomerase RNA template gene (TERC) in a patient with dyskeratosis congenita. Blood. 2005;106:1246–1252. doi: 10.1182/blood-2005-01-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas J.L., Alexander M.K., Sandell L.L., Zakian V.A. Repair of chromosome ends after telomere loss in Saccharomyces . Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Mann C., Gilson E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- Marrone A., Stevens D., Vulliamy T., Dokal I., Mason P.J. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood. 2004;104:3936–3942. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- Marrone A., Walne A., Dokal I. Dyskeratosis congenita: Telomerase, telomeres and anticipation. Curr. Opin. Genet. Dev. 2005;15:249–257. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- O'Connor C.M., Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell. Biol. 2006;26:2029–2036. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C.D., Kolodner R.D. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae . Mol. Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Stellwagen A.E., Diede S.J., Singer M.S., Haimberger Z.W., Johnson C.O., Tzoneva M., Gottschling D.E. The function of a stem–loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Prescott J., Blackburn E.H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes & Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel N., Wise J.A., Swerdlow H., Mak A., Guthrie C. Small nuclear RNAs from Saccharomyces cerevisiae: Unexpected diversity in abundance, size, and molecular complexity. Proc. Natl. Acad. Sci. 1986;83:8097–8101. doi: 10.1073/pnas.83.21.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug A.J., Sobel S.G., Wolin S.L., Cech T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Seto A.G., Livengood A.J., Tzfati Y., Blackburn E.H., Cech T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes & Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen D.E. Plant telomeres. In: de Lange T., et al., editors. Telomeres. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 525–550. [Google Scholar]
- Singer M.S., Gottschling D.E. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.T., Forstemann K., Gasser S.M., Lingner J. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002;3:652–659. doi: 10.1093/embo-reports/kvf133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.T., Arneric M., Sperisen P., Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and non-extendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Jr., Hieter P., Vogelstein B., Kinzler K.W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Vulliamy T.J., Knight S.W., Mason P.J., Dokal I. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol. Dis. 2001;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P.J., Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat. Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- Whitcombe D., Theaker J., Guy S.P., Brown T., Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yi X., Shay J.W., Wright W.E. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D.C., Goodrich K., Cech T.R. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]