Abstract
The β-amyloid peptide (Aβ) is a major component of the Alzheimer's disease (AD)-associated senile plaques and is generated by sequential cleavage of the β-amyloid precursor protein (APP) by β-secretase and γ-secretase. Since BACE1 initiates Aβ generation it represents a valuable target to interfere with Aβ production and treatment of AD. While the enzymatic activity of BACE1 resides in the extracellular domain, the protein also contains a short cytoplasmic tail (B1-CT). This domain serves as a binding site for at least two proteins, the copper chaperone for superoxide dismutase-1 (CCS), and the Golgi-localized, γ-ear-containing, ADP ribosylation factor-binding (GGA1) protein, and contains a single phosphorylation site. However, the precise role of the B1-CT for the overall biological function of this protein is largely unknown. Functional studies focusing on the activity of this domain would strongly benefit from the availability of domain-specific inhibitors. Here we describe the isolation and characterization of RNA aptamers that selectively target the B1-CT. We show that these RNAs bind to authentic BACE1 and provide evidence that the binding site is restricted to the membrane-proximal half of the C terminus. Aptamer-binding specifically interferes with the recruitment of CCS, but still permits GGA1 association and casein kinase-dependent phosphorylation, consistent with selective binding site targeting within this short peptide. Because phosphorylation and GGA1 binding to B1-CT regulate BACE1 transport, these RNA inhibitors could be applied to investigate B1-CT activity without affecting the subcellular localization of BACE1.
Keywords: aptamers, SELEX, Alzheimer's disease, β-secretase, CCS
INTRODUCTION
The deposition of extracellular plaques containing aggregated β-amyloid (Aβ), a 40– to 42–amino acid peptide which is generated by proteolytic processing of the β-amyloid precursor protein (APP) is a major pathological hallmark of Alzheimer's disease (AD). The initial cleavage to generate Aβ is mediated by β-secretase (BACE1) (Haass 2004). BACE1 is a type I transmembrane aspartyl protease which represents a valuable target in AD therapy (Roggo 2002) because BACE1 knockout mice do not show overt abnormalities (Luo et al. 2001; Walter et al. 2001b; Vassar 2005).
The aspartyl protease active site of the enzyme is located in the extracellular domain and consists of the motifs DTGS and DSGT (Fig. 1A). The cytoplasmic tail (CT) of BACE was found to be responsible for its subcellular localization and trafficking (von Arnim et al. 2004; He et al. 2005; Wahle et al. 2005), suggesting that modulation of BACE localization or interference with protein factors that interact with the cytoplasmic domain might also affect APP processing and Aβ formation, either directly or indirectly. At least two protein factors are known to bind directly to the CT of BACE. One of them, the Golgi-localized γ-ear-containing ARF-binding protein (GGA1) binds this domain in a phosphorylation-dependent manner via its VHS (Vps, Hrs, and STAM) domain (Walter et al. 2001a; von Arnim et al. 2004; He et al. 2005; Wahle et al. 2005). The B1-CT contains a single phosphorylation site for Casein kinase 1 at serine S498 (Fig. 1A; Walter et al. 2001a). Phosphorylation at this position is involved in the regulation of intracellular trafficking of BACE1 within the endocytic pathway, presumably by enhancing its interaction with GGA1 proteins required for the retrograde transport to the trans Golgi network (TGN) (Walter et al. 2001a). In contrast, nonphosphorylated BACE1 undergoes recycling to the cell surface (Walter et al. 2001a; He et al. 2005). The second known binding protein of B1-CT is CCS, the copper chaperone for copper–zinc superoxide dismutase 1 (SOD-1) (Angeletti et al. 2005) which is involved in post-transcriptional SOD1 activation by delivering Cu+ to the enzyme. It has been suggested that BACE1 competes with SOD-1 for binding to CCS. The CCS and SOD1 heterodimer becomes linked via a disulfide bond, which is thought to be a prerequisite for SOD1 activation by forming an intramolecular disulfide bond (Wong et al. 2000). These activities require the participation of Cu+ and Zn2+ for proper function of SOD1 that acts as an anti-oxidant enzyme by lowering the steady-state concentration of superoxide, but when mutated, it can also cause disease (Valentine and Hart 2003; Angeletti et al. 2005).
FIGURE 1.
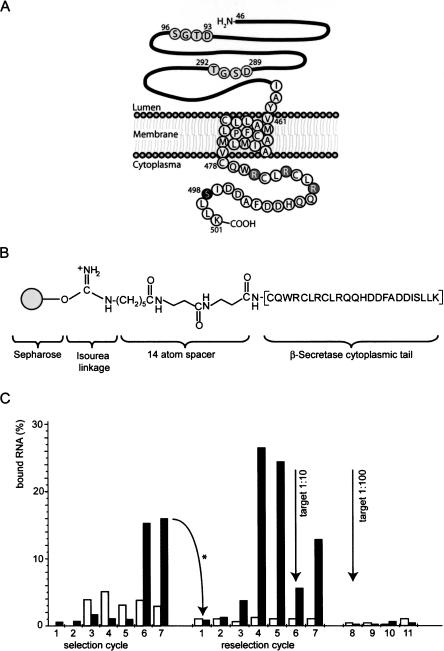
BACE cytoplasmic domain used as selection target and course of selection. (A) Schema of the 501 amino acid full-length transmembrane protein BACE1. The extracellular domain (amino acids 46–460) contains two motifs comprising the active site, located at amino acids 93–96 and 289–292 (black letters circled in gray). The transmembrane domain is comprised of amino acids 461–477; the cytoplasmic domain (amino acids 478–501) consists of 24 amino acids, including a phosphorylation site at S498 (white letter circled in black). The portion proximal to the inner plasma membrane contains the arginine residues (white letters circled in gray), whereas the ultimate C-domain is rich in acidic amino acids. (B) Chemical structure of the Sepharose-immobilized cytoplasmic domain peptide of BACE1. (C) Course of the selection. (White bars) Percentage of RNA bound to the preselection matrix (Tris-blocked Sepharose); (Black bars) percentage of RNA bound to the selection matrix. The RNA from cycle 7 was used as the input for the reselection after mutagenic PCR amplification of cycle 7 DNA (arrow with asterisk). After reselection cycle 5, the selection matrix was diluted 1:10 with preselection matrix, resulting in decrease of RNA immobilization in reselection cycle 6 and further enrichment in cycle 7. Further dilution (1:100) in cycle 8 did not result in enrichment of better binders (reselection cycles 9–11).
These examples suggest a profound role of the BACE1 cytoplasmic domain in cellular processes, but knowledge in this respect is scarce, especially with regard to an involvement of these activities in the development of AD. To gain insight into a possible role of the BACE1 C-terminal domain in these and possible other activities the ability to modulate BACE1 activity in a domain-specific fashion would be highly valuable. Gene knockout or siRNA knockdown strategies are not suitable for elucidating the role of a particular domain in a protein's functional activity. Likewise, deletion mutants of the B1-CT are retained in the endoplasmatic reticulum (ER), making investigations of the functions of the B1-CT difficult. In contrast, specific molecular inhibitors would be better suited for this purpose, the more so as most inhibitors can be applied at defined times and often lead to immediate modulation of the target's activity. However, whereas several inhibitors that bind the extracellular domain of BACE and inhibit its catalytic activity have been described (Roggo 2002; Cumming et al. 2004), no chemical modulator is known that specifically targets the B1-CT.
To fill this gap, we set out to isolate aptamers specific for the CT of BACE that would allow investigating functions of this domain. We describe here the in vitro evolution and characterization of RNA sequences that specifically bind the B1-CT with high affinity. We show that these aptamers are able to selectively block the binding of this domain to CCS while the interaction between B1-CT and GGA1, as well as the phosphorylation at S498, remain unaffected. Importantly, authentic full-length BACE1 from HEK269 cells is also specifically recognized. These inhibitors provide a basis for further functional investigation of the BACE1 C-terminal domain, also in a cellular context.
RESULTS
Selection of B1-CT binding aptamers
We used SELEX (Ellington and Szostak 1990; Tuerk and Gold 1990) to select for aptamers that recognize the cytoplasmic tail of BACE (B1-CT). The 24 amino acid peptide representing the entire cytoplasmic domain of the transmembrane protein BACE1 (Fig. 1A) was attached to cyanogen bromide (CNBr)-activated Sepharose via an N-terminal linker (Fig. 1B). A DNA library with a diversity of 1014 comprising a 40-nt random region flanked by two primer binding sites was in vitro transcribed to yield the respective RNA library. RNA was incubated with the selection matrix and after removal of nonbinding sequences by washing with binding buffer, remaining species were eluted, reverse-transcribed, and used as input DNA for the next transcription and a new selection cycle. Figure 1C illustrates the course of the selection. Black bars indicate the percentage of RNA bound to the selection column, and white bars represent RNA binding to the preselection matrix. B1-CT binding species were enriched after six cycles of selection, reverse-transcribed, cloned, and sequenced. From this first selection the monoclone K11 exhibited the best binding properties in column assays being the only clone that bound better to the B1-CT-Sepharose than the enriched pool from cycle 6 (data not shown). A dissociation constant of 4.1 μM for the binding of K11 to the free B1-CT peptide was determined using surface plasmon resonance (SPR) measurements (Table 1).
TABLE 1.
Sequences of selected aptamers that bind the cytoplasmic domain of BACE1
To improve affinity, we subjected the enriched pool from selection cycle 6 to error-prone PCR (EP-PCR) to generate a doped pool for affinity maturation (M-selection). EP-PCR was performed in 16 × 4 PCR cycles to maintain diversity of the original pool in the input of the first PCR cycles. The mutated pool M16 did not show affinity to the immobilized peptide. The M-selection was carried out for 11 further cycles with increased stringency compared to the original selection. Species that bound under these conditions were enriched after four cycles of reselection with the doped pool (Fig. 1C). The amount of target in the selection was reduced 10-fold after the fifth cycle to increase the stringency of the selection. Binding species were enriched again from cycle 7 on. After further reduction of target in reselection cycle 8, no further enrichment was achieved. Species from cycles 4, 7, and 11 were cloned and sequenced.
In another approach for affinity maturation we subjected the mono-clone K11 to 12 cycles of EP-PCR, thus generating a pool for affinity maturation which still showed ∼70% affinity of the original sequence K11. This pool K11M was also applied for 12 more rounds of selection (the K-Selection), but EP-PCR was performed after every selection cycle. The course of this reselection closely resembled the course of the M-selection. B1-CT binding species were enriched after four cycles of reselection. The amount of BACE was therefore reduced in the fifth cycle. Enrichment was again observed in cycle 9. Further decrease of BACE in the selection did not lead to a new enrichment of binders. Eluted species from cycles 4, 9, and 12 were reverse-transcribed, amplified, cloned, and sequenced.
Reselections with a newly synthesized library based on the sequence of K11 but allowing 30% of mutations per nucleotide did not lead to enrichment of B1-CT binding sequences (data not shown).
Characterization of B1-CT binding aptamers
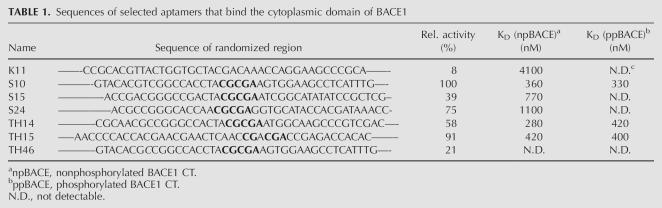
Sequences from both reselections were tested for their binding properties using column assays, and the best sequences were further characterized for their ability to bind the free B1-CT peptide in solution using surface plasmon resonance (SPR). We coupled 5′-biotinylated RNAs to Streptavidine flow-cells and injected different concentrations of B1-CT. Binding curves for a series of measurements are shown for TH14 (Fig. 2C) and S10 (Fig. 2D). Response units in equilibrium versus the respective concentration were plotted (Fig. 2E,F) and KD values were determined from logistic fits. Table 1 sums up binding sequences, their binding activities in column assays, and dissociation constants obtained from SPR measurements. Monoclone K11 from the first selection bound to the free B1-CT peptide with a KD of 4.1 μM. Affinity matured clones show significantly higher affinities: TH14, 280 nM; S10, 360 nM; TH15, 420 nM; S15, 770 nM; S24, 1100 nM. Comparing the dissociation constants of TH14 and S10 to K11 reveals that affinity maturation evolved aptamers that bind to B1-CT with >10-fold improved affinity. No binding to control proteins such as lysozyme, GST, GST-BACE-2, GST-CCS was detectable (data not shown).
FIGURE 2.
The two aptamers with highest binding affinity, TH14 and S10. (A) Secondary structure of aptamer TH14. The 5′-primer binding site is shown in magenta; the 3′-primer binding site, in cyan. The randomized region is in black. (B) Same for the aptamer S10. The nucleotides in bold are conserved between the two loop regions. (C) B1-CT binding to a flow cell derivatized with biotinylated TH14 is concentration-dependent in surface plasmon resonance (SPR) measurements. B1-CT was injected at the indicated concentrations for 120 sec, followed by a buffer wash. (D) Same for aptamer S10. (E) Curve fitting of response vs. concentration for B1-CT binding (black curve) and phosphorylated B1-CT binding (red curve) to immobilized TH14 aptamer revealed the KDs indicated in Table 1. (F) Same experiment done with S10.
Furthermore, we analyzed the selected aptamers for homologies in their primary and secondary structures. Most highly conserved among the random region of the characterized clones is a CGCGA motif (Table 1). Mfold secondary structures predict this motif to be part of a single stranded loop region (Fig. 2A,B). This highly conserved motif is likely to be involved in the interaction with the target peptide.
Aptamers bind to cellular BACE1
We next investigated whether aptamers raised against the 24-amino-acid peptide corresponding to the entire B1-CT (Fig. 1A) also recognize full length cellular BACE1. SPR measurements revealed that S10, TH14, and TH15 bind to GST-B1-CT with reduced affinity compared to the peptide but in a concentration-dependent manner (Fig. 3A). To test binding of full length BACE1 by these aptamers, we immunoprecipitated B1 from HEK269 cell lysate with an antibody against the N terminus of B1 coupled to Protein A agarose and analyzed binding of 5′-[32P]-labeled RNAs to this matrix (Fig. 3C). S10 bound to B1-agarose (9%), whereas blank agarose or antibody-derivatized agarose was not recognized (1%–1.5%). Unselected 5′-labeled pool RNA (cy0) did not bind to B1-Agarose or control matrices. This result strongly indicates that the aptamer S10 also recognizes cellular BACE1, an important prerequisite for functional analyses in cells. We also inverted the setup by using 5′-biotinylated aptamers immobilized on streptavidine magnetic beads to pull down B1 from HEK269 cell lysate. Again S10 interacted with full-length cellular B1, whereas unselected pool RNA (cy0) or blank magnetic beads did not (Fig. 3B), adding further support to the above findings.
FIGURE 3.
Aptamers bind to cellular BACE1. (A) SPR profile of GST-BACE cytoplasmic tail binding to immobilized S10 aptamer (black curve) and nonbinding GST-control protein (light gray). (B) (Top panel) Pull-down of GST-B1-CT with biotinylated RNA aptamers (S10, K11, cy0: unselected pool, no RNA); (bottom panel) pull-down of cellular BACE1 from HEK269 lysate with biotinylated RNAs (S10, cy0: unselected pool, no RNA) coupled to streptavidine coated magnetic beads. Measurements were done in duplicates. (C) Binding of [32P]-labeled RNA to cellular BACE. BACE1 from HEK269 cells is immunoprecipitated with an antibody against the N-terminal domain of BACE1 and coupled to Protein-A agarose. Bars indicate the relative amounts of aptamer (S10) or unselected pool RNA (cy0) bound to Protein A agarose derivatized with cellular full-length BACE (+), underivatized Protein A agarose (neg), or antibody-derivatized Protein A agarose (AK).
Structural characterization and footprinting of selected aptamers
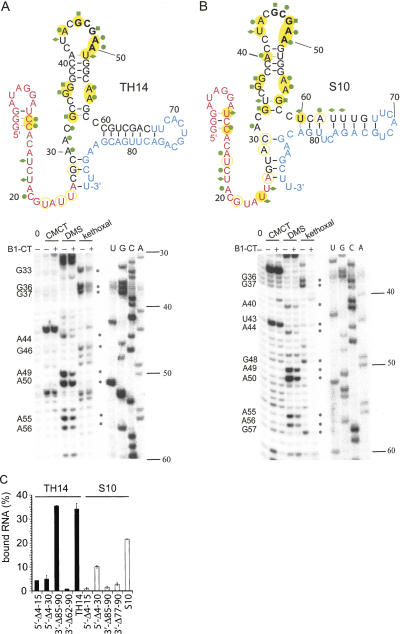
As mentioned above, the selected sequences share a CGCGA motif that is displayed in a loop region. To characterize the best binding aptamers S10 and TH14 in more detail we subjected the respective RNAs to chemical modifications in the absence and presence of the ligand B1-CT and performed primer extension reactions of the modified RNAs using a 5′-[32P]-labeled primer (Fig. 4A,B, below; Burgstaller et al. 1995). We used dimethyl sulphate (DMS) to probe unpaired adenines and cytidines, and kethoxal and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMCT) to investigate unpaired guanosines and uridines, respectively. Reverse transcriptase stops when encountering a modified base, resulting in a product one base short of the modification (Ziehler and Engelke 2000). Location of primer-extension terminations was determined by comparison to standard dideoxynucleotide sequencing reactions using the same primer and the DNA template of the respective aptamer. In this way, Watson–Crick positions of nucleotides not involved in base-pairing or tertiary hydrogen bonding can be determined.
FIGURE 4.
Footprinting of S10 and TH14 aptamers. (A) Secondary structure of TH14 including data from chemical probing (upper panel) as well as primer extension reactions after chemical modifications of TH14 aptamer with CMCT, DMS, or kethoxal in the presence (+) or absence (−) of 10 μM B1-CT (lower panel). Positions that are chemically modified in the absence of B1-CT are marked with symbols (DMS, green dots; kethoxal, green square; CMCT, green diamond). Positions with B1-CT induced changes of the chemical modification pattern are highlighted with yellow circles (closed, decrease; open, increase in signal intensity). Base-pairings stabilized by binding of B1-CT are marked in gray. Probing gel labels are 0, untreated RNA; U,G,C,A, sequencing lanes. (B) Same for the aptamer S10. (C) Binding of [32P]-labeled 5′- and 3′-deletion mutants of S10 and TH14 to B1-CT coupled to CNBr-activated Sepharose. Deleted nucleotides are referred to as Δ; the position of the nucleotides in the sequence is indicated.
Chemical probings confirmed the proposed structures of the two aptamers (Fig. 4A,B). In both TH14 and S10 the stem region (G38–C41 and G51–C54) was not modified (except for a weak DMS signal for A40 of S10), whereas most positions in the loop region (C42–A50) showed strong signals. The bulge region with unpaired G36, G37 and A55, A56 could also be confirmed by chemical probings for both aptamers. When probings were carried out in the presence of 10 μM B1-CT, most of the signals in the conserved loop and bulge regions (G36, G37, A44, G46, A49, A50, A55, A56) were significantly reduced. This indicates that these positions are either directly involved in interactions with the target peptide or that the peptide induces the formation of tertiary interactions in the aptamer. Some nucleotides of the proposed stem regions were also modified in the absence of B1-CT, and their signal was decreased when B1-CT was present in the reaction, indicating that formation of these stems is induced or stabilized in the presence of the ligand B1-CT. These base pairs are depicted in gray in Figure 4. Data from enzymatic probings were also in accordance with the chemical probing data and the proposed secondary structures of S10 and TH14 (Supplemental Figure S2 at http://famulok.chemie.uni-bonn.de/publications/current/index.html). Only in the case of the proposed stem formed between U60–U67 and G73–A77/G81, A82 we found modifications by some of the single-strand specific enzymes, indicating that this stem appears to be less stable than the corresponding one in TH14 (C60–U68 and G76–G84) or that this stem may form only partially. Alternatively, its formation in S10 may be induced or stabilized in the presence of B1-CT.
We also constructed deletion mutants of the aptamers S10 and TH14 lacking nucleotides at the 5′- or 3′-ends and tested their binding ability in column assays in comparison with the wild-type aptamers (Fig. 4C). The only deletion that was fully tolerated was a truncation of 5 nt at the 3′-end of TH14, indicating that although the GCGAA loop and the bulged regions may be primarily involved in interactions with the target, the entire sequence is needed to assure proper folding and functionality of the aptamers S10 or TH14, respectively. Interestingly, the shorter construct S10 5′-Δ4–30 binds better to B1-CT than mutant S10 5′-Δ4–15. This can be due to different secondary structures arising after truncations. Indeed, mfold predicts a structure closely resembling the full length construct for S10 5′-Δ4–30, whereas in S10 5′-Δ4–15 only the GCGAA loop and its adjacent stem are structurally conserved. The observation that most truncations from both the 5′- and the 3′-end of S10 and TH14 lead to almost complete loss of binding is in agreement with results from damage selections, where no minimal motif could be determined (data not shown).
Taken together, our data suggest that the stem–loop CUACGCGAA, comprising the conserved GCGAA motif as well as the conserved G36, G37, A55, A56 bulge, are both involved in ligand binding or ligand-induced tertiary interactions. However, these structural elements are not sufficient to ensure binding of B1-CT as deletion mutants of >5 nt show severely reduced binding affinities.
B1-CT phosphorylation and BACE/GGA1 interaction are not inhibited by anti-B1-CT aptamer
The BACE1 cytoplasmic domain can be phosphorylated at S498 within the acidic clustered dileucine (ACDL) motif by Casein kinase 1 (CK1). This modification plays a role in the regulation of subcellular trafficking of BACE1 within the endocytic pathway (Walter et al. 2001a).
To test the affinity of our aptamers to the phosphorylated version of B1-CT (ppB1-CT) we performed SPR measurements with a phosphorylated B1-CT peptide as described above and determined the dissociation constants from concentration dependent response using logistic fit (Fig. 2E,F). Interestingly, the dissociation constants of the aptamers for the phosphorylated peptide (S10, 330 nM; TH14, 420 nM; TH15, 400 nM) were in the same range as for the nonphosphorylated form (Table 1). This result suggests that the binding region for the aptamer within B1-CT is located apart from the phosphorylation site at S498. Indeed, the amino acid sequence of B1-CT harbors several basic arginine residues within its N-terminal part, whereas acidic aspartate residues are clustered in the C-terminal region, in close vicinity to S498 (Fig. 1A). It has often been observed that tight binding aptamers preferentially evolve to recognize binding epitopes that contain positively charged amino acid residues, presumably utilizing electrostatic interactions. Therefore it is plausible that the aptamers described here bind the B1-CT in the domain proximal to the plasma membrane. The finding that aptamers do not recognize the phosphorylation status of B1-CT is in accordance with this binding mode.
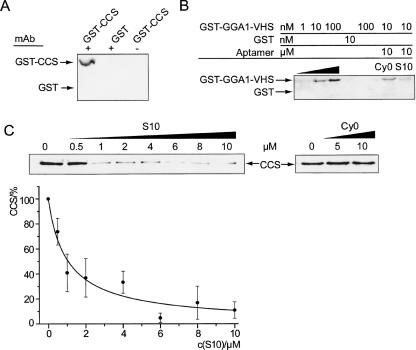
The acidic clustered dileucine (ACDL) motif that also contains the phosphorylation site within the B1-CT serves as a binding site for the Golgi-localized γ-ear-containing ARF-binding protein (GGA1). GGAs have been shown to bind directly to this motif via its VHS (Vps, Hrs, and STAM) domain and phosphorylation at S498 enhances the B1-CT/GGA1 interaction. We tested whether our aptamers interfere with the binding of B1-CT to the VHS-domain of GGA1 by using pull-down experiments with B1-CT Sepharose and GST-GGA1-VHS (Fig. 5B). We confirmed the direct interaction between B1-CT and GST-GGA1-VHS (Wahle et al. 2005) but found only a weak reduction of this interaction upon addition of a thousand-fold excess of the S10 aptamer (Fig. 5B). Furthermore, in vitro phosphorylation of B1-CT with CK1 is not inhibited (Supplemental Figure S1 at http://famulok.chemie.uni-bonn.de/publications/current/index.html). These results are in accordance with the inability of the aptamer to discriminate between phosphorylated and nonphosphorylated forms of the B1-CT. Consequently, the aptamer binds to a different region in the B1-CT than the one responsible for the interaction with GGA1.
FIGURE 5.
Specific inhibition of protein binding to B1-CT by S10. (A) CCS specifically interacts with BACE cytoplasmic tail. B1-CT derivatized Sepharose (+) or preselection matrix (−) was incubated with GST-CCS or GST as control. Precipitated protein was analyzed with anti-CCS and anti-GST antibodies. (B) Interaction of B1-CT with GST-GGA1-VHS remains unaffected by B1-CT binding aptamers. B1-CT derivatized Sepharose (lanes 1–4,6–7) or preselection matrix (lane 5) was incubated with GST-GGA1-VHS (lanes 1–3,5–7) or GST (lane 4). Addition of 10 μM cy0 (lane 6) or 10 μM S10 RNA (lane 7) did not significantly affect the interaction of B1-CT with GST-GGA1-VHS. Note that the intensity of lanes 6 and 7 should be compared to lane 2 (each with 10 nM GST-GGA1-VHS). (C) Specific inhibition of GST-CCS binding to B1-CT. Assay as shown in A but in the presence of increasing amounts of S10 aptamer (upper left panel) or unselected pool RNA cy0 (upper right panel). Curve fitting of relative CCS binding vs. concentration of S10 aptamer (lower panel) revealed a KI of 0.9 μM.
B1-CT binding aptamer inhibits BACE/CCS interaction
The membrane-proximal N-terminal part of the B1-CT is thought to interact with CCS, which is the copper chaperone for superoxide dismutase-1 (SOD-1) (Angeletti et al. 2005). We confirmed this interaction using B1-CT coupled to CNBr-activated Sepharose to pull down GST-CCS (Fig. 5A). We then tested the effect of aptamer S10 on the B1-CT/CCS interaction and found that S10 specifically inhibits this interaction in a concentration-dependent manner, whereas up to 10 μM unselected pool RNA (cy0) shows no effect (Fig. 5C). We determined an inhibition constant (KI) of 1 μM for the inhibition of the B1-CT/CCS interaction by S10 aptamer. Thus, S10 binds to a region in the N-terminal part of B1-CT and is capable of inhibiting the B1-CT/CCS interaction in vitro, whereas the interaction of B1-CT with GGA1 and CK-1 remains unaffected. We have thus generated aptamers that selectively bind a defined epitope of BACE1 within its short cytoplasmic domain. The inhibitors specifically recognize BACE1 in a cellular context. They can be applied for further investigation of the role of BACE1, especially in respect to its effects on SOD1 activation.
DISCUSSION
Since BACE1 initiates Aβ generation it represents a valuable target to interfere with Aβ production and prevention/treatment of Alzheimer's disease (Ghosh et al. 2002; Roggo 2002). Understanding the molecular mechanisms that regulate its cellular metabolism, transport, and activity, therefore, is crucial for that purpose, particularly with respect to gaining further insights into the role of the different domains of BACE1. While the enzymatic activity of BACE1 resides in the extracellular domain, the cytoplasmic domain seems to be responsible for its subcellular trafficking. It has been shown that GGAs bind to B1-CT and the subcellular trafficking of BACE1 is modulated by GGA1 in a phosphorylation state-dependent manner. In addition, B1-CT is known to bind to CCS (Angeletti et al. 2005), which is involved in post-transcriptional SOD1 activation. These examples suggest a profound role of the BACE1 cytoplasmic domain in cellular processes, but knowledge in this respect is scarce, especially with regard to an involvement of these activities in the development of AD. Functional studies focusing on the activity of this domain would strongly benefit from the availability of domain-specific inhibitors. Therefore, the RNA aptamers described here that selectively target the BACE1 cytoplasmic domain represent a first step toward this goal.
Our RNA aptamers specifically bind the cytoplasmic domain of BACE1 with dissociation constants as low as 240 nM. These affinities match with those obtained for other nonconstrained, non-nucleic acid binding peptides for which aptamers have been selected (Nieuwlandt et al. 1995; Weiss et al. 1997; Blind et al. 1999; Proske et al. 2002b). To evolve these tight-binding aptamers we used affinity maturation of the originally selected binders. We mutated both the enriched pool and the best binding monoclone K11 by EP-PCR (Cadwell and Joyce 1992) and reselected under more stringent conditions. Off-rate selection has proven an efficient means to evolve tighter binders (Burke et al. 1997; Zahnd et al. 2004). In our case, the KD value of the original best binder K11 (4.1 μM) was reduced >10-fold. The best of our new BACE1 binding aptamers, S10 and TH14, share a structurally identical domain of 27 nt in conserved loop and bulge motifs, whereas semiconserved nucleotides contribute to the formation of a stem that defines the CUACGCGAA loop, which harbors the highly conserved GCGAA sequence. The unpaired loop and bulge region are likely to be involved in interaction with B1-CT or B1-CT-induced tertiary interactions, as supported by footprinting data in the presence and absence of the ligand (Fig. 5A,B).
Importantly, selected aptamers discriminate between binding epitopes within the 24 amino acid cytoplasmic domain. We showed that the aptamer interferes with the recruitment of CCS with an inhibition constant of 1 μM (Fig. 5C). This value is in the range of the determined KD value (360 nM) for the respective S10/B1-CT interaction, considering that it was determined with a different method, and thus suggests that the aptamer and CCS share a similar or overlapping epitope within B1-CT. Consistently, S10 does not significantly block the association of B1-CT with GGA1 even when applied in a thousand-fold excess (Fig. 5B), nor does it inhibit the phosphorylation of B1-CT by CK-1 (Fig. S1). These results are in accordance with the notion that the aptamer does not recognize the ultimate C-terminal epitope, a region that is rich in acidic amino acids but, rather, the membrane proximal half of the cytoplasmic domain. Further evidence comes from the observation that the aptamers do not discriminate between unphosphorylated and phosphorylated B1-CT, consistent with the fact that the phosphorylation site is located within the acidic dileucine motif. A possible explanation for this epitope specificity within the 24 amino acid peptide may be the clustering of acidic or basic residues in the respective regions within the peptide. Indeed, the membrane-proximal epitope contains three arginine residues and fewer acidic amino acids, whereas the C-terminal half does not contain any basic amino acids but a cluster of acidic ones. Tight-binding aptamers have been observed before to preferentially evolve to epitopes that contain positively charged amino acid residues, presumably utilizing electrostatic interactions (Nieuwlandt et al. 1995; Kimoto et al. 1998; Blind et al. 1999). In addition, our anti-B1-CT aptamers add to a series of other aptamers that target proteins implicated in neurodegenerative diseases, in particular aptamers directed against various forms of the prion protein (Weiss et al. 1997; Proske et al. 2002a; Rhie et al. 2003; Sayer et al. 2004).
The finding that the acidic clustered dileucine (ACDL) motif, which is required for regulation of subcellular trafficking, is not targeted by our aptamer suggests that the subcellular trafficking will not be affected by addition of the aptamer, which would provide the possibility to investigate further post-transcriptional roles of the B1-CT in more detail. The membrane-proximal half of B1-CT also comprises three cysteine residues which have been shown to be palmitoylated (Benjannet et al. 2001) in cells. Palmitoylation diminishes shedding of B1 from the cell surface (Benjannet et al. 2001), but it is unclear which fraction of the protein is palmitoylated at a given time point. We have shown that aptamer S10, selected in vitro on a nonpalmitoylated peptide, also recognizes BACE1 from HEK269 cells (Fig. 3B,C). It is unlikely that the aptamer is able to bind the peptide in the presence of three bulky palmitoyl residues attached to the binding epitope. Because this may also be true for the BACE1/CCS-interaction, an interesting question in this context is which amount of cellular BACE1 is effectively available for interaction with CCS. (Hussain et al. 2003) suggested that the proportion of BACE1 trafficking to the cell surface is <10%; therefore, the bulk of the protein is unlikely to be palmitoylated or trapped by binding to transport factors, but is free to interact with CCS. The same reasoning would also apply to the binding of aptamers to cellular BACE-1.
Furthermore, it has also been shown that BACE1 overexpression in CHO cells leads to a decrease in SOD1 activity in extracts made from these cells (Angeletti et al. 2005), and it is possible that CCS binding to BACE1 might reduce the availability of CCS for SOD1 activation. Intracellular application of an aptamer competing with CCS for binding to B1-CT could possibly restore the amount of CCS available for SOD1 activation and thus rescue SOD1 activity. The same epitope that is thought to be the target of CCS is also a high-affinity binding site of a single Cu+ ion. The binding site for Cu+ involves the central cysteine C482 together with one of the two flanking cysteines and a third nonthiol ligand (Angeletti et al. 2005). The precise function of copper binding to B1-CT remains to be elucidated, but defective copper metabolism has long been thought to be implicated in Alzheimer's disease (Hesse et al. 1994; Multhaup et al. 2002; Bayer et al. 2003; Bayer and Multhaup 2005). As the copper binding domain coincides with the aptamer binding domain, the aptamer may also help to elucidate the role of Cu+-binding within the B1-CT.
In general, the finding that a short aptamer can discriminate binding regions within a 3-kDa peptide is remarkable and suggests that the aptamer provides a useful tool for elucidating functions associated with this epitope without affecting other important biological activities elicited by the C-terminal half of the BACE1 cytoplasmic domain.
MATERIALS AND METHODS
Peptides and proteins
A peptide corresponding to residues 478–501 of BACE-1 representing the complete cytoplasmic tail was elongated by an N-terminal spacer of two β-alanines and ε-amino caproic acid and used for selections. The sequence of the 24-mer B1-CT was H2N-X-βA-βA-CQWRCLRCLRQQHDDFADDISLLK (Peptide Specialty Laboratories, Heidelberg) and was >95% pure. The phosphorylated version carried a phosphate group at serine 498.
The GST-fusion proteins GST-B1-CT, GST-BACE2-CT, GST-GGA1-VHS, GST-CCS, and GST were overexpressed in Escherichia coli and purified on glutathione-Sepharose according to the manufacturer's instructions (Amersham). Purified GST-fusion proteins were desalted on G25 columns (Amersham) followed by dialysis into binding buffer (PBS, 1 mM MgCl2 at pH 7.4) to remove free and bound glutathione.
Derivatization of resin
Peptide was coupled to pre-swollen cyanogen bromide (CNBr)-activated Sepharose according to the manufacturer's instructions (Amersham) to give a final concentration of 5–500 μM. The amount of peptide present in solution before and after derivatization was determined by HPLC and detection at 280 nm. A negative selection residue was generated by derivatizing CNBr-activated Sepharose with Tris-(hydroxymethyl)-aminomethane (Tris).
Pool design and construction
A chemically synthesized DNA pool containing a 40-nt completely random sequence tract was used as a starting point for selections (5′-GGGATAGGATCCACATCTACGTATTA–N40-TTCACTGCAGACTTGACGAAGCTT-3′). The primer sequences flanking the randomized region are derived from (Cox and Ellington 2001): 5′ primer, GATAATACGACTCACTATAGGGATAGGATCCACATCTACGT; 3′-primer, AAGCTTCGTCAAGTCTGCAGTGAA (T7 RNA polymerase promoter underlined). A double-stranded DNA pool was generated from the original synthetic oligomer via a large-scale PCR, and an RNA pool was transcribed from a portion (∼1014 sequences) of this pool using T7 RNA polymerase in the presence of [α-32P]-GTP. The transcribed RNA was purified by PAGE.
In vitro selection of aptamers
Selections were carried out as described before (Mayer et al. 2001; Proske et al. 2002a,b; Theis et al. 2004). In particular, internally [32P]-labeled RNA (400 pmol in cycle 1, 100 pmol in subsequent cycles) in 100 μL binding buffer (PBS, 1 mM MgCl2 at pH 7.4) was mixed with the selection resin (25 μL) and incubated at 25°C for 30 min (shaking, 1000 rpm). The binding reaction was poured into a plastic column and washed in 50 μL portions with 150 μL–7000 μL binding buffer. Flowthrough and wash fractions were counted on a scintillation counter and discarded. Bound RNAs were eluted by incubating twice with 100 μL elution buffer (binding buffer plus 6 M guanidinium hydrochloride) at 80°C for 5 min. Eluates were pooled and counted. From the third cycle on a negative selection was carried out before the actual selection step. In this case RNA was mixed with negative selection resin (50 μL) and incubated at 25°C for 30 min. The binding reaction was poured into a plastic column, and the flowthrough and one wash volume were mixed with the affinity resin as described above. Eluted RNAs were twice precipitated with ethanol, reverse-transcribed, and amplified as described before (Mayer et al. 2001; Proske et al. 2002a,b; Theis et al. 2004). Selected RNAs were reverse-transcribed and PCR-amplified. In vitro transcription in the presence of [α-32P]-GTP using T7 RNA polymerase (New England Biolabs) for 10 h at 37°C yielded RNA that was PAGE purified and used for the next selection cycle.
Error-prone PCR (EP-PCR)
EP-PCRs were carried out according to Cadwell et al. (1992). To mutagenize the enriched pool of cycle 7, 200 ng of pool DNA were amplified for four cycles in a 100 μL EP-PCR mixture (10 mM Tris at pH 8.3, 50 mM KCl, 7 mM MgCl2, 1 mM dCTP, 1 mM dTTP, 0.2 mM dATP, 0.2 mM dGTP, 2 μM 5′-pimer, 2 μM 3′-primer, 0.5 mM MnCl2, 0.05 U/μL Taq DNA polymerase [Promega]). Before the final extension at 72°C was completed 10 μL of the EP-PCR reaction were transferred into a new preheated tube containing 90 μL of EP-PCR mixture and again amplified for four PCR cycles. This procedure was repeated 14 times. Thermocycling was between 94°C, (60 sec), 60°C (60 sec), and 72°C (180 sec). Mutagenesis of monoclone K11 was performed by thermal cycling of 20 pg/μL DNA template in 100 μL of EP-PCR mixture for 12 cycles.
Cloning and sequencing
pGEM-T Vector Systems kit (Promega) was used to clone the PCR products, and plasmids were isolated from transformed cells and sequenced by Sequence Laboratories Göttingen GmbH.
Column assays
Radiolabeled RNAs were incubated with 20 μL of B1-CT-derivatized CNBr-activated Sepharose or Protein A agarose (Sigma) derivatized with B1 from immunoprecipitation of HEK269 cells with an antibody against the N terminus of B1 (Walter et al. 2001a). The slurry was transferred into plastic columns and washed with 150–7000 μL binding buffer. Bound RNA was then eluted as described. The amount of bound RNA was determined by scintillation counting and compared to the input and unbound RNA.
Surface plasmon resonance measurements
Surface plasmon resonance (SPR) measurements were carried out on a BIAcore3000. DNA sequences were transcribed in the presence of guanosine monophosphorothioate (GMPS), 5′-biotinylated using iodoacetamido biotin as previously described (Sengle et al. 2000) and purified on 8% denaturing PAA gels. The 5′-biotinylated sequences were coupled to streptavidine (SA) chips (BIAcore) until ∼1000 response units were reached. The same amount of 5′-biotinylated unselected pool RNA was coupled to a reference cell. To avoid bulk effects, this reference surface was continuously subtracted from the signal of the active surface.
Different concentrations of the respective analyte (B1-CT peptide or control peptide/protein) were injected for 120 sec at a flow rate of 30 μL/min and their association and dissociation were monitored. The response units in equilibrium were then plotted against the injected concentrations to determine KD-values.
Enzymatic probing
[32P] 5′- or 3′-labeled RNA was subjected to enzymatic digestion in 10 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM MgCl2 in the presence of 1 μg unlabeled RNA at 20°C for 8 min with RNase T1 (Ambion, 0.2 and 0.02 U/μL), Nuclease S1 (Promega, 2 and 0.2 U/μL), RNase A (Ambion, 0.01 and 0.001 μg/mL), or Nuclease V1 (Ambion, 0.01 and 0.001 U/μL). The reaction was stopped, and the RNA was subjected to ethanol precipitation and then dissolved in loading buffer (45 mM Tris, 45 mM borate, 4 M urea, 10% sucrose, 100 mM EDTA, 0.05% SDS). The RNA fragments were then sized by electrophoresis on a 10% denaturing polyacrylamide gel. Size of fragments was determined by running an alkaline hydrolysis ladder (achieved by heating the labeled RNA in 50 mM NaHCO3, 1 mM EDTA at pH 9.2, at 95°C for 2, 5, and 7 min) and a denaturing RNase T1-digest ladder (generated by denaturation of RNA at 50°C in 20 mM sodium citrate, 1 mM EDTA, 7 M urea at pH 5.0, and subsequent digestion by RNase T1 (0.2, 0.02, and 0.002 U/μL for 10 min at 20°C) to indicate the position of the G residues.
Chemical probing
Modification with DMS or kethoxal: In a final volume of 10 μL, RNA (0.2 μg/μL) was incubated in 50 mM HEPES (pH 7.8), 100 mM KCl, 10 mM MgCl2 without or with 10 μM B1-CT at 25°C for 20 min. Then, 1 μL of a 600 mM solution of DMS in ethanol or 200 mM kethoxal (ICN) in water, respectively, was added, mixed, and incubated for 20 min at 25°C. After precipitation, 5′-end labeled primer was annealed and primer extension performed. Modification with CMCT: In a final volume of 10 μL, RNA (0.2 μg/μL) was incubated in 50 mM potassium borate (pH 8.0), 100 mM KCl, 10 mM MgCl2 with 10 μM B1-CT at 25°C for 20 min. Then 1 μL of a 200 mM solution of CMCT in water was added, mixed, and incubated 20 min at 25°C. After precipitation, 5′-end labeled primer was annealed and primer extension performed.
Primer extension
In a 20-μL reaction, 0.2 μg RNA in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 20 mM DTT, 0.5 mM dNTPs (each) was heated to 65°C for 5 min, then chilled on ice for 1 min. After addition of 1 μL (200 U) of Superscript II Reverse Transcriptase (Invitrogen) the reaction was incubated at 42°C for 50 min, followed by inactivation at 70°C for 15 min. After precipitation the DNA fragments were separated on a 10% denaturing PAA gel at 2000 V and visualized by PhosphorImaging. The sequencing reactions were carried out using the PCR templates with the Sequenase Version 2.0 PCR Product Sequencing kit (USB).
Pull-down assays with immobilized aptamer
5′-Biotinylated aptamers were coupled to SA-magnetic beads and incubated with GST-B1-CT, GST, or lysate from HEK269 cells overexpressing BACE-1 in binding buffer supplemented with 1 mg/mL BSA, 0.5% Triton-X, and 0.5% NP-40 or CCS binding buffer supplemented with RNasin, 1 mg/mL BSA, and 3 mM MgCl2. After washing with the same buffer, bound protein was eluted with Lämmli buffer, separated on 10% SDS-PAGE, and detected after Western blotting with polyclonal anti-GST-antibody (Amersham) or anti-BACE antibody (Walter et al. 2001a), respectively.
Deletion mutants of aptamers S10, TH14
Deletion mutants were generated from DNA templates of S10 or TH14, respectively, by PCR with primers corresponding to the desired mutations using Pfu DNA Polymerase (Stratagene) according to the manufacturer's recommendations.
BACE phosphorylation
Different concentrations of B1-CT-peptide were incubated at 25°C in PBS, 5 mM MgCl2, 5 mM DTT, 0–10 μM Aptamer and different concentrations of Casein kinase-1 (CK-1) (NEB) in a total volume of 25 μL. The reaction was started by addition of 10 μM ATP including 3 μL [γ-32P]-ATP. Samples of 3 μL were taken at defined time points, inactivated by addition of 5 μL Lämmli buffer and 3 min incubation at 95°C. Samples were separated on 15% Tricine-SDS-gels from unincorporated ATP. Gels were then dried and analyzed by PhosphorImaging and advanced image data analysis (AIDA).
BACE/GGA1 interaction
Twenty microliters (20 μL) B1-CT-Sepharose (50 μM) or blank Sepharose was incubated with GST-GGA1-VHS (0.01 μM) or GST (0.01 μM) and 0–10 μM RNA in binding buffer supplemented with 1 mg/mL BSA, 0.5% Triton-X, and 0.5% NP-40 for 30 min at 25°C in an overhead tumbler. After washing with the same buffer, bound protein was eluted with Lämmli buffer, separated on 10% SDS-PAGE, and detected with polyclonal anti-GST-antibody (Amersham) after Western blotting.
BACE/CCS interaction
Twenty microliters (20 μL) of BACE-Sepharose (50 μM) or blank Sepharose was incubated with 2 μM GST-CCS and 0–10 μM RNA in CCS binding buffer. After elution with Lämmli buffer and separation on a 10% SDS-PAGE, the proteins were detected with anti-CCS antibody (Santa Cruz) and polyclonal anti-GST-antibody (Amersham) after Western blotting.
ACKNOWLEDGMENTS
We thank Jonathan D. Gitlin (Washington University) for a gift of the expression plasmid for GST-CCS, Nicole Kuhn and Thomas Höver for excellent technical assistance, Günter Mayer and Anton Schmitz for discussions, and Beatrix Süss (Erlangen) for a sample of kethoxal. This work was supported by grants from the DFG, SFB 645, to J.W. and M.F. A.R. thanks the Austrian Academy of Sciences for a grant from the Doctoral Scholarship Program.
Footnotes
Reprint requests to: Michael Famulok, LIMES Program Unit Chemical Biology & Medicinal Chemistry, Universität Bonn, Gerhard-Domagk-Strasse 1, 53121 Bonn, Germany; e-mail: m.famulok@uni-bonn.de; fax: 49-228-735388.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.126306.
REFERENCES
- Angeletti B., Waldron K.J., Freeman K.B., Bawagan H., Hussain I., Miller C.C., Lau K.F., Tennant M.E., Dennison C., Robinson N.J., et al. BACE1 cytoplasmic domain interacts with the copper chaperone for superoxide dismutase-1 and binds copper. J. Biol. Chem. 2005;280:17930–17937. doi: 10.1074/jbc.M412034200. [DOI] [PubMed] [Google Scholar]
- Bayer T.A., Multhaup G. Involvement of amyloid β precursor protein (AβPP) modulated copper homeostasis in Alzheimer's disease. J. Alzheimers Dis. 2005;8:201–215. doi: 10.3233/jad-2005-8212. [DOI] [PubMed] [Google Scholar]
- Bayer T.A., Schafer S., Simons A., Kemmling A., Kamer T., Tepest R., Eckert A., Schussel K., Eikenberg O., Sturchler-Pierrat C., et al. Dietary Cu stabilizes brain superoxide dismutase 1 activity and reduces amyloid Aβ production in APP23 transgenic mice. Proc. Natl. Acad. Sci. 2003;100:14187–14192. doi: 10.1073/pnas.2332818100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjannet S., Elagoz A., Wickham L., Mamarbachi M., Munzer J.S., Basak A., Lazure C., Cromlish J.A., Sisodia S., Checler F., et al. Post-translational processing of β-secretase (β-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-β production. J. Biol. Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- Blind M., Kolanus W., Famulok M. Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. Proc. Natl. Acad. Sci. 1999;96:3606–3610. doi: 10.1073/pnas.96.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller P., Kochoyan M., Famulok M. Structural probing and damage selection of citrulline- and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–4776. doi: 10.1093/nar/23.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.H., Hoffman D.C., Brown A., Hansen M., Pardi A., Gold L. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem. Biol. 1997;4:833–843. doi: 10.1016/s1074-5521(97)90116-2. [DOI] [PubMed] [Google Scholar]
- Cadwell R.C., Joyce G.F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cox J.C., Ellington A.D. Automated selection of anti-protein aptamers. Bioorg. Med. Chem. 2001;9:2525–2531. doi: 10.1016/s0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- Cumming J.N., Iserloh U., Kennedy M.E. Design and development of BACE-1 inhibitors. Curr. Opin. Drug Discov. Devel. 2004;7:536–556. [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Ghosh A.K., Hong L., Tang J. β-Secretase as a therapeutic target for inhibitor drugs. Curr. Med. Chem. 2002;9:1135–1144. doi: 10.2174/0929867023370149. [DOI] [PubMed] [Google Scholar]
- Haass C. Take five–BACE and the γ-secretase quartet conduct Alzheimer's amyloid β-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Li F., Chang W.P., Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J. Biol. Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- Hesse L., Beher D., Masters C.L., Multhaup G. The β A4 amyloid precursor protein binding to copper. FEBS Lett. 1994;349:109–116. doi: 10.1016/0014-5793(94)00658-x. [DOI] [PubMed] [Google Scholar]
- Hussain I., Hawkins J., Shikotra A., Riddell D.R., Faller A., Dingwall C. Characterization of the ectodomain shedding of the β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) J. Biol. Chem. 2003;278:36264–36268. doi: 10.1074/jbc.M304186200. [DOI] [PubMed] [Google Scholar]
- Kimoto M., Sakamoto K., Shirouzu M., Hirao I., Yokoyama S. RNA aptamers that specifically bind to the Ras-binding domain of Raf-1. FEBS Lett. 1998;441:322–326. doi: 10.1016/s0014-5793(98)01572-5. [DOI] [PubMed] [Google Scholar]
- Luo Y., Bolon B., Kahn S., Bennett B.D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., Gong Y., et al. Mice deficient in BACE1, the Alzheimer's β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Mayer G., Blind M., Nagel W., Bohm T., Knorr T., Jackson C.L., Kolanus W., Famulok M. Controlling small guanine-nucleotide-exchange factor function through cytoplasmic RNA intramers. Proc. Natl. Acad. Sci. 2001;98:4961–4965. doi: 10.1073/pnas.091100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multhaup G., Scheuermann S., Schlicksupp A., Simons A., Strauss M., Kemmling A., Oehler C., Cappai R., Pipkorn R., Bayer T.A. Possible mechanisms of APP-mediated oxidative stress in Alzheimer's disease. Free Radic. Biol. Med. 2002;33:45–51. doi: 10.1016/s0891-5849(02)00806-7. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt D., Wecker M., Gold L. In vitro selection of RNA ligands to substance P. Biochemistry. 1995;34:5651–5659. doi: 10.1021/bi00016a041. [DOI] [PubMed] [Google Scholar]
- Proske D., Gilch S., Wopfner F., Schatzl H.M., Winnacker E.L., Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem. 2002a;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Proske D., Hofliger M., Soll R.M., Beck-Sickinger A.G., Famulok M. A Y2 receptor mimetic aptamer directed against neuropeptide Y. J. Biol. Chem. 2002b;277:11416–11422. doi: 10.1074/jbc.M109752200. [DOI] [PubMed] [Google Scholar]
- Rhie A., Kirby L., Sayer N., Wellesley R., Disterer P., Sylvester I., Gill A., Hope J., James W., Tahiri-Alaoui A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 2003;278:39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- Roggo S. Inhibition of BACE, a promising approach to Alzheimer's disease therapy. Curr. Top. Med. Chem. 2002;2:359–370. doi: 10.2174/1568026024607490. [DOI] [PubMed] [Google Scholar]
- Sayer N.M., Cubin M., Rhie A., Bullock M., Tahiri-Alaoui A., James W. Structural determinants of conformationally selective, prion-binding aptamers. J. Biol. Chem. 2004;279:13102–13109. doi: 10.1074/jbc.M310928200. [DOI] [PubMed] [Google Scholar]
- Sengle G., Jenne A., Arora P.S., Seelig B., Nowick J.S., Jaschke A., Famulok M. Synthesis, incorporation efficiency, and stability of disulfide bridged functional groups at RNA 5′-ends. Bioorg. Med. Chem. 2000;8:1317–1329. doi: 10.1016/s0968-0896(00)00080-8. [DOI] [PubMed] [Google Scholar]
- Theis M.G., Knorre A., Kellersch B., Moelleken J., Wieland F., Kolanus W., Famulok M. Discriminatory aptamer reveals serum response element transcription regulated by cytohesin-2. Proc. Natl. Acad. Sci. 2004;101:11221–11226. doi: 10.1073/pnas.0402901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Valentine J.S., Hart P.J. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R. β-Secretase, APP and Aβ in Alzheimer's disease. Subcell. Biochem. 2005;38:79–103. [PubMed] [Google Scholar]
- von Arnim C.A., Tangredi M.M., Peltan I.D., Lee B.M., Irizarry M.C., Kinoshita A., Hyman B.T. Demonstration of BACE (β-secretase) phosphorylation and its interaction with GGA1 in cells by fluorescence-lifetime imaging microscopy. J. Cell Sci. 2004;117:5437–5445. doi: 10.1242/jcs.01422. [DOI] [PubMed] [Google Scholar]
- Wahle T., Prager K., Raffler N., Haass C., Famulok M., Walter J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol. Cell. Neurosci. 2005;29:453–461. doi: 10.1016/j.mcn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Walter J., Fluhrer R., Hartung B., Willem M., Kaether C., Capell A., Lammich S., Multhaup G., Haass C. Phosphorylation regulates intracellular trafficking of β-secretase. J. Biol. Chem. 2001a;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- Walter J., Kaether C., Steiner H., Haass C. The cell biology of Alzheimer's disease: Uncovering the secrets of secretases. Curr. Opin. Neurobiol. 2001b;11:585–590. doi: 10.1016/s0959-4388(00)00253-1. [DOI] [PubMed] [Google Scholar]
- Weiss S., Proske D., Neumann M., Groschup M.H., Kretzschmar H.A., Famulok M., Winnacker E.L. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.C., Waggoner D., Subramaniam J.R., Tessarollo L., Bartnikas T.B., Culotta V.C., Price D.L., Rothstein J., Gitlin J.D. Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. 2000;97:2886–2891. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnd C., Spinelli S., Luginbuhl B., Amstutz P., Cambillau C., Pluckthun A. Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 2004;279:18870–18877. doi: 10.1074/jbc.M309169200. [DOI] [PubMed] [Google Scholar]
- Ziehler W.A., Engelke D.R. Current protocols in nucleic acid chemistry. Wiley-VCH; Weinheim, Germany: 2000. Probing RNA structure with chemical reagents and enzymes; pp. 6.1.1–6.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]