Abstract
The cloverleaf structure in the 5′-untranslated region of enterovirus RNA that regulates viral RNA replication contains an evolutionarily conserved YNMG tetraloop closed by a Y-G base pair. This loop is believed to interact specifically with the viral protease 3C. To further characterize the specificity of this interaction, the tetraloop and two flanking base pairs of the poliovirus RNA were randomized, and viable viral clones were obtained using in vivo SELEX. Among many different mutants with the canonical YNMG sequences to be described elsewhere, a large-plaque-forming clone contained a deviating uGCUAg sequence. The NMR structure of a small hairpin capped with uGCUAg that we present here shows that the GCUA tetraloop adopts a novel fold, which is highly similar to that of the YNMG tetraloop with common stacking properties and hydrogen-bond interactions including an unusual syn conformation of the adenosine. Thermodynamic studies show moderate stabilities of hairpins with canonical YNMG and the novel GCUA loops, which, together with the similarity of spatial structures, illustrates that the tetraloop structure itself is crucial for the RNA–protein interaction required for the viral replication. A re-evaluation of the ribosomal secondary structure database reveals a hairpin containing a GCUA loop, which covaries with YNMG and is involved in a tertiary interaction, and in the 50S ribosomal subunit from Haloarcula marismortui the structurally comparable apex of stem–loop 35a is a recognition site for protein L2. These observations show a more general occurrence and importance of the so-far unrecognized GYYA hairpin loops.
Keywords: RNA structure, RNA tetraloop, poliovirus, in vivo SELEX, RNA–protein recognition, NMR
INTRODUCTION
RNA tetranucleotide loops, also referred to as “tetraloops,” are the most common loops in biologically active RNAs and have been the most extensively studied by theoretical, molecular biology, biochemical, and biophysical approaches over the past two decades. The pioneering quintessential phylogenetic studies by Gutell, Woese, and collaborators (Gutell et al. 1985; Woese et al. 1990) have revealed that out of all loops of 16S rRNA, >50% were tetraloops, with GNRA and UNCG being the most abundant followed by CUUG (N corresponds to any nucleotide, and R is either A or G). Since then, tetraloops were recognized as frequent and important elements in many other RNAs as well (Poritz et al. 1988; Pace et al. 1989; Michel and Westhof 1990). Thermodynamic studies revealed that hairpins with these tetraloops are also very stable (Antao et al. 1991), and because the majority of the UNCG and GNRA loops are closed by a C-G base pair, which also contributes to a higher stability (reversing the closing base pair of a UUCG loop to G-C destabilizes tetraloop formation by a Δ(ΔG 37°) of 2.3 kcal/mol [Antao et al. 1991; Hilbers et al. 1994]), the sequences of the two families were subsequently refined to cUNCGg and cGNRAg (the lowercase letters represent the base pair closing the loop).
The most frequently occurring loops in 16S rRNA, i.e., cUUCGg, cGAAAg, and cGCAAg, were the first RNA structures solved at atomic resolution by NMR and revealed tightly packed loops with extensive stacking and dense hydrogen-bond networks, explaining their unusual stability (Cheong et al. 1990; Heus and Pardi 1991). Because UNCG and GNRA loops are sometimes evolutionarily interchangeable (Gutell et al. 1985; Woese et al. 1990), it was initially suggested (Uhlenbeck 1990) that they evolved primarily because of their high stability to serve, for instance, in the proper sequential folding of larger RNAs. However, more specific roles have also been ascribed to these tetraloops by demonstrating that they serve as functionally important specific recognition sites in RNA–protein interactions (e.g., Cai et al. 1998; Legault et al. 1998) and are involved in translation initiation and fidelity (van Knippenberg 1986), in ribosome inactivation (Ackerman et al. 1988), and can mediate long-range tertiary RNA–RNA interactions (Murphy and Cech 1994; Cate et al. 1996).
More recently, the cUNCGg family was extended to cYNMGg (where Y is either U or C and M is either C or A), based on a very elegant study (Proctor et al. 2002) aimed at obtaining a complete library of stable tetraloops. The study involved a combination of in vitro selection (SELEX) and temperature gradient electrophoresis (TGGE). Starting with a 6-nt random RNA hairpin loop library, the already known cUNCGg and gCUUGc sequences were selected, but others were also found, i.e., cUNAGg, cCNCGg, and cCNAGg, with almost the same stability as the cUNCGg hairpin loops. Stable tetraloops with GNRA sequences were not detected, presumably because, as suggested by the investigators, the DNA GNRA tetraloops (contrary to the DNA TNCG tetraloops) can be exceptionally stable as well (Antao and Tinoco 1991; Hilbers et al. 1994), and might have interfered with the PCR reaction used in SELEX. A re-evaluation of a more recent 16S and 23S rRNA database revealed that the cUNAGg, cCNCGg, and cCNAGg hairpin sequences are at least as abundant as the gCUUGc sequence (Proctor et al. 2002). Further support for an extended cYNMGg family came from CD and NMR spectra of the novel hairpins, which were similar to those of the classic cUUCGg hairpin, suggesting a common structure. The high-resolution NMR structures of a uCACGg tetraloop (Du et al. 2003; Ohlenschlager et al. 2004), that occurs within the cloverleaf structure of the 5′-untranslated region (5′-UTR) of the Coxsackievirus B3 (CVB3), confirmed the structural resemblance between CNCG and UNCG loops. In this paper we present the NMR structural study of a novel functionally important tetraloop, GCUA, present in the 5′-UTR cloverleaf structure of a mutant poliovirus RNA, further addressing the structural similarity of different hairpin loops.
Enteroviruses, such as polioviruses and Coxsackieviruses, contain a small (<8000 bases) single-stranded RNA genome, consisting of a 5′-UTR, a single open reading frame (ORF), and a 3′-untranslated region (3′-UTR), which includes a 3′-terminal poly(A) tail (Semler and Wimmer 2002). The 5′-UTR is thought to consist of six separate domains. At the very 5′-end, the first domain folds into a cloverleaf secondary structure and can form a ribonucleoprotein complex with viral and cellular proteins, which is important for both (+) and (−) strand synthesis and terminal primer protein VPg uridylylation (Lyons et al. 2001).
The 5′-cloverleaf structure is highly conserved among enteroviruses and rhinoviruses and contains four subdomains: stem A, and stem–loops B, C, and D (Fig. 1A). Whereas stem–loop B is the binding partner for cellular proteins called poly (rC)-binding proteins (PCBP) (Gamarnik and Andino 1997; Parsley et al. 1997), in vitro and in vivo studies (Andino et al. 1990, 1993; Walker et al. 1995) have shown that stem–loop D is specifically recognized by the viral protein 3C (the viral protease) or, perhaps more relevant for the in vivo situation, 3CD (the common precursor for 3C and the viral polymerase 3D). This ternary 5′-cloverleaf–PCBP–3CD complex is believed to constitute the switch that determines the destiny of a viral RNA to serve either as an mRNA in translation or to go into replication (Gamarnik and Andino 1998).
FIGURE 1.
Secondary structure models of the poliovirus 5′-cloverleaf (A) and the RNA hairpin (B) used for the NMR studies.
The 4-nt apical D-loop appears to be a major determinant in the 3C (or 3CD) recognition. A temperature-sensitive mutant with a 4-nt insertion in the D-loop could be reverted by point mutations in the so-called TGK motif of 3C (Andino et al. 1990). Using in vitro binding assays with purified CVB3 3C protein and cloverleaf RNAs, Zell et al. (2002) showed that whereas the CVB3 3C protein forms stable complexes with its cognate cloverleaf containing a uCACGg D-loop sequence, and cloverleaves of other viruses with 4-nt apical loops D, it does not bind to cloverleaves with 3-nt or 5-nt apical loops. Moreover, shortening the CVB3 loop by 1 nt abolished binding, and insertion of 1 nt into the HRV2 3-nt loop enabled binding (Zell et al. 2002). These observations, together with a phylogenetic study showing a rather degenerate yNNNGg consensus, prompted the investigators to suggest that the structure and not the sequence of the D-loop is important for binding (Zell et al. 2002). Subsequent structural studies of the Coxsackievirus domain D with a uCACGg loop not only confirmed this hypothesis but also extended the family of UNCG-like loops to yYNMGg (Du et al. 2003; Ohlenschlager et al. 2004).
To further characterize residue specificity of the D-loop–3CD interaction, we used an in vivo SELEX procedure, which, in principle, can sample the entire functional loop sequence space. The poliovirus tetraloop D and two flanking base pairs were randomized, yielding 48 = 65,536 variants, and inserted into the infectious viral genome. The viable clones were isolated and investigated. The results of this study will be presented elsewhere. It should be noted, however, that along with numerous YNMG variants, a uGCUAg sequence was unexpectedly observed. This was surprising, indeed, because one could expect that this loop should share structural properties with the GNRA loop, which markedly differs from the YNMG structure, in particular by the presence of a sheared G-A base pair (Heus and Pardi 1991). Despite many biochemical, biophysical, and structural studies on RNA tetraloops, GNYA loops have escaped attention so far, probably because they are much less frequent in the ribosomal RNA and group I/II intron secondary structure models, which have typically been used for the phylogenetic studies.
Since there was no reference structure of a GNYA tetraloop at the start of these studies, we decided to determine its structure and thermodynamic properties. We find that the GCUA loop adopts a novel fold, highly similar to the YNMG motif, and exhibits a moderate stability. These results strongly suggest that it is the structure rather than stability that is primarily responsible for the selection of this loop in the viable viral clones. Importantly, the phylogenetic and structural comparison shows precedence for a more general role for GYYA loops in RNA folding and protein recognition.
RESULTS
Resonance assignment and structure determination
For the NMR studies, we used the non-(13C/15N)-labeled, chemically synthesized 14-nt RNA hairpin, composed of the selected poliovirus mutant apical D-loop, GCUA, and five flanking base pairs as depicted in Figure 1B. The small-size molecule gave excellent spectral dispersion, providing sufficient classic NOE distance and torsion angle information, which supplemented with modern implementation of RDC data allowed for a high-resolution structure determination without the need of a 13C/15N-labeling. Figure 2 gives some examples of the quality of the spectra. Proton resonance assignments were achieved using standard protocols, starting from sequential imino to imino and aromatic to H1′/H2′ sequential walks. A strong mutual NOE between the U5 and G10 imino protons, resonating at 11.8 and 10.85 ppm, provided evidence for the presence of a U-G wobble pair. No imino proton resonance was found resonating at ∼10.8 ppm, the typical position for a G imino proton in a sheared G-A base pair of the GNRA-like loop structure. A very large intraresidual H1′–H8 cross-peak was observed for A9 (Fig. 2A), indicating a syn conformation. Other structural data consistent with a syn conformation were the presence of A9H2–A9H3′, and U8H1′, and the absence of A9H8–U8H1′ NOE cross-peaks. These data indicated that the loop structure was different from that of the GNRA and was closed by a U-G wobble base pair. Besides the common twin amino proton resonances of cytidines in the 6.8–7.2-ppm region of the 2D NOESY recorded in H2O, also broad resonances were found around 6.5 ppm, which could be attributed to the G6 and A9 amino protons. NOE cross-peaks involving these resonances, notably a G6H1′–A9N6H contact, and implementation of RDCs further supported a G-A(syn) noncanonical base pair between the first and the last loop nucleotides.
FIGURE 2.
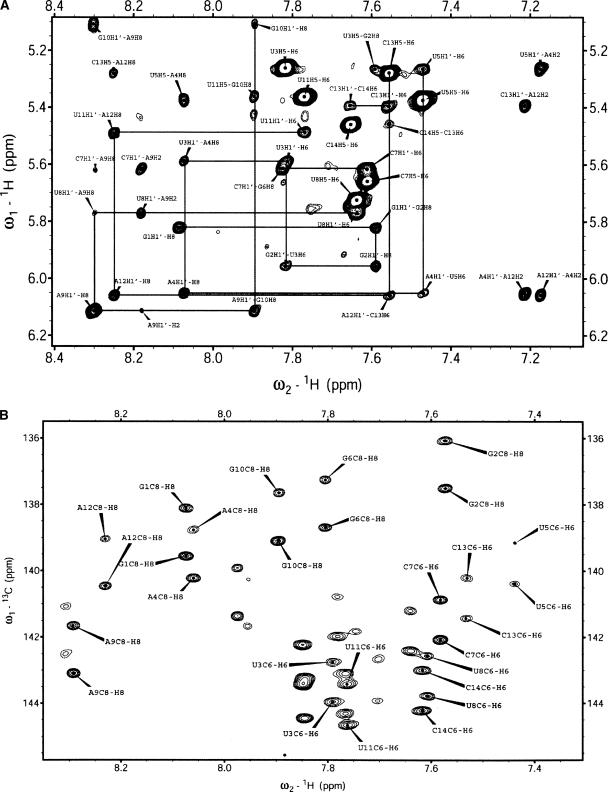
(A) Portion of the 2D NOESY (800 MHz, 300 msec mixing time) recorded in D2O, showing the aromatic proton to H1′ region. Sequential H1′ to aromatic protons are traced out. (B) Portion of the 13C-coupled natural abundance 2D 1H–13C HSQC showing the aromatic C8-H8/C6-H6 region used for collection of RCDs.
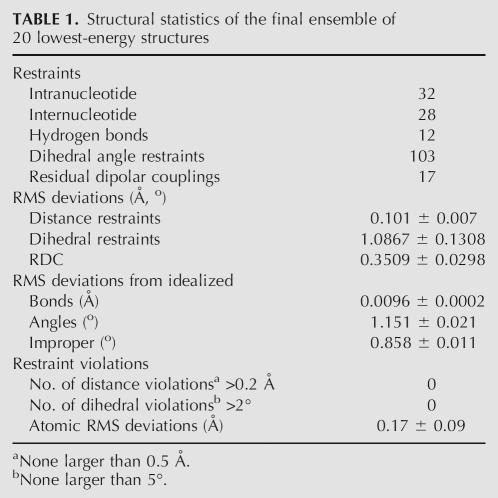
Analysis of the DQF-COSY and TOCSY spectra revealed that all residues adopt N-type sugar conformations, except for the loop C7 and U8 residues, where the intermediate–large 8–10-Hz H1′–2′ couplings indicated equilibria with predominant S-type sugars. C1′ and aromatic carbon resonances were assigned using a natural abundance HSQC spectrum. With the excellent spectral dispersion and narrow linewidths (as a result of using a nonlabeled sample), RDCs were easily collected from a natural abundance 13C-coupled 2D HSQC (Fig. 2B), using cryoprobe technology at 800 MHz. Statistics for the structure calculations are summarized in Table 1.
TABLE 1.
Structural statistics of the final ensemble of 20 lowest-energy structures
Structure of the uGCUAg tetraloop
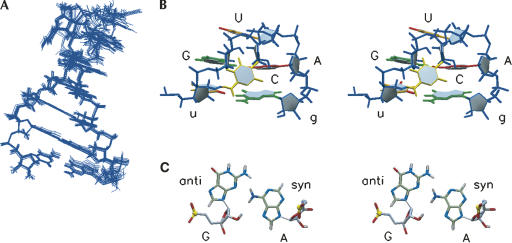
The structure of the model hairpin consists of an A-form helix capped by a well-defined apical uGCUAg loop structure with pairwise RMSD values of 0.5 and 0.7 Å for the ensembles fitted to all residues, or only the loop and closing base-pair residues, respectively (Fig. 3). U5 and G10 clearly form a wobble base pair that stacks upon the lower A-U base pair with common helical parameters (Heus and Hilbers 2003). Enhanced stacking on the lower A-U base pair due to underwinding of the U-G base pair (25° vs. normal 33°) slightly widens the major groove. The first and last nucleotides of the tetraloop form an unusual G6-A9(syn) base pair, where the adenine amino proton is hydrogen-bonded to the guanosine N3 and the amino proton of the guanosine can possibly form a hydrogen bond with the N1 of adenine (Fig. 3C). This unusual G-A trans sugar edge/Watson-Crick base-pair conformation has not been described in the isostericity matrices of noncanonical base pairs yet (Leontis et al. 2002). Although no assumption or explicit hydrogen-bond restraints were introduced to force the hydrogen bonds in the G-A(syn) base pair, the GN3-AN6 heavy atom distances are within hydrogen-bond range (<2.8 Å) in all calculated structures and the GN2-AN1 distances (<3.8 Å) in 25% of the structures. The G6 2′-hydroxyl oxygen and U8 pro-R-phosphoryl oxygen are also within hydrogen-bond distance (<2.6 Å heavy atom distance in all structures), suggesting an additional hydrogen bond that further stabilizes the tetraloop structure. Reversal of the backbone direction is between the third and the fourth loop residues, as defined by the opposite orientations of the sugars. The second nucleotide of the loop, C7, swings its base into the minor groove with the H5–H6 edge facing the sugar moiety of G6 and the Watson-Crick edge exposed to solvent. The third nucleotide of the loop, U8, stacks upon G6 in the major groove with the H5–H6 pointing into the direction of the U8–5′-phosphate and the Watson-Crick edge exposed to solvent. Stacking interactions and observed hydrogen bonds satisfactorily explain the many loop NOEs, absence of U7 and U8 imino proton resonances, and partial resolution of the G6 and A9 amino protons.
FIGURE 3.
(A) Overlays of the 20 lowest-energy hairpin structures fitted to all residues. (B) Stereo view into the minor groove of the apical uGCUAg loop of the lowest energy structure. The A-residue bases are in red, the G-residue bases are in green, the C-residue bases are in yellow, and the U-residue bases are in orange. Uracil C–O bonds are in red. The sugar-phosphate backbone is in blue. (C) Stereo top view down the helical axis of the lowest energy structure, showing configuration of the G6-A9(syn) base pair.
Thermodynamic studies
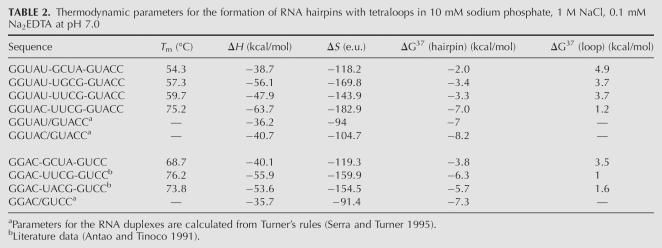
Thermodynamic parameters were obtained from the optical melting data for several minihairpins containing the poliovirus D-loop sequence, uUGCGg, and the loop sequence that was selected by in vivo SELEX, uGCUAg, flanked by the 5-bp stem used for the NMR construct. As references, we also determined thermodynamic data for the UUCG loop with C-G or U-G closing base pairs and the GCUA loop flanked by a different 4-bp stem, for which thermodynamic data have been reported earlier by Tinoco and coworkers (Antao and Tinoco 1991; Antao et al. 1991). All melting curves were obtained in 10 mM Na phosphate, 1 M NaCl, and 0.1 mM Na2EDTA. For these conditions, the nearest-neighbor interactions have been tabulated by Turner and coworkers (Serra and Turner 1995), allowing calculation of stem stabilities. Thus, the ΔG 37° values for loop formation shown in Table 2 were determined by subtracting the estimated stem stabilities from the hairpin values. Minihairpins with a UUCG loop closed by a C-G base pair, considered to be the classical unusually stable RNA loop, show high T m’s of 75.2°C and 76.2°C, and the UUCG loop destabilizes the hairpin by only 1–1.2 kcal/mol. Hairpins with down to 7°C lower T m’s and destabilizing ΔG 37° up to 2 kcal/mol have been classified as unusually stable as well (Antao and Tinoco 1991; Antao et al. 1991; Hilbers et al. 1994; Proctor et al. 2002). The poliovirus UGCG and novel GCUA loops, closed by a U-G base pair, clearly fall outside this range, exhibiting much lower T m’s and higher destabilizing ΔG 37°. As can be inferred from Table 2, the lower stability is primarily caused by changing the closing base pair from C-G to U-G, which has a profound influence on the hairpin stability, lowering the T m by >10°C. Using the available data, the T m of a 5-bp hairpin capped with a uUACGg sequence, present in five enteroviruses, is estimated to be 56.1°C, putting it in the class of moderately stable hairpins as well. The majority of the naturally occurring apical D-YNMG loops in the enterovirus cloverleaf structures are closed by a U-G base pair (75%), while the remainder is closed by a C-G base pair. Thus, it is reasonable to assume that the majority of these D-stem–loops are not unusually stable.
TABLE 2.
Thermodynamic parameters for the formation of RNA hairpins with tetraloops in 10 mM sodium phosphate, 1 M NaCl, 0.1 mM Na2EDTA at pH 7.0
DISCUSSION
The GCUA loop is structurally a member of the YNMG rather than the GNRA family
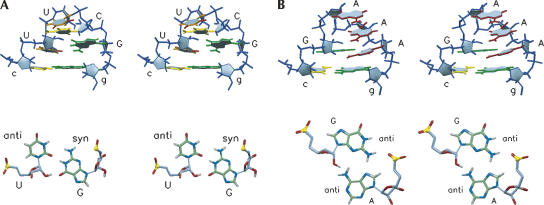
The structure of the classical UUCG loop is the most stable and best-characterized member of the YNMG family and has been determined at atomic resolution both by NMR and X-ray crystallography (Allain and Varani 1995; Ennifar et al. 2000). The structural details of the GCUA loop described here are the same as in the UUCG loop. The UUCG loop structure (PDB 1F7Y) (Fig. 4A) is characterized by a U-G(syn) reverse wobble (U-G trans sugar edge/Watson-Crick) base pair; folding of the second and third loop nucleotides into the minor groove and major groove sides, respectively; and stacking of the third nucleotide, C or U, on top of the U-G reverse wobble. On the other hand, the GAAA loop, i.e., the best structurally characterized member of the GNRA family (Jucker et al. 1996; Batey et al. 2001), clearly has a different fold with a sheared G-A (trans sugar edge/Hoogsteen) base pair, all nucleotides in the anti conformation, and stacking extending from the 3′-side up to the second (N) nucleotide, after which a sharp turn between the first and the second nucleotide reverses the chain direction (PDB 1HQ1) (Fig. 4B).
FIGURE 4.
(A) Stereo view into the minor groove of the uUUCGg loop, and top view down the helix of the loop U-G(syn) base pair. (B) Stereo view into the minor groove of the cGAAAg loop, and top view down the helix of the loop G-A base pair. Coloring is as in Figure 3.
A comparison of Figures 3 and 4 reveals that the GCUA loop has the same topology as the UUCG loop and is markedly different from the GAAA loop. In particular, the stacking pattern with the second and third bases on the opposite sides of the backbone, and presence of a trans sugar edge/Watson-Crick base pair with a syn nucleotide provide for easily recognizable features indicating the structural equivalence. In a recent study, Gorlach and coworkers (Ihle et al. 2005) solved the structure of the bovine enterovirus 1 (BEV1) apical D-loop with a uGUUAg sequence, the sole naturally occurring enterovirus cloverleaf structure with a deviating GNYA sequence. However, no G-A(syn) base pair but rather a displaced pseudo–base pair without direct hydrogen bonds was found for the BEV1 GUUA loop (Ihle et al. 2005). Although we do not have a direct explanation for this difference, it might be a subtle sequence dependency of the second nucleotide (the second cytidine in the GCUA structure descends lower into the minor groove than the second uridine in the GUUA structure). Despite this subtle difference, the overall structures of the two loops are very similar with the characteristic folding of the second and third loop nucleotides and the fourth nucleotide syn.
Notwithstanding a great overall resemblance between the GCUA and UUCG loops, small differences are present. In the UUCG loop the amino group of the third loop nucleotide, C, forms a hydrogen bond with the second phosphate-proR-oxygen, which is absent in the GCUA loop. In the YNMG loop structures, the U-G or C-G base pairs can substitute for each other, preserving the structural motif because they are isosteric trans sugar edge/Watson-Crick base pairs. The GCUA loop also contains a trans sugar edge/Watson-Crick (G-A) base pair; however, it is not exactly isosteric with the Y-G base pairs due to a lateral shift in hydrogen-bond positions. Thus, in the UUCG loop, the 2′-hydroxyl of the first loop nucleotide, U, forms a hydrogen bond with the opposite guanosine O6-moiety, whereas in the GCUA loop, the 2′-hydroxyl of the first loop nucleotide, G, is hydrogen-bonded to the uridine pro-R-phosphoryl oxygen. G-A trans sugar edge/Watson-Crick base pairs more isosteric to Y-G, including a G2′O–ANH6 hydrogen bond, have been found in a distorted uGAAA(syn)Gg pentaloop of Haloarcula marismortui LSU RNA (Klein et al. 2004) and a (G-C)-A(anti) base triple in a frameshifting pseudoknot (Su et al. 1999). In the GCUA loop, the A(syn) is twisted more into the major groove then the G(syn) in the UUCG loop and does not lie directly over the next residue H1′, which is in accordance with the normal chemical shift position of the G10 H1′ resonance. In the case of the UUCG loop, a large upfield shift of ∼1.5 ppm is observed, created by a large current effect of the loop G base on the next residue H1′ (Varani et al. 1991). Also, the G6 nucleotide is twisted more into the major groove, shifting its 2′-hydroxyl to the proper position for donating a hydrogen bond. Overwinding of G6 and A9 narrows the cross-strand C1′–C1′ distance to 8.5 Å in the GCUA loop and leaves the A9 N7-moiety less accessible than the corresponding G N7 in the UUCG loop. Finally, the second loop nucleotide, C, descends more steeply into the minor groove, also rendering the minor groove side of the noncanonical G-A(syn) base pair less accessible to solvent. These subtle differences have consequences for the D-loop 3CD protein interaction and can be used to provide further understanding for the 3CD recognition. It is, for instance, important to note that the similarity between the UUCG and GCUA loops is larger on the major groove side than on the minor groove side.
The YNMG and GCUA loops closed by a U-G base pair are not unusually stable
The presence of YNMG and GNRA tetraloops closed with a C-G base pair found in the ribosomal RNAs has been correlated with unusual stability (Uhlenbeck 1990; Antao and Tinoco 1991; Antao et al. 1991; Hilbers et al. 1994; Proctor et al. 2002). To understand whether functional equivalence of the YNMG and the novel GCUA loops in the viral 5′-cloverleaf structures are due to structural or stability reasons or both, we determined the thermodynamic properties of several minihairpins containing wild-type and mutant D-loop sequences (Table 2). The results show that U-G base-pair-closed YNMG loops, representing the majority of D-loops in the enteroviral and rhinoviral 5′-cloverleaf structures, as well as the novel uGCUAg loop give rise to hairpins, which can be classified as moderately stable with T m’s ranging from 54.3 to 57.3°C, while 25% of the D-loops are YNMG loops closed by a C-G base pair that yield unusually stable hairpins with T m’s around 70°C. Thus, the unusual stability believed to be the key feature of the large number of YNMG and GNRA loops in the ribosomal RNAs is not the reason for the presence of YNMG and GCUA sequences in the enterovirus D-loops.
Phylogenetic comparison
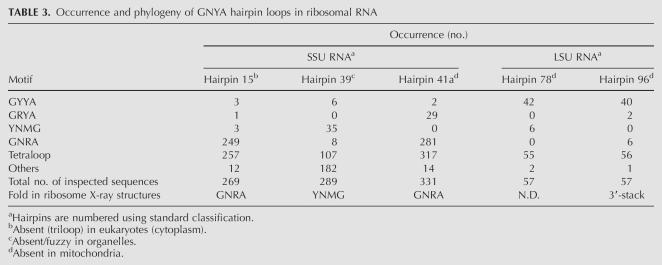
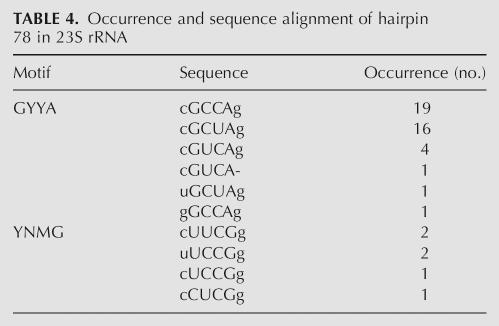
The results described here clearly demonstrate a great structural resemblance between the YNMG and GCUA hairpin loops. This raises the question why nature appears to have selected against GNYA loops, which are virtually absent in the secondary structure models of the ribosomal and group I/II intron RNAs (http://www.rna.icmb.utexas.edu; Cannone et al. 2002). A survey of phylogenetic secondary structure models of 415 small subunit (SSU) rRNAs, 81 large subunit (LSU) rRNAs, and 20 group I and six group II introns reveals that GNYA loops are only present in hairpins 15, 39, and 41 of SSU rRNA, and hairpins 78 and 96 of LSU rRNA (Table 3). The vast majority of the hp15 sequences contain a GNRA tetraloop with only a few examples of YNMG or GNYA, and hp15 has a GNRA fold in the crystal structure of the ribosome (Schluenzen et al. 2000; Wimberly et al. 2000). This suggests accidental occurrence of the YNMG and GNYA sequences in hp15 and that the function of hp15 can be fulfilled by structures not necessarily having a GNRA-like fold. Hp39 with six GYYA examples shows only a low abundance of YNMG and GNRA loops, while the majority have a different sequence showing no clear preference for or significance of either a YNMG or GNRA loop. Hp41a has a GNRA fold involved in a tertiary A-minor interaction in the crystal structures of the ribosome (Nissen et al. 2001). Again, the vast majority of these loops are represented by GNRA, with no YNMG and only two GYYA examples. There are, however, 29 examples of a GRYA loop, which, given the structural importance of the GNRA fold to form an A-minor interaction, suggests that the GRYA loops might adopt a GNRA fold as well. The LSU hp78 is of particular interest for this discussion (Table 4). Here, the majority is represented by a GYYA (GCCA, GCUA, or GUCA) sequence, which covaries with YNMG. Most interestingly, the loop sequences also covary with a proposed tertiary interaction involving the 5′-nucleotide of loop 77: hp78’s with GYYA loops are proposed to interact with a U at the 5′-side of loop 77, while hp78’s with YNMG loops are proposed to interact with a C. This regularity highly suggests a structural role for these loops at this site, in line with our proposal that they have a common fold. Unfortunately, neither hairpin 78 nor its interacting nucleotide is resolved in the available crystal structures of the large ribosomal subunit (Harms et al. 2001; Klein et al. 2004). However, on the basis of the UUCG and GCUA loop structures, a (U-G)-C or (G-A)-U base triple with the third base making a major groove interaction is most likely. Hp96 also contains a large number of GYYA loops, in this case covarying with GNRA. This is unlike the situation with hp78. However, inspection of the crystal structures reveals a distorted loop structure, uncommon to either GNRA or YNMG, with the four loop nucleotides forming a continuous stack from the 3′-side of the helix (Harms et al. 2001; Klein et al. 2004).
TABLE 3.
Occurrence and phylogeny of GNYA hairpin loops in ribosomal RNA
TABLE 4.
Occurrence and sequence alignment of hairpin 78 in 23S rRNA
As mentioned above, a likely reason for the low abundance of GNYA loops in the ribosomal and group I/II introns is that YNMG loops evolved there primarily because of their unusual stability to serve, for instance, as nucleation sites for the folding of larger RNAs. On the other hand, GCUA loops, when present, might have evolved predominantly for structural reasons. The available structural and phylogenetic data—that is, (1) GCUA and GUUA loops are structurally similar to YNMG loops, and (2) a YNMG loop with a possible structural role covaries with GCUA, GUCA, and GCCA—provide evidence for extending the YNMG family with GYYA.
Implications for 3CD binding
A wide body of evidence is available that shows that a direct interaction between the cloverleaf apical D-loop and 3CD protein is essential for poliovirus replication. Earlier phylogenetic, mutational, and structural (inferred from the high-resolution NMR) data have mapped the RNA-binding site to the apical D-loop with a common YNMG-like structure (Andino et al. 1990; Zell et al. 2002; Du et al. 2003; Ohlenschlager et al. 2004). The GCUA loop structure can be used to further evaluate the specificity of binding.
Despite the compact structure of the loop with numerous hydrogen-bond interactions, many hydrogen-bond acceptors and donors remain available for protein binding (Figs. 3,5A). On the major groove side, the amino, imino, O6, and N7 of G6; the N3 of A9; and the three Watson-Crick sites of U8 (O2, O4, and N3) form a cluster of accessible sites. On the minor groove side, the three Watson-Crick sites of C7 (amino, O2, and N3) are available, and possibly the A9 N7, although this site seems to be sequestered by the overwinding, and the C7 base that descends steeply into the minor groove in most of the structures. Also, the various backbone phosphorous, sugar oxygens and 2′-hydroxyls are exposed to solvent.
FIGURE 5.
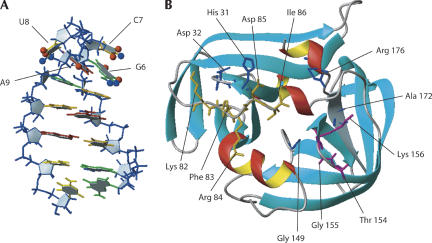
(A) Side view of the RNA hairpin with GCUA tetraloop, with possible hydrogen-bond acceptors and donors of the tetraloop indicated by red and by blue spheres, respectively. (B) Ribbon diagram of the crystal structure of 3C with the presumed RNA-binding site facing the viewer. RNA bases and protein amino acids implicated in RNA–protein binding are annotated. The TGK motif is in magenta, the KFRDIR motif is in gold, and other residues implicated in binding are in blue.
Two motifs of the 3C protein have been implicated in RNA binding, the so-called 154TGK156 and 82KFRDI86 motifs (Andino et al. 1990, 1993; Walker et al. 1995). In our in vivo SELEX study, some small plaque viruses containing a GNYA loop acquired a compensatory mutation after some passages, T154 to I, again showing a direct relation between the apical D-loop and TGK motif (D.V. Bakhmutov, A.P. Gmyl, E.V. Khitrina, M.M. Erokhin, J. Zoll, W.J.G. Melchers, H.A. Heus, and V.I. Agol, in prep.). Studies of engineered 3C mutants revealed several residues outside the known TGK and KFRDIR motifs, which might be involved in RNA binding (Andino et al. 1993). The protein residues implicated in RNA binding are indicated in Figure 5B. On the available high-resolution structure of 3C obtained by X-ray crystallography (Mosimann et al. 1997), the residues map to the presumed RNA-binding site that involves a cluster of the TGK and KFDIR motifs, and amino acids from the domain linker, the amphipatic helices C and D, and several β-hairpin turns on the protein surface.
The crystal structure of 3C, together with the NMR structures of the apical D-loops, provide for a framework to further understand the specificity of binding. For instance, it has been suggested that, although the second nucleotide of the YNMG loops that folds into the minor groove is not relevant for the isolated hairpin loop structures, its sequence conservation (with 52% A and 23% G) might indicate possible involvement in binding (Du et al. 2004). However, the in vivo SELEX study yielded a GCUA loop, and the BEV1 D-loop contains a uridine at the second loop position. Thus, the N-position may be occupied by all four possible residues, which adopt an anti conformation as deduced from the NMR structures. This gives a variety of hydrogen-bond donors or acceptors at comparable sites, making a specific role of the second (N) base less likely. Using a larger CVB3 D-stem construct including the three noncanonical pyrimidine base pairs, specific residues have been implicated in binding on the basis of NMR line broadening and chemical shift changes upon protein 3C addition (Ohlenschlager et al. 2004). Specifically, it was suggested that the minor groove side of the tetraloop together with the major groove side at the C54–C71 region (Fig. 1A) form a bipartite interdependent recognition site for 3C. However, in the GCUA structure, overwinding of the G6 and G9 residues and steep descent of the second variable nucleotide cover possible backbone hydrogen-bond donors and acceptors on the minor groove side of the tetraloop, which is more in favor of the minor groove side being silent in recognition. Also, the similarity between the UUCG and GCUA loops is larger on the major groove side than on the minor groove side. Therefore, the importance for binding of the major groove side of the tetraloop, with a cluster of more accessible hydrogen-bond donors and acceptors, should not be overlooked. The 872uGAAAGg878 pentaloop present in hp35a of H. marismortui LSU RNA closely resembles the GCUA loop. Here also a G-A trans sugar edge/Watson-Crick base pair is formed, and the fifth base, G879, is bulged out, leaving a tetraloop with similar stacking properties as the GCUA loop, be it that the second loop base, A874, is pointing away from the minor groove to form a tertiary stacking contact with A857. Ribosomal protein L2 makes several specific contacts with hp35a in the LSU crystal structure (Klein et al. 2004). Arg2 forms hydrogen bonds with G871-O6, U872-O4, and G878-O6; Arg8, with G873-O6; and Asp198, with the G873 imino and amino protons. All these contacts are on the major groove side of the hairpin loop and two flanking base pairs. Possibly similar interactions can be fulfilled by comparable residues of the stem–loop D apex, in agreement with the importance of its major groove side for 3C recognition.
Figure 5 gives an impression of the spatial relationship between the compact tetraloop that might snugly fit into a tight binding pocket of the 3C protein, but also reveals that the size of the D-loop region implicated in binding (ranging from the tetraloop down to C54) probably exceeds the size of the presumed RNA-binding site of 3C. Clearly, more studies are needed to address these issues.
In summary, the structural data presented here further explain the sequence requirements of the apical D-loop and support the notion that the tetraloop structure is the primary determinant for 3C recognition. This can be used to direct future studies on the role of the 5′-cloverleaf–3CD interaction in enterovirus RNA replication. The novel structure prompted us to re-evaluate the secondary structure database, and the phylogenetic comparison revealed another example of a GYYA loop with a structural role, possibly involved in a triple interaction. Also, the structurally comparable RNA stem–loop 35a from H. marismortui 50S ribosomal subunit is a major recognition site in L2 protein binding. These observations show precedence for a more widespread occurrence and importance of the GCUA fold we describe here. The structures of the GCUA and GCAA loops are very different. This exemplifies again the extreme structural flexibility of RNA, where a 1-nt change can make a huge structural difference, making it virtually impossible to propose a reliable structure from sequence alone, when noncanonical interactions are involved.
MATERIALS AND METHODS
Sample preparation
The 14-mer RNA oligonucleotide was purchased from ABI GmbH. The final concentration of the RNA samples was 0.5 mM. The pH was adjusted to 6.8 (meter reading). Weakly aligned NMR samples were prepared by adding 27 mg/mL filamentous phage Pf1 (Hansen et al. 1998) purchased from ASLA Ltd.
NMR spectroscopy
NMR spectra were acquired on Varian Inova 600 MHz and 800 MHz spectrometers. For the assignment of exchangeable imino and amino protons, 2D NOESY spectra and natural abundance 1H–15N HMQC were recorded in 90% H2O, 10% D2O at 15°C and 800 MHz using a jump return pulse for water suppression (Plateau and Guéron 1982). Other spectra were recorded at 25°C in D2O at 600 and 800 MHz. Nonexchangeable protons were assigned using 2D NOESY (50, 100, and 300 msec mixing times) (Jeener et al. 1970), DQF-COSY (Shaka and Freeman 1983), and TOCSY (Griesinger et al. 1988). Residual dipolar couplings (RDCs) were collected by measuring heteronuclear 1H–13C one-bond couplings in the absence or presence of filamentous phage Pf1 using a f1-proton coupled 2D-HSQC or the IPAP approach (Ottiger et al. 1998) at 600 MHz with cryoprobe technology. Typical acquisition times for the IPAP spectra at natural abundance were ∼24 h with 48 scans per FID, 1024 t 2 data points, and 320 complex t 1 data points. Different data sets were collected involving the aromatic H8, H6, and H2, and the H1′ and H5 with the INEPT-delay set to 2.5 and 2.9 msec, respectively. All data were analyzed with Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco).
Structural restraints
Distances involving the nonexchangeable protons were calculated from NOE cross-peak intensities of the 2D NOESYs with 50, 100, and 300 msec mixing times using the modified isolated spin pair approximation approach (Barsukov and Lian 1993). The calculated distances were used with ±20% error bounds. Distance information involving exchangeable protons was obtained from 2D NOESY spectra recorded in H2O. Hydrogen-bonding restraints were treated as NOE distance restraints and imposed on residues involved in Watson-Crick base-pairing, evidenced by the imino proton spectra and observed NOEs and chemical shifts.
Torsion angle restraints for the sugar moieties were obtained from DQF-COSY and TOCSY experiments, and interpreted as N or S-type sugar puckers when they were small (<3 Hz) or large (>8 Hz), respectively. Dihedral torsion restraints were applied as previously described (Lescrinier et al. 2003). For the stem residues, the ε dihedral angle was restrained to 225° (±60°). The α and ζ dihedral angles were constrained from chemical shift considerations of the phosphorous resonance to 0° (±120°). RDCs were used with 20% error bounds.
Structure calculations
Structure calculations were performed with the torsion angle dynamics protocol implemented within XPLOR-NIH (Stein et al. 1997; Schwieters et al. 2003) and included simulated annealing starting from extended strands. Out of starting sets of 100 randomly extended structures, 20 acceptable structures were obtained with no dihedral angle violations >5° and no NOE violations >0.5 Å (Table 1) and subjected to refinement using RDCs. Initial values for the axial and rhombic components of the alignment tensor were estimated from the distribution of the RDC values (Clore et al. 1998; Bax et al. 2001). The subsequent grid search yielded optimal values of Da = −50 Hz and R = 0.4 Hz. Da was varied from −35 Hz to −55 Hz, and R was varied from 0 Hz to 0.6 Hz in steps of 1 Hz and 0.1 Hz, respectively. The correlation coefficient for the back-calculated RDCs increased from 0.46 to 0.997 after inclusion of the RDCs in the structure calculations. Color figures were generated using MOLMOL (Koradi et al. 1996). Helical parameters were calculated using the programs 3DNA (Lu and Olson 2003) or freehelix98 (Dickerson and Chiu 1997; Dickerson 1998).
UV melting curves and thermodynamic parameters
UV absorbance melting curves were recorded at both 260 and 280 nm simultaneously on a Cary-300 spectrometer. Samples (A 260 = 0.7–1.5 cm−1; corresponding to ∼7–15 μM) were prepared in 10 mM sodium phosphate, 1 M NaCl, and 0.1 mM Na2EDTA. The samples were heated a few minutes to 90°C and quickly cooled to 1°C. The melting curves were measured at a heating rate of 0.5°C/min from 15°C to 90°C with data collected every 0.25°C increment. The melting curves were run in triplicate at two concentrations and showed identical profiles indicating reversible folding and single unimolecular transitions corresponding to unfolding of hairpins. Melting temperatures (T m) and thermodynamic parameters were derived assuming a two-state model using the Cary-WIN software. Enthalpy values and other thermodynamic parameters were calculated from the first derivative of the absorbance with respect to temperature, using the equation (Gralla and Crothers 1973):
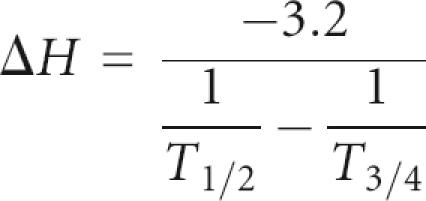 |
In this equation, T 1/2 is the temperature at the maximum, which is equal to the melting temperature T m, and T 3/4 is the temperature at the upper half-height of the derivative of the melting curve.
Atomic coordinates
Coordinates of the ensemble of lowest energy structures have been deposited in the RCSB Protein Data Bank (accession code 2EVY).
ACKNOWLEDGMENTS
This research was supported by grants of the Council of Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW 700.50.021 and NWO-RFBR 047.017.023) and the European Communities (INTAS). The Moscow group was also supported by the Russian Foundation for Basic Research and the Program for Support of Leading Scientific Schools.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.113106.
REFERENCES
- Ackerman E.J., Saxena S.K., Ulbrich N. α-Sarcin causes a specific cut in 28S rRNA when microinjected into Xenopus oocytes. J. Biol. Chem. 1988;263:17076–17083. [PubMed] [Google Scholar]
- Allain F.H., Varani G. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J. Virol. 1990;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao V.P., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1991;21:5901–5905. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao V.P., Lai S.Y., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1991;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsukov I.L., Lian L.Y. Structure determinations from NMR data I. Analysis of NMR data. In: Roberts G.C.K., editor. NMR of macromolecules. A practical approach. Oxford University Press; New York: 1993. pp. 315–357. [Google Scholar]
- Batey R.T., Sagar M.B., Doudna J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- Bax A., Kontaxis G., Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- Cai Z., Gorin A., Frederick R., Ye X., Hu W., Majumdar A., Kettani A., Patel D.J. Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex. Nat. Struct. Biol. 1998;5:203–212. doi: 10.1038/nsb0398-203. [DOI] [PubMed] [Google Scholar]
- Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The comparative RNA Web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3 doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.E., Doudna J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn A.M., Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- Dickerson R.E. DNA bending: The prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R.E., Chiu T.K. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Du Z., Yu J., Andino R., James T.L. Extending the family of UNCG-like tetraloop motifs: NMR structure of a CACG tetraloop from coxsackievirus B3. Biochemistry. 2003;42:4373–4383. doi: 10.1021/bi027314e. [DOI] [PubMed] [Google Scholar]
- Du Z., Yu J., Ulyanov N.B., Andino R., James T.L. Solution structure of a consensus stem loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C., Nikonov S., Dumas P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D.M. Free energy of imperfect nucleic acid helices. III. Small internal loops resulting from mismatches. J. Mol Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- Griesinger C., Otting G., Wüthrich K., Ernst R.R. Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc. 1988;110:7870–7872. [Google Scholar]
- Gutell R.R., Weiser B., Woese C.R., Noller H.F. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Mueller L., Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Heus H.A., Hilbers C.W. Structures of non-canonical tandem base pairs in RNA helices: Review. Nucleosides Nucleotides Nucleic Acids. 2003;22:559–571. doi: 10.1081/NCN-120021955. [DOI] [PubMed] [Google Scholar]
- Heus H.A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Hilbers C.W., Heus H.A., van Dongen M.J.P., Wijmenga S.S. The hairpin elements of nucleic acid structure: DNA and RNA folding. In: Eckstein F., Lilley D.M.J., editors. Nucleic acids and molecular biology. Vol. 8. Springer Verlag; Berlin: 1994. pp. 56–104. [Google Scholar]
- Ihle Y., Ohlenschlager O., Hafner S., Duchardt E., Zacharias M., Seitz S., Zell R., Ramachandran R., Gorlach M. A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA. Nucleic Acids Res. 2005;33:2003–2011. doi: 10.1093/nar/gki501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeener J., Meier B.H., Bachmann P., Ernst R.R. Investigation of exchange processes by two dimensional NMR spectroscopy. J. Chem. Phys. 1970;71:4546–4553. [Google Scholar]
- Jucker F.M., Heus H.A., Yip P.F., Moors E.H., Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Legault P., Li J., Mogridge J., Kay L.E., Greenblatt J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Leontis N.B., Stombaugh J., Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescrinier E.M.H.P., Tessari M., van Kuppeveld F.J.M., Melchers W.J.G., Hilbers C.W., Heus H.A. Structure of the pyrimidine-rich internal loop in the poliovirus 3′-UTR: The importance of maintaining pseudo-2-fold symmetry in RNA helices containing two adjacent non-canonical base-pairs. J. Mol. Biol. 2003;331:759–769. doi: 10.1016/s0022-2836(03)00787-3. [DOI] [PubMed] [Google Scholar]
- Lu X., Olson W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T., Murray K.E., Roberts A.W., Barton D.J. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 2001;75:10696–10708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Murphy F.L., Cech T.R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlenschlager O., Wohnert J., Bucci E., Seitz S., Hafner S., Ramachandran R., Zell R., Gorlach M. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Pace N.R., Smith D.K., Olsen G.J., James B.D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA—A review. Gene. 1989;82:65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Plateau P., Guéron M. Exchangeable proton NMR without baseline distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- Poritz M.A., Strub K., Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Proctor D.J., Schaak J.E., Bevilacqua J.M., Falzone C.J., Bevilacqua P.C. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry. 2002;41:12062–12075. doi: 10.1021/bi026201s. [DOI] [PubMed] [Google Scholar]
- Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Semler B.L., Wimmer E. Molecular biology of picornaviruses. ASM Press; Washington, DC: 2002. [Google Scholar]
- Serra M.J., Turner D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- Shaka A.J., Freeman R. Simplification of NMR spectra by filtration through multiple quantum coherence. J. Magn. Reson. 1983;51:169–173. [Google Scholar]
- Stein E.G., Rice L.M., Brünger A.T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- Su L., Chen L., Egli M., Berger J.M., Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O.C. Tetraloops and RNA folding. Nature. 1990;346:613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- van Knippenberg P.H. Structural and functional aspects of the N6,N6 dimethyladenosines in 16S ribosomal RNA. In: Hardesty B., Kramer B., editors. Structure, function and genetics of ribosomes. Springer-Verlag; New York: 1986. pp. 412–424. [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr. Structure of an unusually stable RNA hairpin. Biochemistry. 1991;30:3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Walker P.A., Leong L.E., Porter A.G. Sequence and structural determinants of the interaction between the 5′-noncoding region of picornavirus RNA and rhinovirus protease 3C. J. Biol. Chem. 1995;270:14510–14516. doi: 10.1074/jbc.270.24.14510. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen D.E., Clemons W.M., Jr., Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Woese C.R., Winker S., Gutell R.R. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops”. Proc. Natl. Acad. Sci. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Sidigi K., Bucci E., Stelzner A., Gorlach M. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA. 2002;8:188–201. doi: 10.1017/s1355838202012785. [DOI] [PMC free article] [PubMed] [Google Scholar]