Abstract
Ribosome-inactivating proteins, such as the pokeweed antiviral protein (PAP), inhibit translation by depurinating the conserved sarcin/ricin loop of the large ribosomal RNA. Depurinated ribosomes are unable to bind elongation factor 2, and, thus, the translocation step of the elongation cycle is inhibited. Though the consequences of depurination are well characterized, the ribosome conformation required for depurination to take place has not been described. In this report, we correlate biochemical and genetic data to conclude that pokeweed antiviral protein depurinates the sarcin/ricin loop when the A-site of the ribosomal peptidyl-transferase center is unoccupied. We show that prior incubation of ribosomes with puromycin, an analog of the 3′-terminus of aminoacyl-tRNA, inhibits both binding and depurination by PAP in a concentration-dependent manner. Expression of PAP in the yeast strain mak8-1 results in little depurination unless the cells are lysed, a process that would promote loss of aminoacyl-tRNA from the ribosome. The mak8-1 strain is known to exhibit a higher affinity for aminoacyl-tRNA compared with wild-type cells, and therefore, its ribosomes are more resistant to PAP in vivo. These data contribute to the mechanism of action of pokeweed antiviral protein; specifically, they have uncovered the ribosomal conformation required for depurination that leads to subsequent translation inhibition.
Keywords: pokeweed antiviral protein, ribosome inactivating protein, translation elongation, translocation, A-site, aminoacyl-tRNA
INTRODUCTION
Pokeweed antiviral protein (PAP) is an N-glycosidase isolated from the pokeweed plant (Phytolacca americana) (Endo et al. 1988; Bolognesi et al. 1990). PAP is a member of a larger group of proteins referred to as ribosome-inactivating proteins, and all are characterized by their ability to depurinate the highly conserved sarcin/ricin loop of the large ribosomal RNA. This depurination inhibits the elongation cycle of protein translation and is considered the reason for toxicity of many of these proteins (Gessner and Irvin 1980; Irvin 1995). Ribosome inactivating proteins are presumably synthesized by many plants as a form of defense against pathogens, and their selective toxicity has led to their application in the generation of pathogen-resistant transgenic plants and their use as biological weapons and as therapeutic agents against cancer (for reviews, see Irvin and Uckun 1992; Neilsen and Boston 2001; Wang and Hudak 2003; Park et al. 2004).
During the elongation cycle of translation, the elongation factor eEF1A (EF-Tu in prokaryotes), with GTP, brings an aminoacyl-tRNA to the A-site of the ribosome. GTPase activity is stimulated by aminoacyl-tRNA anticodon recognition of the mRNA codon and hydrolysis of GTP, then releases eEF1A following delivery of the aminoacyl-tRNA (Pape et al. 1998). Peptide bond formation is catalyzed by the rRNA (Nissen et al. 2000; Schmeing et al. 2002) and causes the transfer of the peptide from the peptidyl-tRNA in the P-site of the ribosome to the A-site, thereby extending the peptide chain by one amino acid (for review, see Merrick and Nyborg 2000). The nascent chain, located now at the A-site, is translocated to the P-site, and this movement is mediated by elongation factor eEF-2 (EF-G in prokaryotes) at the expense of GTP hydrolysis (Rodnina et al. 1997). The deacylated tRNA leaves the ribosome via the E-site, a new aminoacyl-tRNA is brought to the A-site, and the cycle is repeated (for reviews, see Ramakrishnan 2002; Andersen et al. 2003; Joseph 2003).
Several reports have cited that it is the translocation step of elongation that is inhibited upon depurination of the rRNA (Sperti et al. 1976; Gessner and Irvin 1980; Osborn and Hartley 1990). These observations are supported by work with various ribosome-inactivating proteins indicating that eEF-2 binding is inhibited by depurination (Montanaro et al. 1975; Brigotti et al. 1989). Therefore, PAP and other ribosome-inactivating proteins inhibit translation by depurinating the sarcin/ricin loop and thereby preventing eEF-2 binding needed for the movement of the peptidyl-tRNA from the A-site to the P-site of the ribosome.
Though the consequences of rRNA depurination are understood, the precise stage of the elongation cycle at which the ribosome is susceptible to depurination is not known. Through the use of both genetic and biochemical studies, we show that the ribosome is depurinated when the peptidyl-transferase A-site is empty, that is, when neither an aminoacyl- nor peptidyl-tRNA is bound at this site. Previously, we reported that a mutant strain of yeast, mak8-1, which harbors two point mutations in the ribosomal protein L3 (W255C, P257T), was resistant to depurination by PAP (Hudak et al. 1999). In the present study, we correlate the observed in vivo resistance to known increased affinity for aminoacyl-tRNAs of this strain. We also show that this resistance is lost upon disruption of cells or isolation of ribosomes, events that would promote loss of aminoacyl-tRNA from the ribosome. In addition, we show that puromycin, an analog of the 3′ terminus of aminoacyl-tRNA, protects the ribosome from depurination. Moreover, incubation of ribosomes with puromycin prevents PAP binding to these ribosomes. The requirement for an empty peptidyl-transferase A-site provides the first evidence that identifies the discrete step in the elongation cycle that is targeted by a ribosome inactivating protein and contributes to understanding the interaction of these toxic proteins with ribosomes.
RESULTS
Expression of PAP in yeast cells
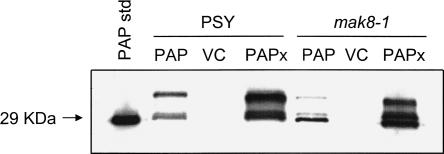
PSY (wt L3) and mak8-1 (mutant L3) cells used in this study were transformed with plasmids encoding wtPAP, PAPx (an active site mutant of PAP), or the empty vector (vector control, VC). The cells were induced to transcribe PAP for 4 h, and an immunoblot analysis was performed using a PAP-specific polyclonal antibody (Fig. 1). wtPAP and PAPx were expressed in both PSY and mak8-1, with PAPx being expressed to a higher level than wtPAP in both cell types. This difference in expression levels in yeast cells has been shown previously (Hudak et al. 1999). The higher molecular mass proteins, larger than the mature size of PAP at 29 kDa, likely represent the precursor forms of PAP, observed previously in yeast (Hur et al. 1995). The results indicate similar levels of expression of PAP in PSY and mak8-1 cells.
FIGURE 1.
Immunoblot analysis of PAP expression in PSY and mak8-1 yeast cells. PSY and mak8-1 cells were induced to transcribe wtPAP and PAPx in SD-Leu 2% galactose medium for 4 h. Total cellular protein (10 μg) was separated by 12% SDS-PAGE, transferred to nitrocellulose, and probed with PAP-specific polyclonal antibody (1:5000). Cells expressing no PAP (vector control) were used as a negative control for PAP expression. Purified PAP (100 ng) was used as a standard.
Depurination of PSY and mak8-1 ribosomes in vivo and in vitro
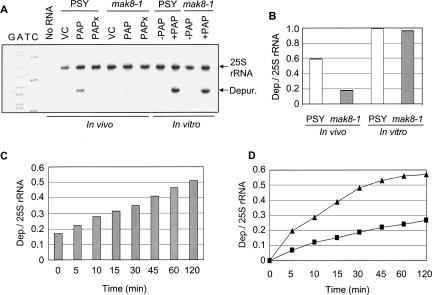
Previous studies using aniline treatment demonstrated that ribosomes of mak8-1 cells expressing PAP were not depurinated (Hudak et al. 1999). Primer extension analysis used in the present study is a more sensitive technique for detecting small amounts of depurination. To determine the level of depurination of ribosomes by PAP in vivo, PSY and mak8-1 cells were induced to transcribe wtPAP, followed by RNA extraction and primer extension analysis (Fig. 2A). The 25S rRNA of ribosomes in PSY cells was depurinated, but interestingly, a small amount of depurination was also observed in mak8-1 cells. Therefore, primer extension analysis showed depurination of mak8-1 ribosomes in vivo, that was previously undetected, although at a significantly lower level than that observed for PSY ribosomes. As expected, the catalytically inactive PAPx did not depurinate the rRNA of either cell type.
FIGURE 2.
Depurination of 25S rRNA in PSY and mak8-1 cells in vivo and in vitro. (A) For in vivo analysis, PSY and mak8-1 cells were induced to transcribe wtPAP and PAPx in SD-Leu 2% galactose medium for 4 h. Total yeast RNA was purified from induced cells and subjected to primer extension. For in vitro analysis, ribosomes (30 μg) from uninduced mak8-1 and PSY cells were incubated with purified PAP (30 ng) for 30 min at 30°C. Ribosomal RNA was extracted and subjected to primer extension analysis. Vector controls (VC) were cells that had been transformed with an empty vector. 25S rRNA is a control for total RNA in each sample and “Depur.” indicates the degree of depurination of the sarcin/ricin loop. Deoxynucleotide sequencing of the 25S rDNA with the depurination primer indicates the site of the missing purine. The gel is a representative of four independent experiments. (B) Densitometric analysis was performed on the level of depurination in each sample by taking the ratio of depurination to 25S rRNA band densities. (C) Depurination of 25S rRNA of mak8-1 cells immediately following lysis versus incubation after cell lysis. mak8-1 cells were induced to transcribe wtPAP for 4 h in SD-Leu 2% galactose medium. Total RNA was extracted from the lysate of cells immediately after induction (time zero) and from lysate incubated at 30°C for the times indicated after lysis. RNA collected from each sample was analyzed by primer extension. Densitometric analysis was performed by taking the ratio of depurination to 25S rRNA band densities. (D) Time course of ribosome depurination in lysates of PSY and mak8-1 cells. Total cellular lysate (400 μg) was incubated with PAP (20 ng) and aliquots were removed at the indicated time points. Following incubation, total RNA was extracted and the level of rRNA depurination was determined by primer extension analysis. Densitometric analysis was performed on each sample by taking the ratio of depurination to 25S rRNA band densities. Triangles indicate the level of depurination in PSY lysate and squares indicate the level of depurination in mak8-1 lysate.
Next, the sensitivity of PSY and mak8-1 ribosomes to depurination was tested in vitro using isolated ribosomes from these cells. PAP was incubated with the isolated ribosomes, followed by rRNA extraction and primer extension analysis (Fig. 2A). The ribosomes of both cell types showed increased sensitivity to depurination when incubated with a concentration of PAP that was calculated to equal the concentration expressed in vivo. Densitometric analysis indicated that the level of depurination seen in vitro for mak8-1 ribosomes was notably higher relative to in vivo depurination, indicating that the sensitivity of mak8-1 ribosomes to depurination had increased upon isolation from the cells (Fig. 2B). An increase in depurination was also seen for PSY ribosomes, though less than observed for mak8-1, relative to in vivo levels of depurination.
Effect of cell lysis on susceptibility of mak8-1 ribosomes to depurination
Primer extension analysis indicated that ribosomes of mak8-1 were more susceptible to depurination in vitro than in vivo (Fig. 2A). For the in vitro analysis, ribosomes were isolated from cells and incubated with purified PAP, whereas in vivo analysis relied on depurination in intact cells expressing PAP. To determine if isolation of ribosomes caused the removal of certain factors preventing in vivo depurination, mak8-1 cells were induced to transcribe wtPAP, were lysed, and the level of depurination was determined over time. Total cellular RNA of an aliquot at time zero was immediately isolated following lysis. Aliquots of the remaining sample were removed at the indicated time points and total cellular RNA was isolated from each. The level of rRNA depurination for all aliquots was determined by primer extension analysis. Increased depurination was observed over time in the incubated cell lysate (Fig. 2C), suggesting that lysis of the cells increased the accessibility of PAP to mak8-1 ribosomes.
The resistance of the mak8-1 strain to depurination relative to the PSY strain suggested that the rate of depurination in mak8-1 cells would be slower compared with PSY cells. To investigate this possibility, purified PAP was added to lysates of mak8-1 and PSY cells and the level of depurination was determined over time. Figure 2D illustrates that the rate of depurination of mak8-1 lysates was lower than in PSY lysates. The slopes of the lines, 0.03 and 0.08 for mak8-1 and PSY, respectively, correlate well with the published values of affinity of mak8-1 and wild-type ribosomes for aminoacyl-tRNA. Specifically, the association constant of mak8-1 ribosomes for aminoacyl-tRNA is 6.0 × 107 M−1 compared with 0.7 × 107 M−1 for wild-type ribosomes (Petrov et al. 2004). We hypothesize that the greater affinity of mak8-1 ribosomes for aminoacyl-tRNA leads to the observed decreased depurination in these cells relative to PSY.
Effect of puromycin on depurination by PAP in vitro
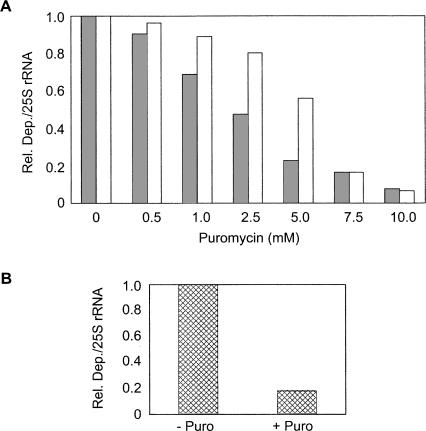
To investigate whether the difference in the affinity of mak8-1 ribosomes for aminoacyl-tRNA was responsible for the observed resistance of this strain to depurination in vivo, the effect of puromycin on PAP activity was analyzed. Puromycin is a structural analog of the 3′ terminus of aminoacyl-tRNA and binds to the A-site of the peptidyl-transferase center (Hansen et al. 2003). Ribosomes, isolated from PSY and mak8-1 cells, were incubated with increasing concentrations of puromycin, followed by incubation with purified PAP. Ribosomal RNA was then isolated and analyzed by primer extension. The results indicate that the level of depurination of PSY and mak8-1 ribosomes decreased with increasing puromycin concentration (Fig. 3A). Therefore, incubation of ribosomes with puromycin prior to the addition of PAP inhibited depurination of both PSY and mak8-1 ribosomes. However, depurination of mak8-1 ribosomes was inhibited to a greater extent than PSY ribosomes incubated with the same concentration of puromycin, which is consistent with the increased affinity of mak8-1 ribosomes for aminoacyl-tRNA (Petrov et al. 2004). Furthermore, these results suggest that peptidyl-transferase A-site occupancy correlates inversely with PAP sensitivity.
FIGURE 3.
Effect of puromycin on depurination by PAP. (A) Ribosomes (50 μg) of PSY (white bars) and mak8-1 (gray bars) cells were incubated with increasing concentrations of puromycin at 4°C for 30 min. PAP (50 ng) was then added to each sample, followed by incubation at 30°C for 30 min. rRNA was extracted for primer extension analysis and results were analyzed by densitometry. The bar graph was generated using the relative ratio of depurination band intensity to 25S rRNA band intensity for each sample. (B) A rabbit reticulocyte lysate system containing template transcript was incubated for 30 min at 30°C prior to the addition of puromycin (10 mM) to half of the lysate (+ Puro). Lysate, with and without the addition of puromycin, was incubated for an additional 30 min prior to the addition of PAP (5 ng) to both of the lysate samples. Following 1 h of incubation, total RNA was extracted for primer extension analysis. Densitometric analysis was performed on each sample by taking the relative ratio of depurination to 25S rRNA band densities. The graphs are representative of three independent experiments each.
Effect of puromycin on PAP activity in an in vitro translation system
To determine if translating ribosomes were also less susceptible to depurination by PAP in the presence of puromycin, a similar experiment was conducted using the rabbit reticulocyte lysate translation system. Analysis of the depurination levels in lysate with and without puromycin addition indicates that ribosomes likely participating in translation are also protected from depurination by puromycin (Fig. 3B). This protection by puromycin could either be direct, due to puromycin binding to the peptidyl-transferase A-site, or indirect, due to peptide release from the peptidyl-tRNA at the A-site. The effect on depurination by the addition of puromycin to reticulocyte lysate does not differentiate between these two possibilities, and an indirect effect may contribute to levels of depurination in vivo. However, the complementary experiment to show the effect of puromycin concentration (Fig. 3A) involved the addition of puromycin to ribosomes that were salt-washed and previously incubated with puromycin to free ribosomes of peptidyl-tRNA prior to subsequent incubation with puromycin. Therefore, the observed decrease in depurination of isolated ribosomes with increased concentration of puromycin was not likely due to loss of peptide chains from ribosomal A-sites, but more likely due to the physical occupation of the peptidyl-transferase A-site by puromycin.
To determine if the decreased depurination of rRNA observed in the presence of puromycin was due to a direct effect of the inhibitor on PAP itself, PAP was incubated in vitro with Brome mosaic virus (BMV) RNAs in the presence or absence of puromycin. We have shown previously that PAP depurinates BMV RNAs and that subsequent incubation of the viral RNAs with aniline results in cleavage of the RNAs at the sites of depurination (Hudak et al. 2000). However, inclusion of puromycin during incubation did not visibly alter the effect of PAP on BMV RNAs; therefore, puromycin does not appear to directly affect the depurination ability of PAP (data not shown).
Binding of PAP or PAPx to ribosomes in vitro
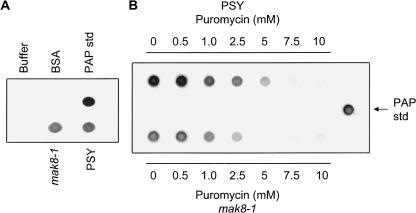
In a previous report, PAP binding to mak8-1 ribosomes in vivo was not detected by immunoblot analysis of isolated ribosomes, suggesting only a low level of binding (Hudak et al. 1999). In this study, the low level of depurination seen in vivo in mak8-1 cells correlates with the undetected level of PAP binding to these ribosomes noted previously. However, since a greater amount of depurination of mak8-1 ribosomes was observed in vitro compared to in vivo, the binding of PAP to these ribosomes was examined by Far Western assay. The direct application of protein to nitrocellulose allowed for the test of interaction without denaturation of the proteins. PAP bound to the ribosomes of both cell types, and the degree of PAP binding to mak8-1 ribosomes, was similar to its binding to PSY ribosomes (Fig. 4A). Therefore, the binding of PAP to ribosomes of mak8-1 was higher in vitro compared with previous in vivo results and correlates with increased depurination of mak8-1 ribosomes in vitro compared with in vivo.
FIGURE 4.
Far Western analysis of in vitro binding of PAP to PSY and mak8-1 ribosomes. (A) Ribosomes, isolated from PSY and mak8-1 cells (50 μg), were dotted on nitrocellulose membrane and incubated with 500 ng of purified PAP. The nitrocellulose was probed for PAP with a PAP-specific polyclonal antibody (1:5000). BSA (10 μg) and wash buffer (100 μL) were used as negative controls for PAP binding, and purified PAP (50 ng) was used as a positive control for antibody specificity. (B) Far Western analysis of the effect of puromycin on PAP binding to ribosomes. PSY and mak8-1 ribosomes (50 μg) were incubated with increasing concentration of puromycin at 4°C for 30 min. The samples were dotted on nitrocellulose membrane, followed by incubation with 500 ng PAP. The membrane was probed with PAP-specific polyclonal antibody (1:5000). Purified PAP (50 ng) was dotted on the membrane as positive control for antibody specificity. Each blot is representative of three independent experiments.
Effect of puromycin on PAP binding to ribosomes
Since puromycin inhibited the depurination of ribosomes by PAP, the effect of this antibiotic on binding of PAP to ribosomes of PSY and mak8-1 cells was also tested by the Far Western assay. Isolated ribosomes were incubated with increasing concentrations of puromycin and the samples were dotted on nitrocellulose membrane and probed for PAP (Fig. 4B). A decrease in the binding of PAP to ribosomes of both PSY and mak8-1 cells was observed with an increasing concentration of puromycin. Therefore, puromycin decreased the ability of PAP to bind to the ribosomes. Interestingly, binding of PAP to mak8-1 ribosomes diminished at a lower concentration (5 mM) of puromycin than PAP binding to PSY ribosomes, which remained clearly visible at this concentration. Decreased binding of PAP to mak8-1 ribosomes incubated with 5 mM puromycin is in good agreement with decreased depurination of these ribosomes relative to PSY ribosomes in the presence of puromycin.
DMS protection of puromycin treated ribosomes
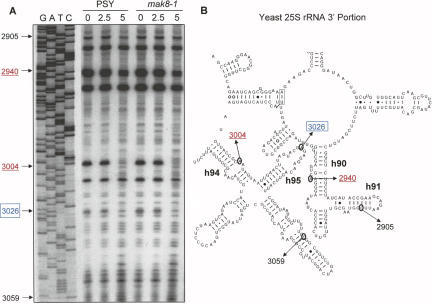
To determine if binding of puromycin to ribosomes caused changes to the rRNA structure or accessibility at the sarcin/ricin loop that may have attributed to the inhibition of depurination by PAP, both PSY and mak8-1 ribosomes were incubated with puromycin, followed by probing with the methylating agent dimethylsulphate (DMS). Subsequent primer extension, using the depurination primer, illustrates the methylation pattern of ∼150 nt of domains V and VI of the yeast 25S rRNA including the sarcin/ricin loop (Fig. 5A). Deoxynucleotide sequencing of the 25S rRNA gene with the same depurination primer identifies the nucleotides within the footprinting gel. Methylation of nucleotides by DMS causes premature termination of the reverse transcriptase one nucleotide 3′ of the methylation site. The extension patterns illustrate that puromycin altered the structure of the rRNA near the sarcin/ricin loop of both PSY and mak8-1 ribosomes. Specifically, puromycin protected the RNA from DMS attack most notably at nucleotide A3004 and to a much lesser extent at nt A2940. As illustrated in the partial secondary structure of the 25S rRNA, nt A2940 is within helix 90 of the peptidyl-transferase center, noted previously to be affected by puromycin binding (Rodriguez-Fonseca et al. 2000) and also to interact with the 3′ end of aminoacyl-tRNA (Sanbonmatsu et al. 2005). Nucleotide A3004, within helix 94, is in close proximity to helix 95, which contains A3026, the adenine removed by PAP (Fig. 5B). A2660, the prokaryotic equivalent of A3026, also interacts with the 3′ end of the aminoacyl-tRNA during accommodation into the A-site (Sanbonmatsu et al. 2005). Hydrogen bonding between nucleotides of the loops of helix 95 and helix 91 (Ban et al. 2000) may bring A3004 in close proximity to helix 90, thereby also affecting this nucleotide by puromycin binding. Therefore, the binding of puromycin to the peptidyl-transferase A-site not only modifies the conformation of rRNA at the peptidyl-transferase center noted previously, but also alters the conformation of the rRNA and/or its accessibility in close proximity to the depurination target of PAP.
FIGURE 5.
Dimethylsulphate protection of the sarcin/ricin loop region of the 25S rRNA. (A) Ribosomes (50 μg) of PSY and mak8-1 cells were either incubated with puromycin (2.5 or 5 mM) or buffer alone for 30 min (0 mM puromycin). All samples were then treated with DMS (35 mM), and rRNA was isolated for primer extension. Samples were separated through a 7 M urea/6% acrylamide gel. Deoxynucleotide sequencing of the 25S rDNA with the same primer used for extension indicates the nucleotides analyzed within domains V and VI of the 25S rRNA. (B) A portion of the secondary structure of the yeast 25S rRNA domains V and VI is shown. Nucleotides altered by methylation in the presence of puromycin are indicated in red underlined font and the adenine target of PAP is shown by blue boxed font. Nucleotides in black font indicate the separation range of the gel.
DISCUSSION
The understanding that ribosome-inactivating proteins inhibit translation elongation and that depurination of the sarcin/ricin loop hinders the binding of elongation factor 2 is well documented (Osborn and Hartley 1990; Brigotti et al. 1989). The role of this factor is to mediate translocation of the peptide chain from the A-site to the P-site of the ribosome. Hence, PAP inhibits the translocation step of elongation. Data presented in this study show that PAP depurinates the ribosome when the A-site of the peptidyl-transferase center is empty; therefore, the conformation of the ribosome that is susceptible to depurination is different from the conformation at which the defective ribosome is stalled due to depurination by PAP.
Evidence for the requirement of an empty peptidyl-transferase A-site is shown by the use of puromycin, an analog of the 3′ terminus of aminoacyl-tRNA that mimics its binding to the A-site of the peptidyl-transferase center. Incubation of ribosomes with puromycin inhibited subsequent depurination by PAP. Moreover, the same concentration of puromycin required to inhibit depurination also decreased PAP binding to ribosomes. Exactly how puromycin binding to the A-site of the peptidyl-transferase center would affect the binding of PAP to the sarcin/ricin loop is not known; however, footprinting data show that puromycin incubated with ribosomes altered the ability of dimethylsulphate to methylate nucleotides close to the sarcin/ricin loop. Therefore, puromycin binding changes the shape and/or accessibility of the rRNA near the sarcin/ricin loop and may alter the ability of PAP to bind to this site. Moreover, recent published data suggest that the occupancy status of the peptidyl-transferase center is allosterically transmitted to the factor-binding site, consisting of the GTPase-associated center and the sarcin/ricin loop. For example, structural probing experiments revealed a defined chain of nucleotides connecting the P-loop of the 23S rRNA to the elongation factor binding site, supporting the hypothesis that the occupancy status of the P-site determines the conformation of the movable GTPase-associated center relative to the sarcin/ricin loop, to either enhance or inhibit EF-G binding (Sergiev et al. 2005). In addition, extensive mutational analysis of the ribosomal protein L3 indicated that amino acid changes displaying the more pronounced mutant phenotypes were those in close proximity to functional regions of the 25S, the peptidyl-transferase A-site and the sarcin/ricin loop. Therefore, L3 may serve to help communicate the tRNA occupancy status of the peptidyl-transferase center to the sarcin/ricin loop (Meskauskas et al. 2005). Our finding that puromycin binding to the peptidyl-transferase A-site inhibits PAP depurination of the sarcin/ricin loop supports the hypothesis that occupancy of the A-site can be structurally signaled to the sarcin/ricin loop.
The requirement for a free peptidyl-transferase A-site is also supported by data showing inhibition of depurination in mak8-1, a yeast strain bearing a chromosomal mutation in the ribosomal protein L3 (Wickner et al. 1982). We have demonstrated previously that mak8-1 ribosomes are resistant to depurination by PAP when the protein is expressed in this strain (Hudak et al. 1999). However, the current study shows that the level of depurination increases when ribosomes of mak8-1 are incubated with PAP in vitro or when PAP-expressing cells are lysed and incubated at room temperature, conditions that would cause the release of aminoacyl-tRNA from the ribosome. In addition, the lack of PAP binding to ribosomes of mak8-1 cells in vivo (Hudak et al. 1999) was initially puzzling, as the two point mutations of L3 that characterize the mak8-1 strain (W255C, P257T) are embedded in the ribosome and are not solvent accessible. Therefore, the two amino acids would not likely be involved in direct binding to PAP. Rather, we hypothesize that the in vitro PAP binding and the level of depurination of mak8-1 ribosomes increases when the process of translation is interrupted and that resistance to binding and depurination is due to an inherent difference in the affinity of the A-site in this strain for aminoacyl-tRNAs. Support for this hypothesis comes from observations of the growth of mak8-1 cells in the presence of translation inhibitors (Petrov et al. 2004). Specifically, the drug anisomycin is known to decrease the affinity of ribosomes for aminoacyl-tRNAs and is used as an indicator of changes specific to the ribosomal A-site (Pestka 1977). The observation that mak8-1 cells were resistant to anisomycin supports the view that the higher affinity for aminoacyl-tRNA of mak8-1 ribosomes antagonizes the effect of anisomycin. Indeed, measurements of the association constants of ribosomes for aminoacyl-tRNA indicate a significantly greater binding affinity of mak8-1 ribosomes compared with those of wild-type cells (Petrov et al. 2004). The increased affinity for aminoacyl-tRNAs would also result in a decreased level of in vivo PAP binding and depurination because the period during which the A-site remains empty would likely be reduced in this strain.
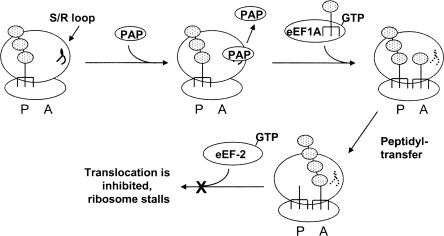
We propose the following model to explain when PAP depurinates the ribosome and its effect on the steps of the elongation cycle (Fig. 6). Prior to the arrival of eEF1A bearing an aminoacyl-tRNA, when only the P-site is occupied, PAP binds to the ribosome and depurinates the sarcin/ricin loop. PAP then dissociates from the ribosome. The newly empty A-site allows binding of an aminoacyl-tRNA, and both sites of the ribosome are now occupied. Peptidyl transfer and peptide bond formation occur, which are known not to be inhibited by PAP (Gessner and Irvin 1980). Depurination by PAP only occurs at the sarcin/ricin loop and does not affect the RNA of the peptidyl-transferase center. Following peptide bond formation, eEF-2 binds to the sarcin/ricin loop and mediates the translocation of the peptidyl-tRNA from the A-site to the P-site. However, depurination of the sarcin/ricin loop inhibits binding of eEF-2, effectively stalling the ribosome with the peptidyl-tRNA at the A-site.
FIGURE 6.
Model to illustrate the step of the elongation cycle at which PAP depurinates the ribosome. Following the translocation step of the peptidyl-tRNA from the A-site to the P-site and prior to the arrival of a new aminoacyl-tRNA to the A-site, PAP binds to the ribosome and depurinates the sarcin/ricin loop. PAP then dissociates from the ribosome. The dashed line represents the depurinated sarcin/ricin loop. The eEF1A ternary complex binds to the ribosome and delivers the aminoacyl-tRNA to the A-site. Peptidyl transfer occurs, mediated by the rRNA. Binding of eEF-2 GTP binary complex required for translocation of the peptidyl-tRNA from the A-site to the P-site is inhibited by depurination (indicated by the X), and the ribosome stalls with the peptidyl-tRNA at the A-site.
The inhibition of translocation due to depurination by PAP is supported by previous studies on the effect of PAP on ribosomal frameshifting. Expression of PAP in yeast has been shown to inhibit Ty1-directed +1 ribosomal frameshifting but not to influence −1 ribosomal frameshifting (Tumer et al. 1998). Frameshifting in the +1 direction, but not in the −1, occurs while only the P-site is occupied by peptidyl-tRNA, which is also the conformation we propose is required for PAP activity.
This model for the effect of PAP on translation elongation is also consistent with other published data. Given the well-documented evidence to support inhibition of eEF-2 binding upon depurination, it logically follows that depurination of the sarcin/ricin loop must occur prior to eEF-2 binding. The possibility that PAP can bind simultaneously with either eEF1A or eEF-2 may be addressed by knowledge of the binding sites of PAP and the elongation factors. Chemical probing of rRNA structure showed that both EF-Tu and EF-G (prokaryotic equivalents of eEF1A and eEF-2, respectively) protected bases in the sarcin/ricin stem–loop from modification, indicating that the loop is involved in factor binding (Moazed et al. 1988). Subsequent in vitro interaction studies showed that EF-G bound to an oligoribonucleotide mimic of the sarcin/ricin stem–loop (Munishkin and Wool 1997). Point mutations of the nucleotides that base pair to form the GAGA tetraloop of the sarcin/ricin loop resulted in a lethal phenotype and prevented binding to ribosomes of an EF-G/GTP binary complex (Chan et al. 2000). Since at least portions of the binding sites of the elongation factors include the sarcin/ricin loop, it is unlikely that the factors and PAP could simultaneously bind to the ribosome. Initially, conflicting reports arose regarding the possibility that PAP inhibits the binding of eEF1A in addition to eEF-2 (Fernandez-Puentes and Vazquez 1977; Grasmuk et al. 1977). These results were due to the effect of elongation factor concentration, as it was subsequently shown that inhibition of translation by PAP could be overcome with increasing eEF1A concentration (Irvin et al. 1980). This observation is relevant to the in vivo effects of PAP, given that eukaryotic cells contain excess amounts of eEF1A relative to eEF-2 and that eEF1A is not a limiting factor for protein synthesis (Slobin 1980). Recent structural analysis of the ribosome also showed that change in position of the GTPase-associated center relative to the sarcin/ricin loop altered the function of EF-G but not that of EF-Tu, suggesting that the structural requirements for activity of each elongation factor also differ (Sergiev et al. 2005). Therefore, even though both factors bind the sarcin/ricin loop, the conformation of the loop and its depurination status may have differing effects on each factor.
Our results with puromycin and the mak8-1 strain support the view that the step of protein synthesis at which ribosomes are depurinated is different from the step at which the symptoms of depurination are manifest; namely, that depurination occurs prior to the arrival of the aminoacyl-tRNA to the A-site, whereas the depurination effect, inhibition of translocation, results in ribosome stalling with the A-site occupied by the peptidyl-tRNA.
MATERIALS AND METHODS
Plasmids and yeast strains
The cDNAs encoding wtPAP and PAPx were cloned into Yep351-based high-copy plasmids with an inducible GAL1 promoter as described previously (Hudak et al. 1999). PAPx is a mutant of wtPAP, bearing a point mutation (E176V), without enzymatic activity and was used as a negative control (Hur et al. 1995). The empty vector, without gene insert following the GAL1 promoter, was also used as a vector control. The three vectors were transformed as described (Ito et al. 1983) into Saccharomyces cerevisiae strains W303 (MATα, ade2-1 trp1-1 ura3-1 leu2-3, 112 his3-11, 15 can1-100) and 1906 (MATα, leu2 mak8-1). The 1906 strain contains two point mutations in the ribosomal protein L3 (W255C and P257T) (Peltz et al. 1999).
Media and growth conditions
Transformed yeast cells were grown in SD-Leu, 2% raffinose, at 30°C and 200 r.p.m. to A 600 = 0.6. To induce transcription of wtPAP and PAPx, the cells were pelleted by centrifugation at 2000g for 5 min, followed by a wash in SD-Leu medium and were resuspended in SD-Leu medium containing 2% galactose. The cells were then incubated at 30°C for the indicated number of hours. Following induction, yeast cells were pelleted at 2000g for 5 min and stored at −20°C for further analysis.
Protein extraction and immunoblot analysis
Cell pellets were resuspended in an equal volume of cold Buffer A (25 mM Tris-HCl, 5% glycerol, 1 mM DTT, 1 mM EGTA, 1 mM PMSF) and lysed by vortexing with cold glass beads. The lysed cells were centrifuged at 6000g for 5 min at 4°C and the supernatants were quantified using the Bradford Assay. An equal amount of protein from each sample was separated by 12% SDS-PAGE and transferred to nitrocellulose. The membrane was blocked in 5% nonfat milk in 1× PBS and probed with PAP-specific polyclonal antibody (1:5000). PAP expression was visualized by chemiluminescence (Western Lightening Chemiluminescence Kit, Perkin Elmer).
Isolation of total yeast RNA and ribosomes
The pellets of induced yeast cells (10 mL cultures, 4 h induction) were resuspended in 250 μL of Complete Buffer A (50 mM NaOAc at pH 5.2, 10 mM EDTA, 1% SDS), and total yeast RNA was isolated as described previously (Parikh et al. 2002). Total RNA was resuspended in 50 μL of DEPC-treated water and quantified by spectrophotometry. Ribosomes were isolated from induced or uninduced yeast cells (100 mL cultures, 4 h induction). Briefly, cell pellets were ground to a fine powder in liquid N2 with a mortar and pestle. Cold (4°C) Buffer A (200 mM Tris-HCl at pH 9.0, 200 mM KCl, 200 mM sucrose, 25 mM MgCl2, 25 mM EGTA, 25 mM 2-mercaptoethanol) was added to the yeast powder and centrifuged at 16,000g for 20 min. The KCl content of the supernatant was increased to 0.5 M and the supernatant was layered over a 3 mL cushion of 1 M sucrose, 25 mM Tris-HCl (pH 7.6), 25 mM KCl, and 5 mM MgCl2. Ribosomes were pelleted by centrifugation at 311,000g for 3.5 h at 4°C. The pellets were resuspended in Buffer B (0.5 M KCl, 25 mM Tris-HCl at pH 7.6, 5 mM MgCl2, 10% glycerol) and treated with puromycin to strip endogenous peptidyl-tRNA from the ribosomes (Triana et al. 1994; Meskauskas et al. 2003). Briefly, ribosomes were resuspended in 1 mL of Buffer B containing 1 mM PMSF and 1 mM DTE. Puromycin and GTP, both at 1 mM final concentrations, were added and the mix was incubated for 30 min at 30°C. Ribosomes were washed by pelleting twice through a 25% glycerol cushion in the same buffer. The washed ribosomes were resuspended in Buffer C (50 mM Tris-HCl at pH 7.5, 50 mM NH4Cl, 5 mM MgCl2, 0.1 mM PMSF, 0.1 mM DTE, 10% glycerol), aliquoted, and stored at −80°C.
Incubation of PAP with yeast ribosomes and isolation of rRNA
Ribosomes (30 μg, 73 nM) were incubated with purified PAP (30 ng, 11 nM) in 1× RIP Buffer (60 mM KCl, 10 mM Tris-HCl at pH 7.6, 10 mM MgCl2) for 30 min at 30°C. The amount of PAP incubated with ribosomes was determined according to previous measurements of the amount of PAP expressed in yeast cells relative to total cellular protein (data not shown). Following incubation, an equal volume of 2× Extraction Buffer (240 mM NaCl, 50 mM Tris-HCl at pH 8.8, 20 mM EDTA, 2% SDS) was added to each sample and vortexed. The samples were extracted with phenol and phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated in 0.3 M NaOAc and 2× volumes 100% ethanol. The samples were pelleted at 14,100g for 30 min at 4°C, washed in 70% ethanol, and resuspended in 30 μL of DEPC-treated water.
rRNA depurination assay
Depurination of total yeast RNA or rRNA was assayed as described previously (Parikh et al. 2002). Briefly, 1 μg RNA was annealed to two [γ-32P]-ATP end-labeled negative strand primers. The depurination primer (5′- AGCGGATGGTGCTTCGCGGCAATG-3′) annealed 73 nt downstream of the depurination site and the 25S rRNA control primer (5′-TTCACTCGCCGTTACTAAGG-3′) annealed ∼100 nt downstream of the 5′ end of the 25S rRNA. Superscript II reverse transcriptase (Invitrogen) was used to extend these primers, and resulting cDNA products for depurination and control fragments were separated on a 7 M urea/6% acrylamide gel. The gel was dried against filter paper under vacuum, and bands were visualized by exposure to X-ray film (Kodak).
Effect of puromycin on depurination by PAP
Ribosomes (50 μg, 120 nM) isolated from PSY and mak8-1 cells were incubated with 0, 0.5, 1.0, 2.5, 5, 7.5, and 10 mM puromycin at 4°C for 30 min in Resuspension Buffer (25 mM Tris-HCl at pH 7.6, 25 mM KCl, 5 mM MgCl2). Following incubation with puromycin, PAP (50 ng, 17 nM) was added to each sample and incubated at 30°C for 30 min. rRNA was extracted for primer extension analysis.
Effect of puromycin on PAP activity in an in vitro translation system
An in vitro translation reaction using rabbit reticulocyte lysate (50 μL, Reticulocyte Lysate IVT Kit, Ambion) was permitted to translate an RNA template supplied with the kit, for 30 min. Puromycin (10 mM) was then added to half the reaction and the two halves continued incubation for another 30 min. PAP (5 ng, 6 nM) was then added to each reaction and incubated for 1 h. Following incubation, total RNA was extracted for primer extension analysis.
Far Western assay of PAP binding to ribosomes
Isolated ribosomes (50 μg, 12 pmol) were diluted in Resuspension Buffer (25 mM Tris-HCl at pH 7.6, 25 mM KCl, 5 mM MgCl2) to a total volume of 50 μL and applied to a nitrocellulose membrane using a dot-blot apparatus (BioRad). PAP (50 ng, 1.7 pmol) was applied as a positive control for antibody specificity and BSA (10 μg) was applied as a negative control. The membrane was blocked with 5% nonfat milk in Blotting Buffer (20 mM HEPES-KOH at pH 7.6, 1 mM DTT, 0.1 mM EDTA, 0.1 mM MgAc, 10% glycerol) for 2 h at room temperature, before incubation with PAP (500 ng) in Blotting Buffer for an additional 2 h. The membrane was washed three times for 5 min in Blotting Buffer at room temperature, followed by air drying at 30°C for 30 min. The membrane was then blocked with 5% nonfat milk in 1× PBS-T for 30 min at room temperature and probed with PAP-specific polyclonal antibody (1:5000). PAP was visualized by chemiluminescence (Western Lightening Chemiluminescence Kit, Perkin Elmer).
To determine the effect of puromycin on PAP binding, ribosomes (50 μg, 12 pmol) of PSY and mak8-1 cells were incubated with 0, 0.5, 1.0, 2.5, 5.0, 7.5, and 10 mM puromycin in Resuspension Buffer for 30 min at 4°C. The samples were then applied to nitrocellulose membrane and incubated with PAP followed by PAP-specific polyclonal antibody (1:5000) as described above.
DMS footprinting of puromycin-treated ribosomes
Ribosomes (50 μg) isolated from PSY and mak8-1 cells were incubated with 2.5 or 5 mM puromycin or in Resuspension Buffer alone for 30 min at 4°C. The modification of rRNA with dimethylsulphate (DMS) and subsequent footprinting analysis were performed essentially as described by Merryman and Noller (1998). Briefly, ribosomes were placed in Reaction Buffer (100 mM NH4Cl, 80 mM HEPES-KOH at pH 7.8, 10 mM MgCl2) and treated with 35 mM DMS for 10 min at 37°C. Reactions were terminated by the addition of 0.5 M beta-mercaptoethanol and 50 mM EDTA, and rRNA was precipitated in 2.5 volumes 95% ethanol. Following resuspension and extraction, the rRNA was used as template for primer extension. This analysis was performed with the depurination primer as described above for rRNA depurination, except that AMV reverse transcriptase (Invitrogen) was used instead of the SuperScript II enzyme.
ACKNOWLEDGMENTS
The authors acknowledge the use of a portion of the secondary structure of yeast 25S rRNA for Figure 5 from the Comparative RNA Web site (http://www.rna.icmb.utexas.edu/). The authors also thank Dr. K.A. White, York University, for critical reading of the manuscript. We thank Dr. Nilgun Tumer, Rutgers University, for a polyclonal antibody against PAP, and Dr. Arturas Meskauskas, University of Maryland, for yeast strains. The authors also gratefully acknowledge infrastructural support provided by the Canadian Foundation of Innovation (CFI) and the Ontario Innovation Trust (OIT). This work is funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada and a Premier's Research Excellence Award to K.A.H. and an Ontario Graduate Scholarship to E.N.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.70306.
REFERENCES
- Andersen G.R., Nissen P., Nyborg J. Elongation factors in protein biosynthesis. Trends Biochem. Sci. 2003;28:434–441. doi: 10.1016/S0968-0004(03)00162-2. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bolognesi A., Barbieri L., Abbondanza A., Falasca A.I., Carnicelli D., Battelli M.G., Stirpe F. Purification and properties of new ribosome-inactivating proteins with RNA N-glycosidase activity. Biochim. Biophys. Acta. 1990;1087:293–302. doi: 10.1016/0167-4781(90)90002-j. [DOI] [PubMed] [Google Scholar]
- Brigotti M., Rambelli F., Zamboni M., Montanaro L., Sperti S. Effect of α-sarcin and ribosome-inactivating proteins on the interaction of elongation factors with ribosomes. Biochem. J. 1989;257:723–727. doi: 10.1042/bj2570723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.L., Sitikov A.S., Wool I.G. The phenotype of mutations of the base-pair C2658.G2663 that closes the tetraloop in the sarcin/ricin domain of Escherichia coli 23 S ribosomal RNA. J. Mol. Biol. 2000;298:795–805. doi: 10.1006/jmbi.2000.3720. [DOI] [PubMed] [Google Scholar]
- Endo Y., Tsurugi K., Lambert J.M. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: The RNA N-glycosidase activity of the proteins. Biochem. Biophys. Res. Commun. 1988;150:1032–1036. doi: 10.1016/0006-291x(88)90733-4. [DOI] [PubMed] [Google Scholar]
- Fernandez-Puentes C., Vazquez D. Effects of some proteins that inactivate the eukaryotic ribosome. FEBS Lett. 1977;78:143–146. doi: 10.1016/0014-5793(77)80292-5. [DOI] [PubMed] [Google Scholar]
- Gessner S.L., Irvin J.D. Inhibition of elongation factor 2-dependent translocation by the pokeweed antiviral protein and ricin. J. Biol. Chem. 1980;255:3251–3253. [PubMed] [Google Scholar]
- Grasmuk H., Nolan R.D., Drews J. Further evidence that elongation factor 1 remains bound to ribosomes during peptide chain elongation. Eur. J. Biochem. 1977;79:93–102. doi: 10.1111/j.1432-1033.1977.tb11787.x. [DOI] [PubMed] [Google Scholar]
- Hansen J.L., Moore P.B., Steitz T.A. Structures of five antibiotics bound at the peptidyl-transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Hudak K.A., Dinman J.D., Tumer N.E. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- Hudak K.A., Wang P., Tumer N.E. A novel mechanism for inhibition of translation by pokeweed antiviral protein: Depurination of the capped RNA template. RNA. 2000;6:369–380. doi: 10.1017/s1355838200991337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y., Hwang D.J., Zoubenko O., Coetzer C., Uckun F.M., Tumer N.E. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: Identification of residues important for toxicity. Proc. Natl. Acad. Sci. 1995;92:8448–8452. doi: 10.1073/pnas.92.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin J.D. Antiviral proteins from Phytolacca. In: Chessin M., et al., editors. Antiviral proteins in higher plants. CRC Press; Boca Raton, FL: 1995. pp. 65–94. [Google Scholar]
- Irvin J.D., Uckun F.M. Pokeweed antiviral protein: Ribosome inactivation and therapeutic applications. Pharmacol. Ther. 1992;55:279–302. doi: 10.1016/0163-7258(92)90053-3. [DOI] [PubMed] [Google Scholar]
- Irvin J.D., Kelly T., Robertus J.D. Purification and properties of a second antiviral protein from Phytolacca Americana which inactivates eukaryotic ribosomes. Arch. Biochem. Biophys. 1980;200:418–425. doi: 10.1016/0003-9861(80)90372-0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukada Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. After the ribosome structure: How does translocation work? RNA. 2003;9:160–164. doi: 10.1261/rna.2163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W.C., Nyborg J. The protein biosynthesis elongation cycle. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 89–125. [Google Scholar]
- Merryman C., Noller H.F. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith C.W.J., editor. RNA: Protein interactions, a practical approach. Oxford University Press; Oxford, UK: 1998. pp. 237–253. [Google Scholar]
- Meskauskas A., Harger J.W., Jacobs K.L., Dinman J.D. Decreased peptidyltransferase activity correlates with increased programmed −1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae . RNA. 2003;9:982–992. doi: 10.1261/rna.2165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A., Pertov A.N., Dinman J.D. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 2005;25:10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Robertson J.M., Noller H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Montanaro L., Sperti S., Mattioli A., Testoni G., Stirpe F. Inhibition by ricin of protein synthesisin vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem. J. 1975;146:127–131. doi: 10.1042/bj1460127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munishkin A., Wool I.G. The ribosome-in-pieces: Binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc. Natl. Acad. Sci. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., Boston R.S. Ribosome-inactivating proteins: A plant perspective. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:785–816. doi: 10.1146/annurev.arplant.52.1.785. [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Osborn R.W., Hartley M.R. Dual effects of the ricin A chain on protein synthesis in rabbit reticulocyte lysate. Inhibition of initiation and translocation. Eur. J. Biochem. 1990;193:401–417. doi: 10.1111/j.1432-1033.1990.tb19353.x. [DOI] [PubMed] [Google Scholar]
- Pape T., Wintermeyer W., Rodnina M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh B.A., Coetzer C., Tumer N.E. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the α-sarcin/ricin loop of rRNA. J. Biol. Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
- Park S.W., Vepachedu R., Sharma N., Vivanco J.M. Ribosome-inactivating proteins in plant biology. Planta. 2004;219:1093–1096. doi: 10.1007/s00425-004-1357-8. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., Hammell A.B., Cui Y., Yasenchak J., Puljanowski L., Dinman J.D. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol. Cell. Biol. 1999;19:384–391. doi: 10.1128/mcb.19.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Inhibitors of protein synthesis. In: Weissbach H., Pestka S., editors. Molecular mechanisms of protein biosynthesis. Academic Press; New York: 1977. pp. 487–562. [Google Scholar]
- Petrov A., Meskauskas A., Dinman J.D. Ribosomal protein L3. Influence on ribosome structure and function. RNA Biol. 2004;1:56–62. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fonseca C., Phan H., Long K.S., Porse B.T., Kirillov S.V., Amils R., Garrett R.A. Puromycin rRNA interaction sites at the peptidyl-transferase center. RNA. 2000;6:744–754. doi: 10.1017/s1355838200000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu K.Y., Joseph S., Tung C.S. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing T.M., Seila A.C., Hansen J.L., Freeborn B., Soukup J.K., Scaringe S.A., Strobel S.A., Moore P.B., Steitz T.A. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Sergiev P.V., Lesnyak D.V., Burakovsky D., Kiparisov S.V., Leonov A.A., Bogdanov A.A., Brimacombe R., Dontsova O.A. Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyl-transferase center and activity of elongation factor G. J. Biol. Chem. 2005;280:31882–31889. doi: 10.1074/jbc.M505670200. [DOI] [PubMed] [Google Scholar]
- Slobin L.I. The role of eukaryotic elongation factor Tu in protein synthesis. The measurement of the elongation factor Tu content of rabbit reticulocytes and other mammalian cells by a sensitive radioimmunoassay. Eur. J. Biochem. 1980;110:555–563. doi: 10.1111/j.1432-1033.1980.tb04898.x. [DOI] [PubMed] [Google Scholar]
- Sperti S., Montanaro L., Mattioli A., Testoni G., Stirpe F. Inhibition of protein synthesis in vitro by crotins and ricin. Effect on the steps of peptide chain elongation. Biochem. J. 1976;156:7–13. doi: 10.1042/bj1560007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana F., Nierhaus K.H., Chakraburtty K. Transfer RNA binding to 80S ribosomes from yeast: Evidence for three sites. Biochem. Mol. Biol. Int. 1994;33:909–915. [PubMed] [Google Scholar]
- Tumer N.E., Parikh B.A., Li P., Dinman J.D. The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae . J. Virol. 1998;72:1036–1042. doi: 10.1128/jvi.72.2.1036-1042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Hudak K.A. Applications of plant antiviral proteins. Genet. Eng. (N.Y.) 2003;25:143–161. doi: 10.1007/978-1-4615-0073-5_7. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Ridley S.P., Fried H.M., Ball S.G. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]