Abstract
Two different models currently exist for the assembly pathway of the spliceosome, namely, the traditional model, in which spliceosomal snRNPs associate in a stepwise, ordered manner with the pre-mRNA, and the holospliceosome model, in which all spliceosomal snRNPs preassemble into a penta-snRNP complex. Here we have tested whether the spliceosomal A complex, which contains solely U1 and U2 snRNPs bound to pre-mRNA, is a functional, bona fide assembly intermediate. Significantly, A complexes affinity-purified from nuclear extract depleted of U4/U6 snRNPs (and thus unable to form a penta-snRNP) supported pre-mRNA splicing in nuclear extract depleted of U2 snRNPs, whereas naked pre-mRNA did not. Mixing experiments with purified A complexes and naked pre-mRNA additionally confirmed that under these conditions, A complexes do not form de novo. Thus, our studies demonstrate that holospliceosome formation is not a prerequisite for generating catalytically active spliceosomes and that, at least in vitro, the U1 and U2 snRNPs can functionally associate with the pre-mRNA, prior to and independent of the tri-snRNP. The ability to isolate functional spliceosomal A complexes paves the way to study in detail subsequent spliceosome assembly steps using purified components.
Keywords: spliceosome, pre-mRNA splicing, A complexes
INTRODUCTION
Many nuclear processes such as transcription initiation by RNA polymerases I and II, DNA repair by nuclear excision, and pre-mRNA splicing require the combined action of numerous factors that are components of complex molecular machines. In many cases, it is not entirely clear how these “machines” assemble onto their substrates. Different models ranging from (1) the stepwise assembly of individual components or subcomplexes of these machines to (2) the interaction of preassembled holocomplexes have been discussed (for review, see Lemon and Tjian 2000; Brow 2002; Nilsen 2002). Experimentally, it can often prove difficult to distinguish between these two possibilities, perhaps as best exemplified by the numerous conflicting reports regarding the assembly pathway of those molecular machines responsible for transcription initiation and DNA repair. In vivo kinetic experiments have only recently provided solid evidence that transcription and DNA repair proteins diffuse in relatively small complexes and assemble in a sequential order at their sites of action (Dundr et al. 2002; Hoogstraten et al. 2002), rather than existing in preassembled holocomplexes as previously proposed (Svejstrup et al. 1995; Maldonado et al. 1996; Hannan et al. 1999).
Different models also currently exist for the assembly pathway of the spliceosome, the large ribonucleoprotein complex that catalyzes intron removal from pre-mRNAs. The spliceosome is composed of the U1, U2, U4/U6, and U5 snRNPs, and a large number of splicing factors (Burge et al. 1999; Will and Lührmann 2006). Spliceosome assembly was initially characterized by an extensive array of in vitro studies using human and yeast cell extracts. Several biochemical approaches, including native gel electrophoresis (Konarska and Sharp 1986; Cheng and Abelson 1987), gradient centrifugation (Brody and Abelson 1985; Frendewey and Keller 1985; Grabowski et al. 1985), and affinity chromatography (Bindereif and Green 1987), revealed a sequential appearance of spliceosomal complexes that differ in their snRNP composition. These findings resulted in the development of a model in which spliceosome assembly occurs by the stepwise interaction of spliceosomal components with the pre-mRNA (for review, see Will and Lührmann 2006). According to this model, spliceosome assembly in mammals is initiated by the ATP-independent interaction of the U1 snRNP with the 5′ splice site, forming the so-called E complex. The U2 snRNP is loosely associated at this stage (Das et al. 2000), and subsequently, in an ATP-dependent manner, it stably interacts with the branch point sequence (BPS) of the pre-mRNA, forming the A complex. At this stage the 5′ and 3′ splice sites are functionally paired (Lim and Hertel 2004). In the next step, the pre-formed 25S U4/U6•U5 tri-snRNP is recruited to the A complex to form the precatalytic B complex. Dramatic structural rearrangements resulting in the destabilization of the U1 and U4 snRNPs generate the catalytically activated spliceosome (B* complex). Subsequently, the first catalytic step of splicing occurs, and the C complex is generated. After the second step of splicing, the mRNA is released and the post-spliceosomal complex—containing the excised intron and the U2, U5, and U6 snRNPs—disassembles, and the snRNPs are recycled to take part in new rounds of splicing.
A similar stepwise assembly pathway was also proposed for the Saccharomyces cerevisiae spliceosome, as equivalent intermediates are detected in vitro (for review, see Burge et al. 1999). Two reports using chromatin immunoprecipitation (ChIP) to investigate spliceosome assembly in yeast in vivo also support the stepwise assembly model. That is, data from these assays indicate that the U1 and U2 snRNPs, the key components of the A complex, bind cotranscriptionally to the nascent pre-mRNA but prior to the U4/U6•U5 tri-snRNP (Görnemann et al. 2005; Lacadie and Rosbash 2005). By combining ChIP assays with in vivo depletion of different spliceosomal snRNPs, additional evidence has recently been provided that the yeast snRNPs are recruited stepwise to the pre-mRNA in vivo (Tardiff and Rosbash 2006). However, in higher eukaryotes, the mechanism of spliceosome assembly in vivo is not known.
Several studies carried out in cell extracts identified interactions that are difficult to reconcile with the temporal order of events depicted in the stepwise assembly model. For example, cross-linking studies in HeLa cell and yeast cell extract demonstrated an early interaction between the U4/U6•U5 tri-snRNP and the 5′ splice site that occurs prior to stable binding of the U2 snRNP to the pre-mRNA branch point (Maroney et al. 2000; Johnson and Abelson 2001). Similarly, it was shown that when the 5′-end of U1 is sequestered, the U4/U6•U5 tri-snRNP can bind an oligoribonucleotide, mimicking the 5′ splice site; as the BPS is lacking, this interaction can occur in the absence of a U2–BPS interaction (Konforti et al. 1993; Konforti and Konarska 1994, 1995).
Furthermore, several observations suggest that the U1 and/or U2 snRNPs interact with the U4/U6•U5 tri-snRNP prior to engaging the pre-mRNA. For example, psoralen cross-linking of RNAs in nuclear extract in the absence of pre-mRNA revealed an endogenous U2–U4–U6 cross-linkable complex (Hausner et al. 1990; Wassarman and Steitz 1992). Furthermore, U2, U4, U5, and U6 snRNPs associate efficiently in the absence of pre-mRNA, either when HeLa cell nuclear extract is incubated at 200–300 mM NH4Cl (Konarska and Sharp 1988) or under splicing conditions when a short oligonucleotide mimicking the 5′ splice site is added to the extract (Hall and Konarska 1992). More recent studies revealed that a large 200S complex containing the U1, U2, U4, U5, and U6 snRNPs can assemble onto a short RNA containing a 5′ splice site in HeLa cell nuclear extract, consistent with the existence of a human U1/U2/U4/U5/U6 penta-snRNP (Malca et al. 2003). In S. cerevisiae, a pre-formed U1/U2/U4/U5/U6 penta-snRNP could be isolated under low salt conditions from yeast cell extract. Significantly, this complex was shown to function as an intact entity in in vitro splicing assays when supplemented with micrococcal nuclease-treated extract (Stevens et al. 2002).
Based on the latter studies, an alternative model for spliceosome assembly was proposed. In this model, the spliceosome preassembles in the absence of the pre-mRNA, and all of the snRNPs (as part of this “holospliceosome”) engage the pre-mRNA concomitantly. In subsequent remodeling steps, initially weak contacts between the spliceosome and the pre-mRNA are progressively stabilized through dynamic RNA/RNA rearrangements, and a catalytically active spliceosome is ultimately formed. Proponents of this model suggest that due to the relatively harsh conditions typically used to investigate spliceosome assembly in vitro, the various assembly intermediates that are observed (i.e., E, A, and B) simply reflect the stepwise stabilization of snRNP–pre-mRNA interactions, rather than a stepwise recruitment of snRNPs. Thus, according to the holospliceosome model, spliceosomal complexes such as the A complex do not represent bona fide functional assembly intermediates, but rather arise by dissociation of the holospliceosome.
Here, we have investigated whether the human A complex represents a genuine, spliceosome assembly intermediate. For this purpose, spliceosomal A complexes were isolated by a two-step purification method, involving tobramycin affinity selection followed by immunoaffinity purification, and subsequently tested for splicing activity in nuclear extract depleted of U2 snRNPs. Significantly, A complexes isolated from either intact HeLa cell nuclear extract or from extract that was depleted of U4/U6 snRNPs (and thus unable to form a penta-snRNP) supported splicing in U2-depleted extract, whereas naked tagged pre-mRNA did not. Furthermore, in the presence of an equimolar amount of purified A complexes, splicing of naked untagged pre-mRNA was not observed, demonstrating that A complexes do not dissociate and form de novo in the U2-depleted extract. Taken together, our data provide evidence that in the human system, at least in vitro, spliceosome assembly does not require the formation of a holospliceosome. Rather, functional spliceosomes can assemble on the pre-mRNA from the U1 and U2 snRNPs and the (U4/U6•U5) tri-snRNP in a stepwise and temporally ordered manner. A complexes purified in this way can potentially be used to reconstitute catalytically active spliceosomes from purified spliceosomal components, and thus enable a detailed dissection of the stepwise assembly pathway of the spliceosome.
RESULTS
Affinity purification of the human spliceosomal A complex
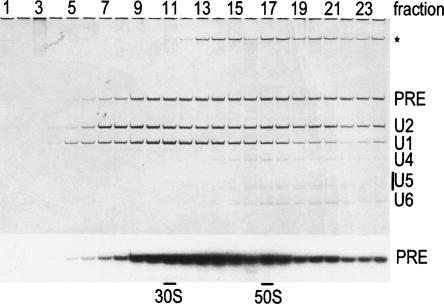
To investigate whether the spliceosomal A complex is a bona fide assembly intermediate, we first affinity-purified this complex from HeLa cell nuclear extract under native conditions and tested whether it is active in splicing. For this purpose, we performed tobramycin affinity selection, as previously described to isolate human A complexes (Hartmuth et al. 2002), but included an additional immunoaffinity selection step to ensure that highly pure complexes were obtained. Briefly, MINX pre-mRNA tagged with a tobramycin RNA aptamer was first immobilized on a tobramycin-derivatized Sepharose matrix and incubated for 45 min under splicing conditions with HeLa cell nuclear extract. Assembled spliceosomal complexes were eluted by competition with tobramycin, and the eluate was subjected to immunoaffinity selection using anti-peptide antibodies against the U2-associated SF3a66 protein. After eluting with antigenic peptide, spliceosomal complexes (i.e., a mixture of A and B) were further separated on a 10%–30% glycerol gradient, and the distribution of RNA across the gradient was analyzed by denaturing PAGE. As shown in Figure 1, complexes containing equimolar amounts of the U1 and U2 snRNAs and the pre-mRNA (i.e., A complexes) peaked in fractions 10 and 11 and were well separated from complexes containing all of the spliceosomal snRNAs (i.e., B complexes), which peaked in fraction 16 (Fig. 1). Mass spectrometry analyses of A complexes purified in this way revealed that their protein composition is very similar to that of the previously reported human A complex (Hartmuth et al. 2002; data not shown; N. Behzadnia, K. Hartmuth, H. Urlaub, and R. Lührmann, in prep.). In the following experiments, A complexes from gradient fraction 10 were used.
FIGURE 1.
Characterization of spliceosomal A complexes purified via tobramycin affinity selection followed by immunoaffinity selection and gradient centrifugation. Complexes were separated on a 10%–30% glycerol gradient. RNA was recovered from each gradient fraction (as indicated above), analyzed by denaturing PAGE, and visualized by silver staining (upper panel) or by autoradiography (lower panel). The identity of the various RNAs is indicated on the right. Contaminating high molecular weight RNAs are indicated by an asterisk. “PRE” indicates the pre-mRNA. 30S and 50S correspond to migration positions of E. coli ribosomal subunits in parallel gradients.
Affinity-purified A complexes are functional
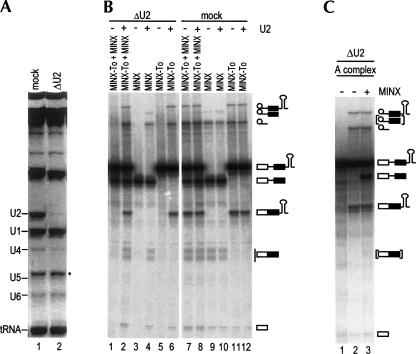
To test whether our purified A complexes are functional, as opposed to being “dead-end complexes,” we performed splicing assays in vitro with HeLa cell nuclear extract that was depleted of the 12S form of the U2 snRNP and thus cannot form a holospliceosome. U2 snRNPs were depleted by incubating the extract with a biotinylated 2′-OMe RNA oligonucleotide complementary to the 5′-end of the U2 snRNA, followed by streptavidin agarose affinity selection (Barabino et al. 1990; Ségault et al. 1995). Analysis of the snRNA composition of the mock-depleted versus U2-depleted extract confirmed that the U2 snRNP was quantitatively removed (Fig. 2A, cf. lanes 1 and 2). U2-depleted (ΔU2) extract did not support the splicing of tobramycin aptamer-tagged MINX or untagged MINX pre-mRNA, or an equimolar mixture of both substrates (Fig. 2B, lanes 1,3,5). In contrast, both pre-mRNAs were efficiently spliced in the mock-depleted extract (Fig. 2B, lanes 7,9,11). Furthermore, preincubation of the ΔU2 extract with purified 12S U2 snRNP restored splicing of both substrates to the level observed with the mock-depleted extract (Fig. 2B, cf. lanes 2,4,6 and 7,9,11, respectively), whereas splicing activity of the mock-depleted extract was unaffected by 12S U2 snRNP addition (Fig. 2B, lanes 8,10,12). These results confirm that the splicing defect is due specifically to the depletion of the 12S U2 snRNP. Thus, the ΔU2 extract should contain all of the factors necessary to chase a preassembled A complex into a catalytically active spliceosome.
FIGURE 2.
Double affinity-purified A complexes are functional. (A) RNA composition of U2 snRNP-depleted (lane 2) and mock-depleted (lane 1) extracts. RNA was separated by denaturing PAGE and detected by staining with ethidium bromide. The identity of the various RNAs is indicated on the left; 5.8S rRNA is indicated by an asterisk. (B) In vitro splicing of mock- vs. U2-depleted extract. An equimolar mixture of 32P-labeled tobramycin aptamer-tagged MINX pre-mRNA (MINX-To) and MINX pre-mRNA (lanes 1,2,7,8), MINX pre-mRNA alone (lanes 3,4,9,10), or MINX-To pre-mRNA alone (lanes 5,6,11,12) were incubated under splicing conditions for 3 h at 30°C in the presence of U2-depleted nuclear extract (lanes 1,3,5), U2-depleted extract complemented with purified 12S U2 snRNP (lanes 2,4,6), mock-depleted extract (lanes 7,9,11), or mock-depleted extract plus purified 12S U2 snRNP (lanes 8,10,12). RNA was recovered and analyzed by denaturing PAGE and visualized by autoradiography. The positions of the pre-mRNAs, the splicing intermediates, and the products are indicated on the right. (C) Splicing activity of purified A complexes. Purified spliceosomal A complexes from gradient fraction 10 (lanes 1,2) or an equimolar mixture of the purified A complex and naked, untagged MINX pre-mRNA (lane 3) were incubated in the presence of U2-depleted extract for 3 h at 0°C (lane 1) or at 30°C (lanes 2,3). RNA was analyzed as in B. Similar results were obtained with A complexes from gradient fractions 9, 11, and 12.
In contrast to naked, untagged MINX pre-mRNA, purified A complexes could be chased in ΔU2 extract into mature spliceosomes that catalyzed both steps of splicing (Fig. 2C, lane 2). To rule out the possibility that U2 snRNPs dissociate from the exogenously added, purified complexes and that A complexes subsequently assemble de novo from components of the ΔU2 extract, we performed a mixing experiment. Equimolar amounts of untagged MINX pre-mRNA and purified A complex (containing tagged MINX pre-mRNA) were incubated under splicing conditions in the ΔU2 extract. Significantly, only spliced mRNA and lariat intermediate from the tagged pre-mRNA, but not from the untagged pre-mRNA, were observed (Fig. 2C, lane 3), even after extended incubation times (i.e., 180 min). Note that the cleaved exon 1 and excised intron lariat generated from both substrates are identical. Therefore, we conclude that A complexes do not form de novo under these conditions and that the spliceosomal A complex purified from HeLa cell nuclear extract is functional.
Spliceosomal A complexes purified from U4/U6-depleted nuclear extract are functional
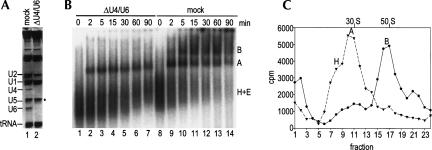
As A complexes were isolated from nuclear extract in which a penta-snRNP could potentially be formed, it is possible that the complexes that we isolated were generated via dissociation of holospliceosomes during the purification procedure. To exclude such a possibility, we tested whether functional spliceosomal A complexes are also formed in a nuclear extract devoid of a penta-snRNP. To this end we first codepleted U4 and U6 snRNPs from nuclear extract by affinity selection with a biotinylated 2′-OMe RNA oligonucleotide complementary to the 3′-end of the U6 snRNA (Blencowe et al. 1989). Analysis of the RNA composition of the depleted versus mock-depleted extract indicated that U4/U6 snRNPs were quantitatively removed (Fig. 3A, lane 2). Thus, the ΔU4/U6 extract cannot support the formation of the U4/U6•U5 tri-snRNP or penta-snRNP. Consistent with the observation that even in the absence of pre-mRNA the U2 snRNP associates with U4/U6 snRNPs in HeLa cell nuclear extract (Wassarman and Steitz 1992), a portion of the U2 snRNP was codepleted together with the U4/U6 snRNP. As expected, the ΔU4/U6 extract did not support splicing (data not shown). However, consistent with previous reports (Barabino et al. 1990; Wolff and Bindereif 1992), ΔU4/U6 extract supported the efficient assembly of the spliceosomal A complex, but not the B complex, as assayed by native gel electrophoresis (Fig. 3B) and glycerol gradient centrifugation (Fig. 3C).
FIGURE 3.
U4/U6-depleted nuclear extract supports the formation of only spliceosomal A complexes. (A) RNA composition of U4/U6-depleted (lane 2) and mock-depleted (lane 1) extracts. RNA was analyzed as in Figure 2A. (B) Spliceosome assembly in U4/U6-depleted extract (lanes 1–7) and mock-depleted extract (lanes 8–14). Spliceosomal complexes were analyzed on a native gel at the indicated times. The positions of the H, A, and B complexes are indicated on the right. (C) Spliceosome assembly in mock- vs. U4/U6-depleted extract. Spliceosomal complexes were allowed to form under splicing conditions at 30°C for 20 min in U4/U6-depleted nuclear extract (triangles) and mock-depleted extract (circles). Subsequently, the complexes were separated on a 10%–30% glycerol gradient. The distribution of the radioactivity across each gradient is shown. Peaks were assigned as A or B complex based on their migration behavior according to Figure 1; the position of H complexes was verified independently (data not shown).
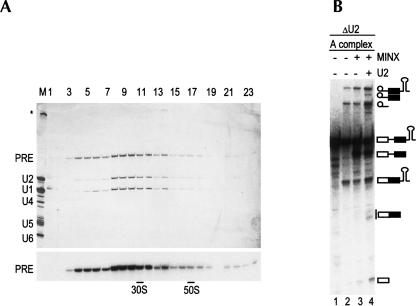
We next purified the A complex from ΔU4/U6 extract using the double affinity selection procedure described above. After eluting with antigenic peptide, spliceosomal complexes were further separated on a 10%–30% glycerol gradient, and the distribution of RNA across the gradient was analyzed by denaturing PAGE. As shown in Figure 4A, complexes containing equimolar amounts of the U1 and U2 snRNAs and the pre-mRNA peaked in fractions 9–11, similar to A complexes isolated from intact HeLa cell nuclear extract (Fig. 1).
FIGURE 4.
Spliceosomal A complexes purified from U4/U6-depleted nuclear extract are functional. (A) RNA composition of spliceosomal complexes purified from U4/U6-depleted nuclear extract. RNA was analyzed as in Figure 2A and visualized by silver staining (upper panel) or by autoradiography (lower panel). (B) Splicing activity of A complexes purified from U4/U6-depleted extract. Purified spliceosomal A complexes (lanes 1,2) or an equimolar mixture of the purified A complex and naked, untagged MINX pre-mRNA (lanes 3,4) were incubated in the presence of U2-depleted extract for 3 h at 0°C (lane 1) or at 30°C (lanes 2,3), or for 3 h at 30°C in U2-depleted extract complemented with 12S U2 snRNPs (lane 4). RNA was analyzed as in Figure 2B.
Significantly, A complexes purified from extract devoid of a holospliceosome could be chased in ΔU2 extract into mature spliceosomes that catalyzed both steps of splicing (Fig. 4A, lane 2). To again rule out de novo assembly of A complexes in the ΔU2 extract, splicing was performed with a mixture of the purified A complex and an equimolar amount of untagged MINX pre-mRNA. Significantly, only spliced mRNA and lariat intermediate from the tagged pre-mRNA present in the purified A complex, but not from the untagged pre-mRNA, were observed (Fig. 4B, lane 3), even after extended incubation times. We note that the amount of mRNA in lane 3 is less than that observed in lane 2 (quantification revealed a decrease of ∼37%). However, significant change in the level of spliced intron lariat was not observed upon addition of the untagged MINX pre-mRNA, indicating that the overall splicing efficiency of the tagged substrate did not decrease but, rather, that the tagged-mRNA was selectively degraded. To demonstrate that the untagged MINX pre-mRNA can be spliced under these conditions, we complemented the ΔU2 extract with purified 12S U2 snRNPs. Indeed, in the presence of exogenously added U2, both the tagged pre-mRNA (present in the purified A complexes), as well as the naked untagged MINX pre-mRNA, were spliced (Fig. 4B, lane 4). We therefore conclude that the A complex formed in the absence of an intact U4/U6•U5 tri-snRNP and penta-snRNP is functional and not a dead-end product. Taken together, these studies demonstrate that functional human A complexes can assemble in a stepwise manner.
DISCUSSION
Much discussion has recently been focused on the assembly pathway of the highly complex, multicomponent splicing machinery. The identification and characterization of a splicing-active penta-snRNP complex in budding yeast (Stevens et al. 2002) challenged the traditional model of spliceosome assembly, in which spliceosomal snRNPs associate in a stepwise, temporally ordered manner. Recent observations suggest that a penta-snRNP may also form in human cell extracts in the absence of the pre-mRNA (Malca et al. 2003). However, definitive proof that such a complex indeed exists and, additionally, functions in splicing is still lacking. To reconcile the apparent existence of a holospliceosome with the fact that numerous spliceosome assembly intermediates are detected using in vitro approaches, such as native gel electrophoresis or gradient centrifugation, it was proposed that the observed assembly intermediates could reflect different states of the spliceosome in which certain components are merely more stably associated with the pre-mRNA. Only the most stable interactions would withstand the stringent conditions used to study the spliceosome, giving rise to the appearance of the various spliceosomal complexes. Thus, according to this view, A complexes detected by various biochemical methods would represent a breakdown product of the holospliceosome and not a bona fide assembly intermediate.
Data presented here clearly demonstrate that this is not the case. That is, we provide evidence that human spliceosomal A complexes assembled in vitro are functional, even under conditions excluding the formation of a holospliceosome. Thus, our studies clearly demonstrate that the formation of a holospliceosome is not a prerequisite for generating catalytically active spliceosomes. Rather, our data provide evidence that, at least in vitro, the U1 and U2 snRNPs can associate with the pre-mRNA, prior to and independent of the tri-snRNP, and form functional prespliceosomal complexes. Previous studies with partially purified human E and A complexes also suggested that these early spliceosomal complexes are functional assembly intermediates (Michaud and Reed 1991). More recent studies performed with MS2-affinity-purified E and A complexes also indicated that both of these complexes are functional; that is, E and A complexes purified from intact HeLa nuclear extract could be chased into catalytically active spliceosomes in nuclear extract depleted of SF3a (Das et al. 2000). In vivo studies using chromatin immunoprecipitation (ChIP) or ChIP combined with in vivo snRNP depletions also support a stepwise assembly of the spliceosome in budding yeast (Görnemann et al. 2005; Lacadie and Rosbash 2005; Tardiff and Rosbash 2006). Data from our laboratory and that of others, as well as data supporting the holospliceosome model, do not rule out that both pathways can exist both in vivo and in vitro; i.e., under some conditions, active spliceosomes might be formed via the holospliceosome pathway, and under others, stepwise recruitment of snRNPs may occur.
HeLa cells are known to contain a several-fold excess of U1 and U2 snRNPs over components of the tri-snRNP. Thus, a large portion of these snRNPs will clearly not be present as part of a penta-snRNP and will be capable of interacting with the pre-mRNA as such. Likewise, U1 snRNPs are present in excess over the other spliceosomal snRNPs in yeast cells. Although our data demonstrate that U1 and U2 can functionally associate with the pre-mRNA prior to the tri-snRNP, they do not demonstrate that there is a fixed, temporal order of snRNP association. Indeed, several studies suggest that there is an early interaction between the U4/U6•U5 tri-snRNP and the 5′ splice site that does not require prior stable binding of the U2 snRNP to the BPS (Maroney et al. 2000; Johnson and Abelson 2001). Our data indicate, however, that an early tri-snRNP/5′-ss interaction is not required for the formation of an active spliceosome.
In both assembly pathway models, spliceosome formation involves a combination of pre-assembly and stepwise recruitment of spliceosomal complexes/factors. Purified yeast penta-snRNPs contained not only all five snRNPs but also numerous non-snRNP-associated splicing factors (Stevens et al. 2002). Nonetheless, they were not capable of forming functional spliceosomal complexes without the addition of soluble factors from yeast extract. Consistent with this observation, mass spectrometry analyses revealed that 15% of known splicing factors in yeast were not present in purified penta-snRNP complexes (Stevens et al. 2002). Although some of these factors may have been stripped off during purification, this observation indicates that several functionally important factors—foremost step II factors such as Slu7 and Prp18, and most DExD/H-box RNA-dependent ATPases—are recruited stepwise to the pre-mRNA, independent of the penta-snRNP. Indeed, previous studies indicated that there is a dynamic exchange of many spliceosomal proteins during the splicing cycle (for review, see Will and Lührmann 2006). In the stepwise assembly model, many spliceosomal proteins are preassembled in the form of the snRNPs, and the U4/U6 and U5 snRNPs also interact with the pre-mRNA as a pre-formed complex. Furthermore, some spliceosomal proteins also appear to preassemble into multisubunit complexes prior to engaging the pre-mRNA (e.g., proteins of the Prp19/CDC5 complex). Thus, the stepwise assembly model also entails the recruitment of preassembled spliceosomal subcomplexes. Given the highly complex nature of the spliceosome, it would be relatively inefficient to individually recruit all of the >100 proteins comprising the splicing machinery to each intron.
Preassembly of a penta-snRNP clearly would afford an initial kinetic benefit, in that there would be a concomitant recruitment of all snRNPs (plus many other factors) to the pre-mRNA. This would alleviate assembly problems that arise if the concentration of one or more spliceosomal proteins is very low, and allow for a more rapid response to regulatory signals. Nonetheless, a penta-snRNP complex must still undergo multiple rearrangements after engaging the pre-mRNA. That is, it is clear from numerous studies that there is an ordered remodeling of not only RNA–RNA interactions but also of those that are protein–protein or protein–RNA in nature, over the course of splicing (Nilsen 1998; Will and Lührmann 2006). Maturation of the spliceosome into a catalytically active entity thus requires numerous remodeling events regardless of whether the vast majority of spliceosomal components are recruited to the pre-mRNA as a pre-formed complex or stepwise in the form of multiple subcomplexes.
Stepwise recruitment of the U1 and U2 snRNPs followed by the tri-snRNP, on the other hand, likely provides more flexibility in terms of regulating the recognition and pairing of the 5′ and 3′ splice sites. This may be particularly advantageous for higher eukaryotes in which alternative splicing events—including those in which there is alternative pairing of 5′ and 3′ splice sites—occur at a very high frequency. That is, essentially no alternative splicing (with only minor exceptions) is observed in both Schizosaccharomyces pombe and S. cerevisiae, whereas 35%–65% of human genes are alternatively spliced (Modrek and Lee 2002; Ast 2004). Thus, it is conceivable that the holospliceosome pathway of spliceosome formation may be less prevalent (or even nonexistent) in higher eukaryotes.
The ability to purify functional spliceosome assembly intermediates represents an important step for analyzing both the structure and function of these complexes. For example, future studies of the higher-order structure of the A complex by electron microscopy should provide much needed information about the 3D organization of the spliceosome during its early stages of assembly. Furthermore, the complexes isolated here can be used to reconstitute in vitro, catalytically active spliceosomes from purified tri-snRNPs and other spliceosomal proteins. These studies may potentially lead to novel insights into the factor requirements for the transition from the spliceosomal A complex to the B complex.
MATERIALS AND METHODS
In vitro transcription and nuclear extract preparation
A transcription template for the MINX pre-mRNA was generated from pMINX plasmid (Zillmann et al. 1988) by PCR. MINX pre-mRNA tagged with the J6f1 tobramycin RNA aptamer was generated as described previously (Hartmuth et al. 2002). Uniformly [32P]-labeled, m7G(5′)ppp(5′)G-capped MINX and MINX-To pre-mRNA were synthesized in vitro by T7 runoff transcription as described previously (Hartmuth et al. 2002). HeLa cell nuclear extract was prepared essentially according to Dignam et al. (1983), except that initial cell lysis was performed according to Bergamini et al. (2000) in MC Buffer (10 mM HEPES-KOH at pH 7.6, 10 mM KOAc, 0.5 mM MgOAc, 5 mM DTT, 1× Complete Protease Inhibitor [Roche]). U2-depleted nuclear extract was prepared by affinity selection with a biotinylated 2′-OMe RNA oligonucleotide complementary to nucleotides 1–20 of U2 snRNA (RNA-Tec) and streptavidin-agarose beads essentially as described previously (Ségault et al. 1995), except that the oligonucleotide was used at a concentration of 14 μM and dialysis was against buffer D (Dignam et al. 1983) without glycerol (Dönmez et al. 2004). Similarly, an oligonucleotide complementary to nucleotides 82–101 of U6 snRNA (RNA-Tec) was used to deplete U4/U6 snRNP from nuclear extract as previously described (Blencowe et al. 1989), except that no ATP and creatine phosphate were added and that the oligonucleotide was used at a concentration of 1.4 μM. Mock-depleted extracts were prepared in an identical manner except that oligonucleotide was omitted. RNA was recovered and analyzed on a 10% polyacrylamide 8.3 M urea gel and visualized by staining with ethidium bromide.
Purification of human spliceosomal A complexes
Rabbit polyclonal antibodies raised against a peptide (amino acids 444–458) of SF3a66 were affinity-purified using a SulfoLink column (Pierce) containing the cognate peptide (Will et al. 2002). For immunoaffinity purification of spliceosomal complexes, 220 μg of affinity-purified antibodies were covalently coupled to 300 μL of protein A-Sepharose (PAS) beads with dimethylpimelimidate. Solid phase splicing reactions and tobramycin affinity selection of spliceosomal complexes were performed as described previously (Hartmuth et al. 2002), except that for a standard purification, 24 × 15-μL aliquots of matrix-bound pre-mRNA were prepared, and each aliquot was incubated for 45 min with 500 μL of a splicing reaction mixture. The latter contained 35% HeLa cell nuclear extract or 40% U4/U6-depleted nuclear extract. The tobramycin eluate (870 μL in total containing 280 nM pre-mRNA) was diluted with one volume of dilution buffer (20 mM Tris-HCl at pH 8.1, 1 mM CaCl2, 3 mM MgCl2, 5.5 mM DTT, 6% glycerol) and incubated for 1.5 h at 4°C with 300 μL of anti-SF3a66 charged PAS beads preblocked with 0.5 mg/mL BSA and 50 μg/mL yeast tRNA. Beads were washed three times with IP buffer (20 mM Tris-HCl at pH 8.1, 1 mM CaCl2, 3 mM MgCl2, 5.5 mM DTT, 75 mM KCl, 3% glycerol), and the bound material was specifically eluted by incubation for 2 h with 320 μL of IP buffer containing 0.6 mg/mL of the cognate peptide. The eluate was loaded onto a 4.1-mL, 10%–30% glycerol gradient containing G75 buffer (20 mM HEPES-KOH at pH 7.9, 75 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) and centrifuged in a Sorvall TH660 rotor at 60,000 rpm for 1 h 47 min at 4°C. Gradients were fractionated manually from top to bottom into 175-μL aliquots. RNA was recovered and analyzed on a 10% polyacrylamide 8.3 M urea gel and visualized by silver staining and autoradiography.
Isolation of 12S U2 snRNPs
A mixture of U1, U2, and U4/U6•U5 tri-snRNPs was isolated from HeLa cell nuclear extract by anti-m3G immunoaffinity chromatography (Bach et al. 1990). U2 snRNPs were subsequently isolated by glycerol gradient centrifugation followed by ion exchange chromatography (Bach et al. 1990) and dialyzed against 20 mM Tris-HCl (pH 7.9), 50 mM KCl, 1.5 mM MgCl2, and 2 mM DTT.
In vitro splicing assays
Standard splicing reactions contained 30% (v/v) U2 snRNP-depleted nuclear extract or mock-depleted extract, 60 mM KCl, 3 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate, and 1 nM naked pre-mRNA (MINX pre-mRNA and/or MINX-To pre-mRNA), purified A complex, or equimolar mixtures of naked MINX pre-mRNA and purified A complex. Reactions were incubated for 3 h at 30°C. For complementation of the U2-depleted extract, purified 12S U2 snRNP was added to the splicing reaction to a final concentration of 120 pmol/μL. The samples were preincubated for 30 min on ice before addition of the pre-mRNA, or an equimolar mixture of pre-mRNA and A complex. For splicing analysis, RNA was recovered and separated on a 10% polyacrylamide 8.3 M urea gel and detected by autoradiography. For analysis of spliceosome assembly, MINX-To pre-mRNA was incubated under splicing conditions as described above for 0–90 min, except that 40% extract was used. At defined time intervals, 0.5 μL of heparin (5 mg/mL) was added to 20-μl aliquots of the splicing reaction. Splicing complexes were separated on a 2% native agarose gel (Das and Reed 1999) and detected by autoradiography. Alternatively, 150 μL of a splicing reaction was loaded without addition of heparin onto a 10%–30% glycerol gradient containing G75 buffer and centrifuged as described above. Gradients were fractionated manually from top to bottom into 175-μL aliquots, and the distribution of [32P]-labeled pre-mRNA across the gradient was determined by Cherenkov counting in a scintillation counter.
ACKNOWLEDGMENTS
We are grateful to Irene Öchsner for excellent technical assistance and Thomas Conrad, Peter Kempkes, and Hossein Kohansal for help in preparing HeLa cell nuclear extract. We thank Gert Weber and Simon Trowitzsch for kindly providing purified 12S U2 snRNPs. N.B. was supported by a Kekulé-Stipendium from the Fonds der Chemischen Industrie. This work was supported by grants from the DFG Forschergruppe, Fonds der Chemischen Industrie, and the Ernst Jung Stiftung to R.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.120606.
REFERENCES
- Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Bach M., Bringmann P., Lührmann R. Purification of small nuclear ribonucleoprotein particles with antibodies against modified nucleosides of small nuclear RNAs. Methods Enzymol. 1990;181:232–257. doi: 10.1016/0076-6879(90)81125-e. [DOI] [PubMed] [Google Scholar]
- Barabino S.M., Blencowe B.J., Ryder U., Sproat B.S., Lamond A.I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990;63:293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Bergamini G., Preiss T., Hentze M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindereif A., Green M.R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987;6:2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Sproat B.S., Ryder U., Barabino S., Lamond A.I. Antisense probing of the human U4/U6 snRNP with biotinylated 2′-OMe RNA oligonucleotides. Cell. 1989;59:531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R.F., et al., editors. The RNA world II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Cheng S.C., Abelson J. Spliceosome assembly in yeast. Genes & Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Das R., Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Zhou Z., Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez G., Hartmuth K., Lührmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985;42:355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Görnemann J., Kotovic K.M., Hujer K., Neugebauer K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Grabowski P.J., Seiler S.R., Sharp P.A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985;42:345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Hall K.B., Konarska M.M. The 5′ splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc. Natl. Acad. Sci. 1992;89:10969–10973. doi: 10.1073/pnas.89.22.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan R.D., Cavanaugh A., Hempel W.M., Moss T., Rothblum L. Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res. 1999;27:3720–3727. doi: 10.1093/nar/27.18.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Urlaub H., Vornlocher H.-P., Will C.L., Gentzel M., Wilm M., Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner T.P., Giglio L.M., Weiner A.M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes & Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Nigg A.L., Heath H., Mullenders L.H., van Driel R., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 2002;10:1163–1174. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- Johnson T.L., Abelson J. Characterization of U4 and U6 interactions with the 5′ splice site using a S. cerevisiae in vitro trans-splicing system. Genes & Dev. 2001;15:1957–1970. doi: 10.1101/gad.895601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M., Sharp P.A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M.M., Sharp P.A. Association of U2, U4, U5, and U6 small nuclear ribonucleoproteins in a spliceosome-type complex in absence of precursor RNA. Proc. Natl. Acad. Sci. 1988;85:5459–5462. doi: 10.1073/pnas.85.15.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti B.B., Konarska M.M. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes & Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- Konforti B.B., Konarska M.M. A short 5′ splice site RNA oligo can participate in both steps of splicing in mammalian extracts. RNA. 1995;1:815–827. [PMC free article] [PubMed] [Google Scholar]
- Konforti B.B., Koziolkiewicz M.J., Konarska M.M. Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell. 1993;75:863–873. doi: 10.1016/0092-8674(93)90531-t. [DOI] [PubMed] [Google Scholar]
- Lacadie S.A., Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lemon B., Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Lim S.R., Hertel K.J. Commitment to splice site pairing coincides with A complex formation. Mol. Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Malca H., Shomron N., Ast G. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 2003;23:3442–3455. doi: 10.1128/MCB.23.10.3442-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E., Shiekhattar R., Sheldon M., Cho H., Drapkin R., Rickert P., Lees E., Anderson C.W., Linn S., Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- Maroney P.A., Romfo C.M., Nilsen T.W. Functional recognition of 5′ splice site by U4/U6•U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R.W., Grunberg-Manago M., editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- Nilsen T.W. The spliceosome. No assembly required? Mol. Cell. 2002;9:8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- Ségault V., Will C.L., Sproat B.S., Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Ryan D.E., Ge H.Y., Moore R.E., Young M.K., Lee T.D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q., Wang Z., Feaver W.J., Wu X., Bushnell D.A., Donahue T.F., Friedberg E.C., Kornberg R.D. Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- Tardiff D.F., Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D.A., Steitz J.A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world III. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Will C.L., Urlaub H., Achsel T., Gentzel M., Wilm M., Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T., Bindereif A. Reconstituted mammalian U4/U6 snRNP complements splicing: A mutational analysis. EMBO J. 1992;11:345–359. doi: 10.1002/j.1460-2075.1992.tb05057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M., Zapp M.L., Berget S.M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1988;8:814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]