Abstract
Some RNA silencing systems in plants, nematodes, and fungi show spreading of silencing along target sequences, termed transitive silencing. Here, we address the question of whether endogenous targets can be silenced by a transitive silencing signal in plants. In transgenic Arabidopsis thaliana plants that harbored a silencing-inducing locus and a transgenic chimeric primary target, silencing of a secondary transgenic target occurred and the expression of the endogenous catalase genes was down-regulated, coinciding with a knock-down phenotype. Strikingly, the efficiency of the catalase silencing appeared to be correlated with the zygosity of the primary target locus and, to a lesser extent, with that of the silencing-inducing locus. These data suggest that silencing of an endogene induced by transgenic secondary small interfering RNAs (siRNAs) might depend on the amount of primary target transcripts that can act as template for the production of an efficient transitive silencing signal.
Keywords: RNA silencing, transitivity, inverted repeat locus, gene family, Arabidopsis thaliana
INTRODUCTION
RNA silencing is a general term used for epigenetic regulatory pathways in which small RNAs of 21–28 nucleotides (nt) mediate either transcriptional gene silencing by changes in chromatin state or post-transcriptional gene silencing (PTGS) through target degradation or translational repression (for review, see Baulcombe 2004). Since the accidental discovery that the introduction of a sense overexpressing transgene can lead to cosuppression of both the transgene and the endogene (Napoli et al. 1990), RNA silencing has been widely used as a tool to down-regulate gene expression. Transgenic RNAs are recruited in the silencing pathway by the production of double-stranded RNA (dsRNA), which is processed by an RNase III-like enzyme (Dicer) into “small interfering” RNAs (siRNAs) that mediate the degradation of target RNAs and possibly also DNA methylation by RNA-induced silencing complex (RISC)-like effector complexes (Mette et al. 1999; Hamilton et al. 2002; Zilberman et al. 2003).
In plants, nematodes, and fungi, not only can trigger-derived siRNAs be detected, but also siRNAs that correspond to regions upstream from the region targeted by the dsRNA trigger. Moreover, in plants and fungi, siRNAs can be found corresponding to the downstream region as well (Sijen et al. 2001; Braunstein et al. 2002; Han and Grierson 2002; Klahre et al. 2002; Sanders et al. 2002; Vaistij et al. 2002; Nicolás et al. 2003; Van Houdt et al. 2003). Secondary siRNAs have been shown to induce silencing of targets without homology with the silencing inducer (for review, see Bleys et al. 2006). This phenomenon, designated transitive silencing, involves an amplification process catalyzed by the RNA-dependent RNA polymerase RDR6 that synthesizes dsRNA by using target RNAs as template, either by priming of primary siRNAs (Lipardi et al. 2001; Sijen et al. 2001) or by a primer-independent mechanism that starts at the 3′ end of the target RNAs (Schiebel et al. 1993; Vaistij et al. 2002; Tang et al. 2003; Petersen and Albrechtsen 2005).
In Caenorhabditis elegans and Mucor circinelloides, transitivity occurs along transcripts derived from both transgenes and endogenes (Sijen et al. 2001; Alder et al. 2003; Nicolás et al. 2003). In plants, transgenes and endogenes seem to behave differently in relation to transitivity. Many studies have demonstrated spreading of silencing along transgene sequences, but failed to do so for endogenous transcripts (Vaistij et al. 2002; Himber et al. 2003; Kościańska et al. 2005; Miki et al. 2005; Petersen and Albrechtsen 2005). The ability of endogenes to serve as template for the RDR6-mediated production of secondary siRNAs has been described just once (Sanders et al. 2002). Because all these transitivity studies only investigated the ability of secondary siRNAs to induce silencing of transgenic secondary targets, it is not known whether they can target plant endogenes as well.
Previously, we have established that in tobacco (Nicotiana tabacum) a post-transcriptionally silenced inverted repeat (IR) transgene locus (X) is able to silence a nonhomologous transgene (Z), when a stepwise homology is created by introducing a chimeric primary target (Y) with one region homologous to the silencing inducer X and a second upstream region homologous to the secondary target Z (Van Houdt et al. 2003). A similar XYZ system has been developed in Arabidopsis thaliana (L.) Heynh., allowing us to demonstrate that a transitive silencing signal produced by an amplification mechanism is able to trigger silencing of endogenous targets, and that the efficiency of silencing correlates with the zygosity of the primary target locus, suggesting a connection with the transcript abundance.
RESULTS AND DISCUSSION
Development of a transitive silencing system in Arabidopsis
First, we checked whether a particular isolated IR transgene locus X21 (P35S-nptII-3′chs; Fig. 1a) in Arabidopsis is able to transmit its in trans silencing capacity to a primary target Y7 (P35S-gus-npt3′chs), resulting in transitive silencing of a secondary target transgene Zc (P35S-gus-npt3′nos), similar to the transitive XYZ system in tobacco (Van Houdt et al. 2003). For details of constructs, plant material, and crosses, we refer to Materials and Methods. Genotype and zygosity of the transgenic loci in the different Arabidopsis plants were schematically annotated as follows: slashes separate the loci X, Y, and Z; XX, YY, or ZZ represent homozygosity for the respective loci; hyphens indicate hemizygosity, double hyphens azygosity, and an asterisk unknown zygosity with the locus either homo- or hemizygous.
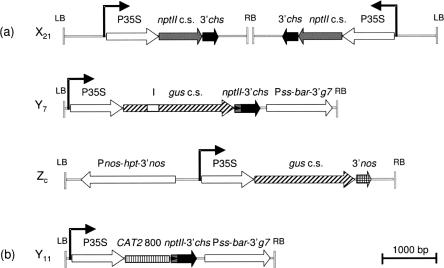
FIGURE 1.
Schematic outline of the T-DNA constructs (drawn to scale), present in locus X21, primary targets Y7 and Y11, and secondary target Zc. (a) Loci X21, Y7, and Zc used to assess transitive silencing of transgenes in A. thaliana; (b) locus Y11 used to assess transitive silencing of endogenes. Silenced locus X21 contains two copies of the DEchs278 T-DNA that are arranged as an IR around the right border. Locus Zc is a single-copy locus. The copy number of the Y7 and Y11 loci is unknown. 3′chs, 3′-UTR of the chalcone synthase gene of Anthirrinum majus; 3′g7, 3′-UTR of the Agrobacterium tumefaciens octopine T-DNA gene 7; 3′nos, 3′-UTR of the nopaline synthase gene; bar, bialaphos acetyltransferase-coding sequence conferring phosphinothricin resistance; CAT2 800, last 800 nt of the A. thaliana CAT2 sequence; gus c.s., gus-coding sequence; hpt, hygromycin phosphotransferase-coding sequence; I, artificial intron; nptII c.s., neomycin phosphotransferase II-coding sequence; P35S, CaMV 35S promoter; Pnos, nopaline synthase promoter; Pss, promoter of the small subunit of ribulose-1,5-bisphosphate carboxylase; LB, left T-DNA border; RB, right T-DNA border.
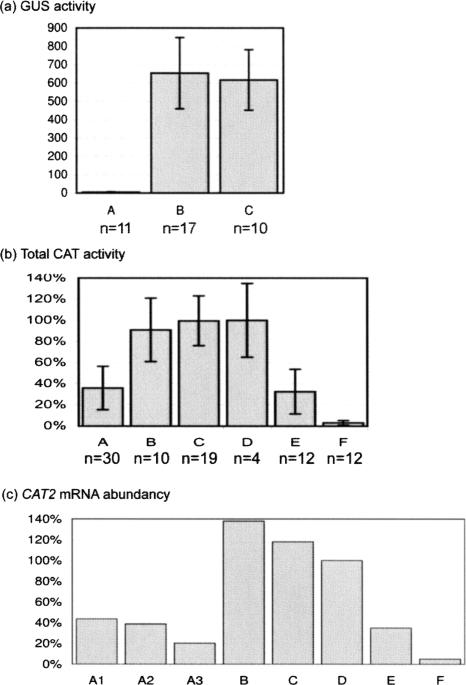
The occurrence of transitive silencing in X21-/Y7-/ZcZc hybrid plants was analyzed through measurement of the GUS activity, because both Y7 and Zc encode a functional β-glucuronidase (GUS) protein. A 130- to 140-fold reduction in GUS activity was observed in 11 X21-/Y7-/ZcZc hybrid plants (Fig. 2a, A) compared to the progeny of the parental plants (17 --/Y7*/ZcZc and 10 X21X21/--/ZcZc plants; Fig. 2a, B and C, respectively). These data indicate that not only in tobacco (Van Houdt et al. 2003), but also in Arabidopsis, in trans-silenced Y transcripts can produce a transitive silencing signal leading to spreading of silencing.
FIGURE 2.
Protein and RNA levels in F1 hybrid plants. (a) GUS activity levels in protein extracts of leaf tissue harvested from transgenic Arabidopsis plants containing different combinations of loci X21, Y7 and Zc. SD is given by error bars. A, X21-/Y7-/ZcZc; B, --/Y7*/ZcZc; C, X21X21/--/ZcZc; n, total number of plants tested; TSP, total soluble protein. (b) Average total CAT activity in wild-type and transgenic Arabidopsis plants containing different combinations of loci X21 and Y11 or containing a CAT2 hairpin construct (Vandenabeele et al. 2004). CAT activity in protein extracts of leaf tissue was measured with a spectrophotometric assay. The average total CAT activity of the different transgenic plants is relative to the average CAT activity of four wild-type (Col-0, D) plants, which is set at 100%. SD is given by error bars. A, X21-/Y11-/CATCAT; B, X21X21/--/CATCAT; C, --/Y11Y11/CATCAT; D, Col-0; E, CAT2HP1; F, CAT2HP2; n, total number of plants tested. (c) CAT2 mRNA abundancy. A quantitative real-time PCR was performed using CAT2-specific primers to determine the CAT2 RNA level relative to that in a wild-type background (100%). A1, A2, and A3, RNA levels in three X21-/Y11-/CATCAT plants; B, RNA level in one X21X21/--/CATCAT plant; C, RNA level in one --/Y11Y11/CATCAT plant; D, RNA levels in one wild-type Col-0; E, RNA level in one CAT2HP1 plant; F, RNA level in one CAT2HP2 plant.
In C. elegans, transitive RNA interference (RNAi) is not limited to transgenic targets (Sijen et al. 2001). In plants, however, secondary siRNAs have been shown to target transgenic and viral sequences (Sanders et al. 2002; Vaistij et al. 2002; Van Houdt et al. 2003), but it has never been investigated whether endogenous sequences can be silenced by a transitive silencing signal. Therefore, we constructed Y11 (P35S-CAT2 800-npt3′chs; Fig. 1b), which harbors an expression cassette with the last 800 nt of the CAT2-coding sequence. The objective was to determine whether the CAT genes in A. thaliana can be transitively silenced by combining the X21 and Y11 loci. The Arabidopsis catalase (CAT) multigene family consists of three members (CAT1, CAT2, and CAT3), with a nucleotide and amino acid sequence identity of 70%–72% and 75%–84%, respectively (Frugoli et al. 1996).
All wild-type (Fig. 2b, D) and parental plants (X21X21/--/CATCAT and --/Y11Y11/CATCAT; Fig. 2b, B and C, respectively) retained a normal CAT activity, whereas the X21-/Y11-/CATCAT F1 hybrid plants (Fig. 2b, A; see Materials and Methods) had an average total CAT activity of 36%. This level of silencing corresponds with that observed in the hairpin RNA (hpRNA)-silenced line CAT2HP1 (Fig. 2b, E; 33% total CAT activity; Vandenabeele et al. 2004), but is less strong than the reduction seen in the CAT2HP2 line (Fig. 2b, F; 3% total CAT activity; Vandenabeele et al. 2004).
To analyze the silencing at the RNA level, we performed a quantitative real-time PCR with a CAT2-specific primer pair (see Materials and Methods). Total RNA was prepared from leaf material, and first-strand cDNA was generated with an oligo(dT)12–18 primer. Figure 2c clearly shows a two- to fivefold lower CAT2 transcript level in X21-/Y11-/CATCAT plants (A1, A2, and A3) than that of the wild-type plant (D). The reduction was similar in the CAT2HP1 plant (Fig. 2c, E), but 20-fold lower in the CAT2HP2 plant (Fig. 2c, F). The parental plants (X21X21/--/CATCAT and --/Y11Y11/CATCAT; Fig. 2c, B and C, respectively) showed a slight increase in the CAT2 RNA level. We can conclude that an in trans-silenced transgenic primary target produces transitive silencing signals that are able to down-regulate the expression of endogenes. This capacity does not depend on the potential of the primary target transcripts to be translated into a functional protein, because the Y11 construct contains no translation start codon.
Influence of the zygosity of the silencing inducer and the primary target on the efficiency of transitive silencing of an endogene
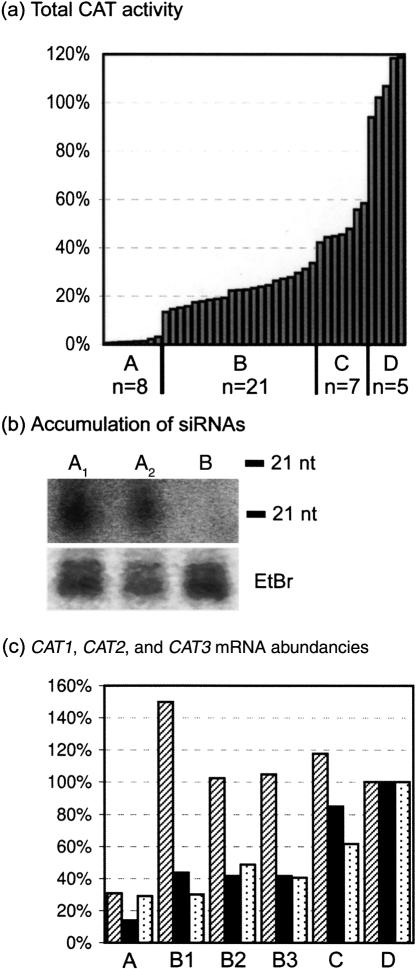
To investigate the stability of endogene suppression via a transitive signal and to determine whether the efficiency of silencing is affected by the homozygous condition of either the silencing-inducing locus X21 or the primary target locus Y11, seeds from a self-fertilized F1 hybrid plant (X21-/Y11-/CATCAT) were grown on medium selective for both X21 and Y11. The two loci segregated independently, and these F2 progeny plants were expected to have genotypes X21X21/Y11Y11/CATCAT, X21-/Y11Y11/CATCAT, X21X21/Y11-/CATCAT, or X21-/Y11-/CATCAT at a 1:2:2:4 ratio. Thirty-six selected F2 progeny plants were analyzed and ranked according to their total CAT activity (Fig. 3a). Segregation analysis of the 36 plants showed a correlation between the zygosity of loci X21 and Y11 and the efficiency of transitive CAT silencing. The eight plants exhibiting very efficient silencing (<3% total CAT activity; Fig. 3a, A) were all plants homozygous for locus Y11 (X21X21/Y11Y11/CATCAT and X21-/Y11Y11/CATCAT), whereas the seven plants with the lowest degree of reduction (between 42% and 59% total CAT activity; Fig. 3a, C) were hemizygous for locus Y11, but homozygous for locus X21 (X21X21/Y11-/CATCAT). The 21 plants with intermediate silencing efficiencies (between 13% and 34% total CAT activity; Fig. 3a, B) were hemizygous for both loci (X21-/Y11-/CATCAT). These results suggest that the zygosity of the primary target locus Y11 and to a lesser extent that of the silencing-inducing locus X21 influence the efficiency of transitive silencing of the CAT genes. We cannot exclude the possibility that some of the observed CAT silencing results from the activation of cosuppression induced by the sense construct Y11, because sense-induced PTGS can occur stochastically in transformed plants. However, the Y11 locus did not induce silencing in any of the 24 --/Y11Y11/CATCAT plants (Fig. 2b, C; Fig. 3a, D), making it is very likely that the efficient silencing observed in X21X21/Y11Y11/CATCAT plants results from transitive signals produced by Y11.
FIGURE 3.
Protein levels, siRNA accumulation, and RNA levels in F2 progeny plants and parental plants. (a) Total CAT activity in F2 progeny plants with segregating X21 and Y11 loci and parental --/ Y11Y11/CATCAT plants. The total CAT activity of the different transgenic plants is relative to the average CAT activity of five wild-type plants (100%; not shown). A, X21-/Y11Y11/CATCAT and X21X21/Y11Y11/CATCAT; B, X21-/Y11-/CATCAT; C, X21X21/Y11-/CATCAT; D, --/ Y11Y11/CATCAT; n, total number of plants per group. (b) Accumulation of CAT2-specific siRNAs in two silenced X21X21/Y11Y11/CATCAT plants (A1 and A2) and absence of siRNAs in one nonsilenced --/Y11Y11/CATCAT plant (B). Low molecular weight RNA was extracted from leaves of 4-wk-old plants. RNA oligomers of 21 nt and 24 nt were used as molecular markers. siRNAs were detected with a hydrolyzed 32P-labeled probe comprising 274 bp in the 3′ end of the CAT2 sequence. With this probe it is impossible to discriminate between siRNAs originating from the transgene Y11 or from the endogenous sequences. The predominant ethidium bromide (EtBr)-stained species in the low molecular weight RNA fraction are shown as loading controls. (c) CAT1, CAT2, and CAT3 mRNA abundancies. Quantitative real-time PCRs were performed with gene-specific primers to determine the RNA levels of CAT1 (shaded bars), CAT2 (black bars), and CAT3 (dotted bars) relative to those in a wild-type background (100%). A, RNA levels in one X21X21/Y11Y11/CATCAT plant; B1, B2, and B3, RNA levels in three X21-/Y11-/CATCAT plants; C, RNA levels in one X21X21/Y11-/CATCAT plant; D, RNA levels in wild-type Col-0.
The positive correlation between the silencing efficiency and the zygosity of the primary target locus suggests that the amount of primary transcripts serving as template for RDR6 might determine the level of secondary siRNAs produced, thus resulting in a certain degree of silencing of the secondary target. Analyses in the transitive XYZ system in tobacco have demonstrated that the accumulation level of secondary siRNAs depends on the zygosity of the primary target locus from which they are derived (García-Pérez et al. 2004). The negative correlation in plants hemizygous for Y11 between the level of silencing and the zygosity of locus X21 indicates that the primary silencing signal also plays a role. Perhaps a higher dose of primary siRNAs might be produced by locus X21 in the homozygous condition, which could trigger a higher rate of RISC-mediated cleavage of Y11 templates. This hypothesis is supported by the fact that a hairpin construct controlled by the strong 35S promoter induces a stronger silencing phenotype than the same construct controlled by the weak nopaline synthase promoter (Chuang and Meyerowitz 2000). In this case, the production of more hpRNAs and thus siRNAs leads to a more efficient degradation and silencing of the endogenous target RNAs.
We also looked at the accumulation of siRNAs that are the key molecules of PTGS. An RNA gel blot with a hydrolyzed 32P-labeled probe comprising 274 bp in the 3′ end of the CAT2 sequence revealed the presence of CAT2-specific siRNAs in X21X21/Y11Y21/CATCAT (Fig. 3b, A1 and A2) plants, whereas in --/Y11Y11/CATCAT plants (Fig. 3b, B) no such molecules were detected.
To assess whether the most efficiently silenced X21X21/Y11Y11/CATCAT progeny plants displayed the characteristic CAT-deficient necrosis phenotype, we performed a high-light (HL) treatment (see Materials and Methods). HL irradiation induces photorespiration that leads to accumulation of H2O2 in CAT-deficient plants, followed by active cell death (Chamnongpol et al. 1998; Dat et al. 2003; Vandenabeele et al. 2004). All X21X21/Y11Y11/CATCAT progeny plants had visible signs of cell death within 8 h of HL irradiation, whereas wild-type control plants did not, even after 48 h of HL (Fig. 4). In conclusion, transitive silencing elicits the same knock-down phenotype as hairpin-induced silencing.
FIGURE 4.
Phenotypes of wild-type and X21X21/Y11Y11/CATCAT plants after exposition to high-light (HL) irradiation (1000 μmol m−2 sec−1). Cell death was visible in CAT-deficient X21X21/Y11Y11/CATCAT plants within 8 h of HL irradiation, while in wild-type control plants this phenotype did not occur, even not after 48 h of HL.
Differential susceptibility of different members of the CAT gene family to transitive silencing signals
The Arabidopsis CAT2 gene targeted by the Y11 construct is part of a gene family. All three members are highly expressed in inflorescences, but in leaves only the CAT2 and CAT3 genes are highly expressed (Frugoli et al. 1996). With the CAT activity assay used above, it is not possible to discriminate between the three CAT proteins, because the total CAT activity is measured, implying that possible differential silencing efficiencies of CAT1, CAT2, and CAT3 cannot be determined in this way. To assay to what extent the three CAT genes were silenced in F2 progeny plants, we performed a quantitative real-time PCR with gene-specific primer pairs. In wild-type samples, the CAT1 and CAT3 mRNAs were ∼500- to 600-fold and 20- to 25-fold less abundant than those of CAT2, respectively (data not shown).
The transcript levels of CAT1, CAT2, and CAT3 in leaf tissue of F2 progeny plants relative to those of a wild-type plant are given in Figure 3c. In this graph, the mRNA abundancies of CAT1, CAT2, and CAT3 in the wild-type plant (Fig. 3c, D) are all three set at 100%. The CAT2 RNA level was sevenfold reduced in a X21X21/Y11Y11/CATCAT sample (Fig. 3c, A), 2.3-fold in three X21-/Y11-/CATCAT samples (Fig. 3c, B1, B2, B3), and not significantly in a X21X21/Y11-/CATCAT sample (Fig. 3c, C), indicating that the reduction in CAT2 RNA levels also correlated with the zygosity of loci X21 and Y11. The transcript levels of CAT3 seemed to be reduced to the same extent as those of CAT2. This observation is very intriguing, because a sevenfold reduction in CAT2 RNAs in the X21X21/Y11Y11/CATCAT sample results in a residual CAT2 RNA level that is higher than the nonsilenced CAT3 steady-state RNA level. Still, the CAT3 RNAs are targeted for degradation to the same degree as those of CAT2, depending on the zygosity of both loci X21 and Y11. We cannot explain this result, but it shows that there are factors other than just the RNA abundance that determine the susceptibility of particular transcripts for siRNA-mediated turnover, for instance, the spatial and temporal gene expression patterns. Similarly, Kerschen et al. (2004) found that 25 different endogenous target sequences possess an inherent degree of susceptibility to RNA silencing induced by hairpin constructs, which was independent from the gene expression levels.
The degree of reduction in CAT2 and CAT3 RNAs is consistent with the dosage dependency observed at the protein level. However, the reduction at the RNA level is not as pronounced as that at the protein level; for instance, in X21X21/Y11Y11/CATCAT plants the CAT2 and CAT3 RNA levels are reduced to sevenfold, whereas the total CAT activity is reduced to 44-fold. This supports previous data (Van Houdt et al. 1997) and implies that the silencing signal can also act at the translational level, either because the detected mRNAs might not be translatable anymore (e.g., no 5′ cap structure) or might be sequestered in cytoplasmic processing bodies (P-bodies) that are found in yeast and mammalian cells (Rossi 2005).
In contrast to CAT2 and CAT3, the CAT1 mRNA abundance is not significantly reduced in plants that are hemizygous for locus Y11 (Fig. 3c, B1,B2,B3), but it is in Y11 homozygous plants (Fig. 3c, A). This difference in susceptibility to silencing between CAT1 and CAT3 is most probably not the consequence of a different degree in homology, because both the sequence identity of the CAT1 and CAT3 genes with the Y11 transcript (78% and 75%, respectively) as well as the number of stretches of at least 21 nt with full complementarity are very similar. One of the contributing factors might be the different accumulation levels of the CAT1 and CAT3 genes, the former being about 25-fold less abundant than the latter.
Conclusions
We demonstrate that secondary siRNAs originating from a transgenic primary target are able to down-regulate the expression of the endogenous CAT gene family in A. thaliana, resulting in a knock-down phenotype. Our experimental setup suggests that the expression of genes in gene families is determined by a subtle equilibrium between the gene expression level and the amount of siRNAs and could reflect an epigenetic gene regulation mechanism mediated by endogenous small RNAs.
MATERIALS AND METHODS
Constructs
The T-DNA-derived plant transformation vector pDEchs287 (referred to as P35S-nptII-3′chs) carried a neomycin phosphotransferase II (nptII) gene (conferring kanamycin resistance) fused to the 3′-untranslated region (UTR) of the Antirrhinum majus chalcone synthase gene (3′chs, 287 nt) and transcribed from the cauliflower mosaic virus 35S promoter (P35S) toward the right T-DNA border. After floral dip transformation with the DEchs287 T-DNA, plants were obtained harboring the IR locus X21 (see below; Fig. 1a). The T-DNA vector pPs35SGUSnpt3′chs (referred to as P35S-gus-npt3′chs) contained two chimeric genes between the T-DNA borders: the bialaphos acetyltransferase-coding (bar) sequence conferring phosphinothricin resistance, fused to the 3′-UTR of the Agrobacterium tumefaciens octopine T-DNA gene 7 (3′g7) and under the control of the promoter of the small subunit of ribulose-1,5-bisphosphate carboxylase (Pss) and a P35S-driven gus transgene fused to the last 122 nt of the nptII gene and the A. majus chs terminator sequence (npt3′chs). After floral dip transformation with the Ps35SGUSnpt3′chs T-DNA, plants were obtained harboring the single locus Y7 (Fig. 1a). A high-throughput Gateway vector, pPs35SGW+npt3′chs, was constructed by inserting the reading frame A (rfA) sequence (1700 nt EcoRV fragment of the vector pGemGWrfA; Karimi et al. 2002) into the vector pPs35Snpt3′chs (cut with PacI and blunted with T4-DNA polymerase). The orientation of the inserted rfA fragment was checked by sequencing. The T-DNA vector pPs35SCAT800npt3′chs (referred to as P35S-CAT2 800-npt3′chs) was obtained through an LR clonase reaction between the vector pPs35SGW+npt3′chs and the entry clone pENTR3′CAT800 (obtained by a BP clonase reaction between pDONR and an attB-flanked PCR product containing the last 800 nt of the A. thaliana CAT2-coding sequence). After floral dip transformation with the Ps35SCAT800npt3′chs T-DNA, plants harboring the single locus Y11 were obtained (see below; Fig. 1b). The T-DNA H610 (H T-DNA in De Buck et al. 1998, 2001) contained a gus gene fused to the 3′-UTR of the nopaline synthase gene (3′nos) under control of P35S and the hygromycin acetyl transferase (hpt) gene conferring resistance to hygromycin. Plants homozygous for the Zc locus (Fig. 1a) with a single copy of the T-DNA H610, were already available. All the T-DNA vectors were inserted into the Agrobacterium tumefaciens strain C58C1RifR (pMP90).
Plant material, crosses, and high-light treatment
All transformants were obtained via the floral dip procedure, performed as described by Clough and Bent (1998). Either wild-type A. thaliana (L.) Heyhn. (ecotype Columbia) or transgenic plants homozygous for the Zc locus (--/--/ZcZc plants) containing a single copy of the T-DNA H610, were used for the dipping experiments. Seeds of the dipped plants were harvested and sown on K1 medium supplemented with kanamycin (50 mg L−1) for selection of the DEchs287 T-DNA or phosphinothricin (10 mg L−1) for selection of Ps35SGUSnpt3′chs and Ps35SCAT800npt3′chs. To obtain plants with a candidate X locus, --/--/ZcZc plants were supertransformed with the DEchs287 T-DNA. Primary transformants (X-/--/ZcZc) were retained with low amounts of the NPTII protein, indicative of silencing of the nptII gene(s) in the X locus, and with a high GUS activity, excluding transcriptional silencing of the gus gene in locus Zc because of 35S promoter homology. These plants were screened for harboring a single locus and were further characterized through DNA gel blot analysis to determine the T-DNA integration pattern. Because the efficient in trans-silencing locus X in the tobacco XYZ system contained an IR about the right T-DNA border (Van Houdt et al. 2003), a supertransformant with the T-DNAs in an IR configuration (locus X21, Fig. 1a), namely X21-/--/ZcZc, was chosen for further handling. To obtain X21-/--/-- plants, seeds from a self-fertilized X21-/--/Zc- plant (obtained after crossing X21-/--/ZcZc with a --/Y-/-- plant, not discussed here) were sown on K1 medium supplemented with kanamycin. The presence of locus Zc was checked through a callus induction test on hygromycin-containing (20 mg L−1) medium. Only one plant out of 52 kanamycin-resistant plants was hygromycin sensitive, suggesting that locus X21 and locus Zc were genetically linked. To obtain candidate Y7 and Y11 plants, --/--/ZcZc plants were supertransformed with the Ps35SGUSnpt3′chs T-DNA, and wild-type plants were dipped with A. tumefaciens containing the T-DNA Ps35SCAT800npt3′chs, respectively. Fluorometric GUS assays and CAT activity assays were performed to select primary transformants with highly expressed Y transgenes, excluding silencing of the gus transgene of Zc or cosuppression of the endogenous CAT genes. Via segregation analysis we ensured the presence of single loci, leading to the selection of transformants --/Y7-/ZcZc (Ps35SGUSnpt3′chs) and --/Y11-/-- (Ps35SCAT800npt3′chs). Transitive silencing was studied by crossing progeny plants of X21X21-/--/ZcZc with those of --/Y7-/ZcZc and --/Y11-/--. The resulting hybrid seeds were grown on medium selective for the presence of X21 and Y7 or Y11. After 2 wk, all plants were transferred to soil and grown under a light regime of 16 h day/8 h night, at 21°C. For high-light (HL) treatment, 4-wk-old plants were exposed to continuous HL irradiation (∼1000 μmol photons m−2 sec−1).
NPTII and GUS quantification
Proteins were extracted from two rosette leaves harvested 6 wk after sowing and frozen in liquid nitrogen. The protein concentrations, NPTII concentrations, and GUS activities were determined as described previously (Breyne et al. 1993; Van Houdt et al. 1997).
Catalase activity assay
Protein extracts were prepared by grinding two rosette leaves, frozen in liquid nitrogen, in 120 μL extraction buffer (60 mM Tris-HCl at pH 6.9, 1 mM phenylmethylsulfonylfluoride, 10 mM dithiothreitol, 20% glycerol). The homogenate was centrifuged (15,000g) twice at 4°C for 10 min to remove insoluble material. The supernatant was used for spectrophotometric catalase analysis according to Clare et al. (1984) after determining the total amount of soluble protein with the BioRad Protein Assay (Bradford 1976) with bovine serum albumin as a standard.
Quantitative real-time PCR analysis
For the quantification of CAT1, CAT2, and CAT3 transcript levels by real-time PCR, total RNA from leaf material of 5-wk-old plants was prepared with TRIzol Reagent (Invitrogen). For each sample, 1 μg of DNase-treated total RNA was reverse-transcribed with the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the instruction manual. First-strand cDNA was generated with an oligo(dT)12–18 primer. Of the first-strand cDNA, 0.2 μL was used as a template in a subsequent quantitative PCR using the qPCR™ Core Kit for SYBR® Green I (Eurogentec). The transcripts were amplified with gene-specific primers: 5′-TGCTCCTCTTACGGTTTGGTTTC-3′ and 5′-ACCCTTCTTTAAGCGTTTCATTGTC-3′ for CAT1 (At1g20630); 5′-GCTGGCAAGCCGTCTGAAC-3′ and 5′-AGCACAGAAGATCCACATGATGAAG-3′ for CAT2 (At4g35090); 5′-GGCCAATCTCCATATAAGCTCAGT-3′ and 5′-GGATTTAACGACCAAGCGATGATAG-3′ for CAT3 (At1g20620); 5′-AACTTGTGCTCATCTGCCATTAGG-3′ and 5′-TGATTCTGCGGAAACACCACTTTAG-3′ for actin2 (At3g60830).
Small-RNA analysis
Small RNAs were detected as described by Van Houdt et al. (2003), with minor changes. Low molecular weight RNA (35 μg) was loaded on the gel, as well as 21-nt and 24-nt RNA oligomers as size controls. The probe, a PCR fragment of 274 bp amplified with the primers 5′-CCGGTTAATTAAATGCTGAGAAGTATCCAACT-3′ and 5′- CCGGTTAATTAACGGCTTGCCAGCTTCTGTCC-3′, was labeled with [α-32P]dCTP using the Rediprime II random prime labeling system (GE-Healthcare) according to the manufacturer's instructions. After removal of unincorporated [α-32P]dCTPs with the Bio-Spin p30 Column (Bio-Rad Laboratories), the probe was hydrolyzed into fragments of ∼50 nt. Hybridization and washes were performed as described at 50°C. Labeled membranes were exposed to a PhosphorImager screen (GE-Healthcare). Predominantly ethidium bromide-stained species of low molecular weight RNA, separated by agarose gel electrophoresis (1 μg per lane) were used as loading controls.
ACKNOWLEDGMENTS
We thank Els Van Lerberge and Ellen Wirawan for technical assistance and Martine De Cock and Karel Spruyt for help with the manuscript and figures, respectively. This work was supported by grants from the European Union Biotech Project (no. QLRT-2000-00078). A.B. is a Research Fellow of the Research Foundation-Flanders and H.V.H. is indebted to the Institute for the Promotion of Innovation by Science and Technology in Flanders for a postdoctoral fellowship.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.108106.
REFERENCES
- Alder M.N., Dames S., Gaudet J., Mango S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNAi silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bleys A., Van Houdt H., Depicker A. Transitive and systemic RNA silencing: Both involving an RNA amplification mechanism? In: Nellen W., Hammann C., editors. Small RNAs: Analysis and regulatory functions. Nucleic Acids and Molecular Biology. Vol. 17. Springer; Berlin: 2006. pp. 119–139. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braunstein T.H., Moury B., Johannessen M., Albrechtsen M. Specific degradation of 3′ regions of GUS mRNA in posttranscriptionally silenced tobacco lines may be related to 5′–3′ spreading of silencing. RNA. 2002;8:1034–1044. doi: 10.1017/s1355838202026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P., De Loose M., Dedonder A., Van Montagu M., Depicker A. Quantitative kinetic analysis of β-glucuronidase activities using a computer-directed microtiter plate reader. Plant Mol. Biol. Rep. 1993;11:21–31. [Google Scholar]
- Chamnongpol S., Willekens H., Moeder W., Langebartels C., Sandermann H., Jr., Van Montagu M., Inzé D., Van Camp W. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic plants. Proc. Natl. Acad. Sci. 1998;95:5818–5823. doi: 10.1073/pnas.95.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.-F., Meyerowitz E.M. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare D.A., Duong M.N., Darr D., Archibald F., Fridovich I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 1984;140:532–537. doi: 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dat J.F., Pellinen R., Beeckman T., Van De Cotte B., Langebartels C., Kangasjärvi J., Inzé D., Van Breusegem F. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 2003;33:621–632. doi: 10.1046/j.1365-313x.2003.01655.x. [DOI] [PubMed] [Google Scholar]
- De Buck S., Jacobs A., Van Montagu M., Depicker A. Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol. Plant Microbe Interact. 1998;11:449–457. doi: 10.1094/MPMI.1998.11.6.449. [DOI] [PubMed] [Google Scholar]
- De Buck S., Van Montagu M., Depicker A. Transgene silencing of invertedly repeated transgenes is released upon deletion of one of the transgenes involved. Plant Mol. Biol. 2001;46:433–445. doi: 10.1023/a:1010614522706. [DOI] [PubMed] [Google Scholar]
- Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;112:327–336. doi: 10.1104/pp.112.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pérez R.D., Van Houdt H., Depicker A. Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J. 2004;38:594–602. doi: 10.1111/j.1365-313X.2004.02067.x. [DOI] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Grierson D. The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol. Genet. Genomics. 2002;267:629–635. doi: 10.1007/s00438-002-0696-z. [DOI] [PubMed] [Google Scholar]
- Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kerschen A., Napoli C.A., Jorgensen R.A., Muller A.E. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004;566:223–228. doi: 10.1016/j.febslet.2004.04.043. [DOI] [PubMed] [Google Scholar]
- Klahre U., Crété P., Leuenberger S.A., Iglesias V.A., Meins F., Jr. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kościańska E., Kalantidis K., Wypijewski K., Sadowski J., Tabler M. Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum . Plant Mol. Biol. 2005;59:647–661. doi: 10.1007/s11103-005-0668-x. [DOI] [PubMed] [Google Scholar]
- Lipardi C., Wei Q., Paterson B.M. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Mette M.F., van der Winden J., Matzke M.A., Matzke A.J.M. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters. in trans. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Itoh R., Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás F.E., Torres-Martínez S., Ruiz-Vázquez R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Albrechtsen M. Evidence implying only unprimed RdRP activity during transitive gene silencing in plants. Plant Mol. Biol. 2005;58:575–583. doi: 10.1007/s11103-005-7307-4. [DOI] [PubMed] [Google Scholar]
- Rossi J.J. RNAi and the P-body connection. Nat. Cell Biol. 2005;7:643–644. doi: 10.1038/ncb0705-643. [DOI] [PubMed] [Google Scholar]
- Sanders M., Maddelein W., Depicker A., Van Montagu M., Cornelissen M., Jacobs J. An active role for endogenous β-1,3-glucanase genes in transgene-mediated co-suppression in tobacco. EMBO J. 2002;21:5824–5832. doi: 10.1093/emboj/cdf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel W., Haas B., Marinković S., Klanner A., Sänger H.L. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H.A., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. A biochemical framework for RNA silencing in plants. Genes & Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F.E., Jones L., Baulcombe D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H., Ingelbrecht I., Van Montagu M., Depicker A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 1997;12:379–392. [Google Scholar]
- Van Houdt H., Bleys A., Depicker A. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 2003;131:245–253. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana . Plant J. 2004;39:45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]