Abstract
MicroRNAs (miRNAs) have been suggested to play important roles in cell proliferation, apoptosis, and differentiation. In this study, we examined miRNA expression profiles during 3T3-L1 pre-adipocyte differentiation. We constructed miRNA libraries from pre- and post-differentiated 3T3-L1 cells, and identified the expression of 77 previously reported miRNAs and 3 new miRNAs. Next, we investigated the expression levels of 102 miRNAs, including those identified in the libraries, during adipogenesis by Northern blot analysis. Sixty-five miRNAs were detected and the expression of 21 miRNAs was up- or down-regulated during adipogenesis. Intriguingly, changes in the miRNA expression pattern were observed at day 9, when lipid droplets were visible, but not at days 1, 2, or 5 after the induction of differentiation. Antisense inhibition of the up-regulated miRNAs did not affect 3T3-L1 pre-adipocyte differentiation. Although these miRNAs may be involved in modulating adipocyte function, mild down-modulations of the up-regulated miRNAs do not appear to affect 3T3-L1 pre-adipocyte differentiation.
Keywords: microRNA, adipocyte, differentiation, 3T3-L1
INTRODUCTION
Recent advances in the Human Genome Project (Lander et al. 2001) and in the experimental and computational prediction of coding and noncoding genes have led to the notion that there are 10-fold more transcription units than predicted coding genes in the human genome (Kapranov et al. 2002). These findings have forced us to revise the notion that RNA is simply an intermediate molecule in protein synthesis. Over the past few years, 20- to–25-nucleotide (nt) noncoding RNAs have been identified in the genomes of plants, invertebrates, and vertebrates, and have been categorized as microRNAs (miRNAs) (Carrington and Ambros 2003; Bartel and Chen 2004).
miRNAs are thought to regulate gene expression by partially base-pairing with their target mRNAs, which leads to mRNA degradation or repression of translation (Ambros 2004; Bartel 2004). In vertebrates, some species of miRNAs are specifically expressed or greatly elevated in a particular organ, which suggests that miRNAs may have some organ- or tissue-specific function (Lagos-Quintana et al. 2002; Sempere et al. 2003; Liu et al. 2004). It has also been reported that miR-181 is preferentially expressed in the B-lymphoid cells of mouse bone marrow, and that hematopoietic lineage differentiation can be altered by the action of miR-181 (Chen et al. 2004). Moreover, miR-15 and miR-16 are located on chromosome 13q14 in a region that is deleted in more than half of B-cell leukemias, and thus miRNA expression patterns may be related to the biological and clinical behavior of this leukemia (Calin et al. 2002). These findings suggest that miRNA plays important roles in vertebrate development and differentiation processes.
Adipose tissue plays a crucial role as an energy reservoir (Klaus 2004) and has recently been identified as an endocrine organ that secretes various biologically active substances, including adiponectin, leptin, and PAI-I (Matsuzawa et al. 2004). Phenotypic changes in adipose tissue may play important roles in the genesis of obesity and subsequent insulin resistance syndrome (Stolar 2002). Clarification of the mechanisms of adipogenesis should contribute to our understanding of the pathogenesis of obesity and insulin resistance syndrome.
In the present study, we focused on the possible involvement of miRNA in adipogenesis using a well-defined pre-adipocyte, 3T3-L1 (Green and Kehinde 1975; Morrison and Farmer 1999a). We found that the expression of 21 miRNAs is modulated in fully differentiated adipocytes. We also examined whether modulation of the expression of these differentially expressed miRNAs influences pre-adipocyte differentiation.
RESULTS
Cloning of microRNA expressed during adipogenesis
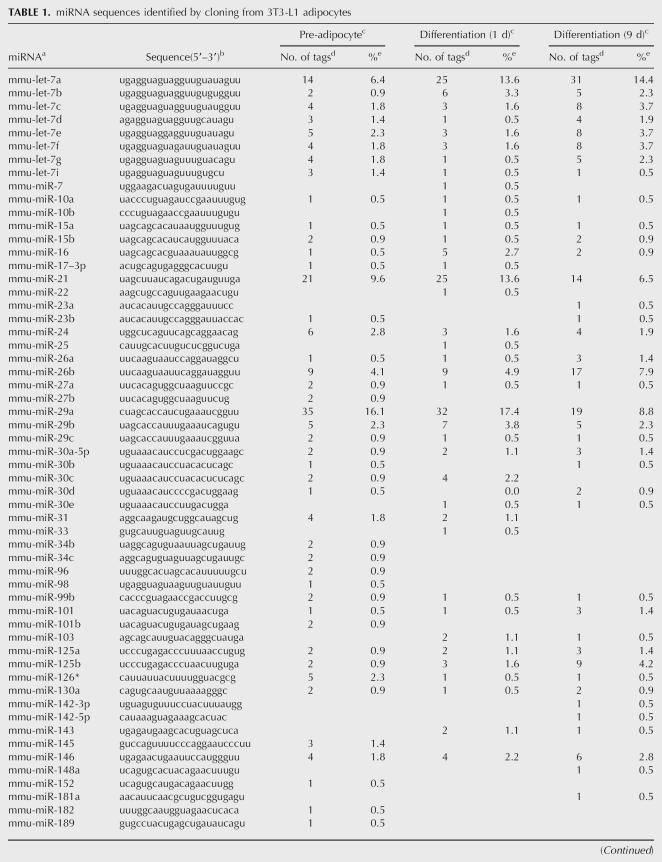
We constructed miRNA libraries from pre-adipocytes and cells at days 1 and 9 after the induction of differentiation. Approximately 10% of tags corresponded to RNAs other than miRNA, and included rRNA, degraded mRNA, tRNA, and snRNA. A total of 622 tags (74% of total tags) appeared to correspond to miRNAs (Table 1). They were grouped into 80 miRNAs, including three unregistered possible miRNAs.
TABLE 1.
miRNA sequences identified by cloning from 3T3-L1 adipocytes
TABLE 1.
Continued
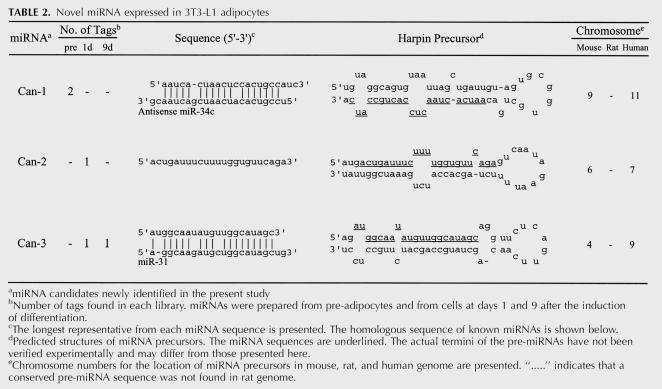
The sequences of these three possible miRNAs (denoted Can-1, Can-2, and Can-3) and their predicted secondary structures are shown in Table 2. These three possible miRNA sequences were found in the mouse genome. The sequences that surrounded these possible miRNAs show a stem-loop structure, which is a prerequisite for miRNA processing. Moreover, these possible pre-miRNA sequences are highly conserved in the human genome but are not present in the currently available rat genome sequence. Can-3 exhibits sequence homology with miR-31. Furthermore, Can-1 exhibits sequence homology with antisense miR-34c, but this is not the opposite strand of a known precursor.
TABLE 2.
Novel miRNA expressed in 3T3-L1 adipocytes
The number of tags for each miRNA in the three libraries is shown in Table 1. Some miRNAs, such as let-7a, miR-21, and miR-29a, were expressed abundantly in 3T3-L1 cells. Because there were very few tags for most miRNAs, it was difficult to assess the expression levels of miRNA based on the number of tags in the library. Thus, a total of 102 miRNAs, consisting of the 80 miRNAs identified in the library and an additional 22 mouse miRNAs, were subjected to Northern blot analysis.
Identification of miRNAs that are up- or down-regulated during adipogenesis
We examined the expression profiles in a highly differentiating subclone (#29) and a nondifferentiating subclone (#3). Sixty-five miRNAs were detected (63.7%). All of the Northern results will be provided upon request (to ekigaku1@ri.ncvc.go.jp) as supplemental data. The remaining 37 miRNAs were not detected under the conditions used.
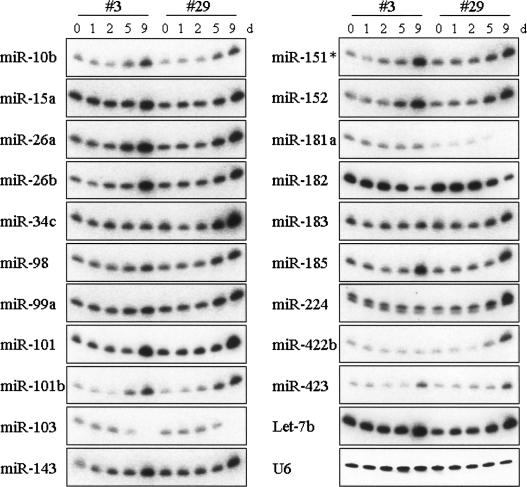
Expression was modulated in 21 species of miRNA during pre-adipocyte differentiation (Fig. 1). Thirteen miRNAs (miR-10b, 15a, 26a, 26b, 99a, 101, 101b, 143, 151*, 152, 185, 423, and let-7b) increased in abundance in both #3 and #29 (Fig. 1). These 13 miRNAs may not be related to an actual differentiation process and may be induced by growth arrest and/or hormonal stimulation.
FIGURE 1.
Temporal expression of miRNAs during adipocyte differentiation. Total RNA was prepared with TRIzol reagent, electrophoresed, blotted, fixed to a membrane, and hybridized to the StarFire probe for each miRNA and U6 RNA (control) as described in Materials and Methods. Representative results of more than two independent experiments are shown. All the other 81 miRNAs were also tested. All results are provided in Supplemental Data 1.
miR-34c and 98 were more abundantly expressed in #29 than in #3 at day 9. Expression of miR-183, 224, and 422b was up-regulated only in #29 at day 9. On the other hand, miR-103, 181a, and miR-182 were down-regulated at day 9. Since #29 accumulated cellular fat droplets at day 9, these #29-specifically modulated miRNAs may be directly involved in the formation of lipid droplets or in the storage of cellular fat.
Antisense inhibition of miRNAs that are up-regulated during adipogenesis
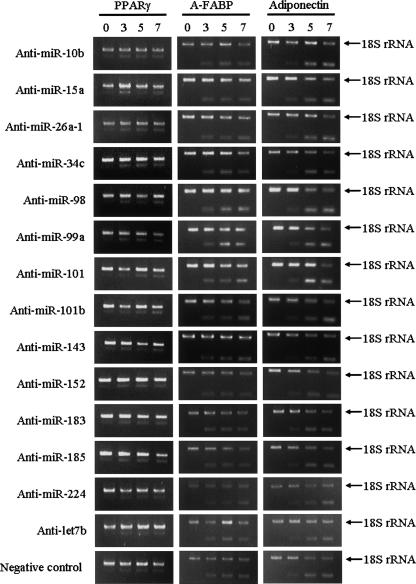
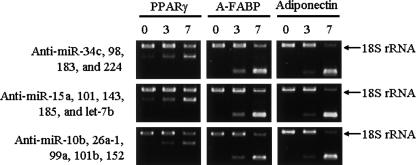
The miRNAs that are differentially expressed during 3T3-L1 pre-adipocyte differentiation may play active roles in differentiation. miR-10b, 15, 26a, 34c, 98, 99a, 101, 101b, 143, 152, 183, 185, 224, and let-7b were up-regulated during 3T3-L1 pre-adipocyte differentiation. We examined the effects of antisense oligo-RNAs (Anti-miR) for these miRNAs. Anti-miRs have been confirmed to block the effects of corresponding miRNAs by a luciferase reporter assay (Cheng et al. 2005). The 3T3-L1 cells were cultured, and differentiation was induced at 2 d after confluency (day 0). At days 3 and 5, the cells were transfected by Anti-miRs. None of the Anti-miR modulated the expression patterns of the marker genes (PPARγ, A-FABP, and adiponectin) compared with the mock-transfected cells (Fig. 2). The efficiency of transfection of Anti-miR was estimated to be >90%, which was assessed by cotransfection of Cy3-labeled negative control (data not shown). The expression patterns of the marker genes were not affected even by the inhibition of combination of several miRNAs (Fig. 3).
FIGURE 2.
RT-PCR analysis of adipocyte marker genes in cells that had inhibited miRNA function. 3T3-L1 cells were cultured, and differentiation was induced under normal conditions. At day 3 and day 5, the cells were transfected by each Anti-miR. Total RNA was isolated from cells at days 0, 3, 5, and 7. The expression patterns of three adipocyte differentiation markers (PPARγ, A-FABP, and adiponectin) were analyzed by competitive RT-PCR analysis using 18s ribosomal RNA as an internal control.
FIGURE 3.
RT-PCR analysis of adipocyte marker genes in cells that had inhibited multiple miRNAs function. 3T3-L1 cells were cultured, and differentiation was induced under normal conditions. At day 3 and day 5, the cells were transfected by each Anti-miR. Total RNA was isolated from cells at days 0, 3, and 7. The expression patterns of three adipocyte differentiation markers (PPARγ, A-FABP, and adiponectin) were analyzed by competitive RT-PCR analysis using 18s ribosomal RNA as an internal control.
DISCUSSION
The deletion of miR-14 in Drosophila results in increased levels of triacylglycerol and diacylglycerol, whereas increases in miR-14 copy number have the opposite effect (Xu et al. 2003). This recent finding suggests that miRNA might be involved in the regulation of fat metabolism, but the gene that corresponds to Drosophila miR-14 has not been found in mammalian genomes. The purpose of the present study was to identify miRNAs, if any, that are differentially expressed during adipogenesis.
We constructed miRNA libraries from pre-adipocytes and cells at days 1 and 9 after the induction of differentiation, and identified 80 miRNAs, including 3 unregistered possible miRNAs. To assess the expression levels of these miRNAs, a total of 102 miRNAs, consisting of the 80 miRNAs identified in the library and an additional 22 mouse miRNAs, were subjected to Northern blotting. Although the expression of 21 miRNAs changed dramatically during differentiation, intriguingly most changes in miRNA expression were observed at day 9, rather than at day 1, 2, or 5 after the induction of differentiation. Similar results have been reported in the TPA-induced differentiation of HL-60 cells (Kasashima et al. 2004) and in the neuronal differentiation of primary rat cortical cells (Kim et al. 2004). It has been shown that the differentiation of pre-adipocytes into adipocytes is regulated by transcription factors such as PPARγ and C/EBPα, which play a crucial role in the early stages of adipocyte differentiation (Morrison and Farmer 1999b). We confirmed by RT-PCR that the expression of PPARγ and C/EBPα is up-regulated during differentiation in #29, but not in #3 (data not shown). The fact that dramatic modulation of miRNA expression was observed at day 9 but not at early phases of differentiation suggests that miRNAs may modulate adipocyte function after differentiation rather than initiate differentiation.
Recently, the down-regulation of miR-181 and up-regulation of miR-15 were reported to be involved in B-cell differentiation (Chen et al. 2004) and B-cell leukemia (Calin et al. 2002), respectively. Furthermore, the expression of both let-7 and miR-34 are temporally regulated during Drosophila metamorphosis (Sempere et al. 2004). Esau et al. (2004) recently demonstrated that miR-143 is involved in human adipocyte differentiation and may act through the target gene ERK5. Up-regulation of miR-143 was also observed in 3T3-L1 cells during adipocyte differentiation in the present study. As with the other up-regulated miRNAs, expression of miR-143 was mostly up-regulated at day 9. Esau et al. (2004) reported that expression of miR-143 was elevated at days 7 and 10 in human adipocytes, but not at days 1 and 4, similar to the present results. Esau et al. (2004) also listed 22 miRNAs differentially expressed in human adipocytes during differentiation. However, the same miRNAs were not identified in the present study, except for miR-143, suggesting that the types of miRNA involved in adipocyte function may differ between human adipocytes and 3T3-L1 cells.
The antisense inhibition of miR-10b, 15, 26a, 34c, 98, 99a, 101, 101b, 143, 152, 183, 185, 224, and let-7b, all of which were up-regulated during adipogenesis, did not affect adipocyte differentiation in terms of marker gene expression and the accumulation of lipid droplets. Moreover, the combined inhibition of several miRNAs also did not affect adipocyte differentiation. However, it is possible that more thorough inhibition might be needed to affect differentiation.
We tried to establish cell lines that overexpressed miR-181a and miR-182, which were down-regulated during 3T3-L1 pre-adipocyte differentiation. Although we can express mature miR-181a and miR-182 by expression vectors under transient conditions, we could not obtain stable cell lines that overexpressed mature miR-181a or mature miR-182. According to the current literature, exportin-5 appears to be rate-limiting for miRNA processing, and the overexpression of a miRNA may have a detrimental influence on cellular functions (Grimm et al. 2006). Thus, we are now unable to assess the significance of miR-181a and miR-182 in 3T3-L1 pre-adipocyte differentiation.
In conclusion, the present results suggest that miRNAs may have various roles in modulating adipocyte function after 3T3-L1 pre-adipocyte differentiation, but do not appear to have active roles in 3T3-L1 pre-adipocyte differentiation itself.
MATERIALS AND METHODS
Cell culture and induction of differentiation of 3T3-L1 pre-adipocytes
3T3-L1 cells were maintained in DMEM supplemented with 10% calf serum and allowed to reach confluency. Cells were differentiated at 2 d post-confluency (designated as day 0) by the addition of DMEM (10% fetal bovine serum) containing 1 μg/mL insulin, 1 μM DEX, and 0.5 mM MIX (Green and Kehinde 1975). After 2 d, cells were grown in media containing only insulin for another 2 d. Subsequently, media was changed every 2 d until the cells were well differentiated (day 9). Total RNA was isolated from cells of days 0, 1, 2, 5, and 9, using TRIzol (Invitrogen) according to the manufacturer's protocol.
Single cell clone
Single cell clone selection was carried out by the limited dilution method in 96-well dishes to obtain subclones of 3T3-L1 with high differentiation capacity. Fifty-two clones were then screened for the ability to differentiate into adipocytes. We selected two cell clones (#3 and #29) that showed different features in adipogenesis. Clone #29 exhibited a high differentiation potential at day 9 (lipid droplets were observed in 100% of the cells) and #3 exhibited no detectable staining in any of the cells. No obvious differences in morphology or growth rate were observed between the two clones before differentiation.
Cloning of microRNA
Cloning of miRNAs was performed essentially as described by (Lagos-Quintana et al. 2002). We used parental 3T3-L1 cells before single cell cloning to construct libraries. Total RNA (500 μg) was separated on a 15% denaturing polyacrylamide gel, and small RNA fractions of between 18 and 25 bases were selected. A 5′-phosphorylated 3′-adaptor oligonucleotide (5′-pUUUaaccgcgaattccagx: uppercase, RNA; lowercase, DNA; p, phosphate; 3′-Amino-Modifier C-7; Integrated DNA Technologies, Inc.) and a 5′-adaptor oligonucleotide (5′-acggaattcctcactAAA: uppercase, RNA; lowercase, DNA) were directionally ligated to the small RNA. The adaptor-ligated RNA was amplified by RT-PCR with the following primers: 3′-primer (5′-CAGCCAACAGGCACCGAATTCCTCACTAAA) and 5′-primer (5′-GACTAGCTTGGTGCCGAATTCGCGGTTAAA). The PCR products were then digested with Ban1 and concatamerized using T4 DNA ligase. Concatamers within a size range of 500–1000 bp were separated on agarose gel and directly ligated by pCR3.1-TOPO vector (Invitrogen). Plasmid inserts were analyzed by PCR using primers for vector sequences and then directly subjected to sequencing. Sequences from cloned miRNAs were grouped by sequence identity and compared with a public database, the miRNA registry (http://www.sanger.ac.uk/Software/Rfam), to identify these clones. Unknown sequences, including sequences that were homologous to particular miRNAs, were used for searches with the Basic Local Alignment Sequence Tool (BLAST) algorithm, available at the National Center for Biotechnology Information (NCBI). Flanking sequences of the novel miRNAs were used to predict secondary structure using the RNAfold program of the Vienna RNA package. All of the new miRNAs were submitted to the miRNA Registry Web site for official annotation.
Northern blot analysis
Fractionated total RNA was transferred to a Zetaprobe membrane (BioRad) and cross-linked to the membrane by UV irradiation. The resulting blots were then probed with several antisense StarFire probes (Integrated DNA Technologies) that had been labeled with [α-32P]dATP. Prehybridization and hybridization were carried out as described by Sempere et al. (2003).
Transient transfection of Anti-miRs
Anti-miR-10b, 15, 26a-1, 34c, 98, 99a, 101, 101b, 143, 152, 183, 185, 224, let-7b, and negative control #1 were purchased from Ambion. These RNA-based inhibitors are chemically modified to increase their stability and to improve their activity and have been confirmed to antagonize corresponding miRNAs by luciferase reporter assay (Cheng et al. 2005). 3T3-L1 cells were cultured, and differentiation was induced under the same conditions as described above. At day 3 and day 5, the cells were transfected separately with each Anti-miR using siPORT Amine reagent (Ambion) according to the manufacturer's protocol. To confirme the efficiency of transfection, Cy3-labeled negative control #1 (Ambion) was also transfected. Total RNA was isolated from cells at days 0, 3, 5, and 7, using TRIzol (Invitrogen) according to the manufacturer's protocol.
RT-PCR of adipocyte differentiation markers
To assess the differentiation of 3T3-L1 cells, the expression patterns of three adipocyte differentiation markers (PPARγ, adipose-type fatty acid binding protein (A-FABP), and adiponectin) were analyzed by competitive RT-PCR analyses using 18s ribosomal RNA as the internal control (QuantumRNA 18s Internal Standards Kit; Ambion). The following amplimers were used for RT-PCR analyses: PPARγ-S (5′-cagcatttctgctccacactatgaag) and PPARγ-A (5′-agcaaggcacttctgaaaccg), AFABP-S (5′-ggatttggtcaccatccggtc) and AFABP-A (5′-cataaactcttgtggaagtcacgcc), and Adipo-S (5′-agggaacttgtgcaggttggat) and Adipo-A (5′-tgagcgatacacataagcggct).
ACKNOWLEDGMENTS
We thank Ayako Nagahashi, Miho Miyazaki, Megumi Ikeuchi, and Naoko Kubo for their technical assistance. This study was supported by the Program for the Promotion of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation, and the Uehara Memorial Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7228806.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cheng A.M., Byrom M.W., Shelton J., Ford L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C., Kang X., Peralta E., Hanson E., Marcusson E.G., Ravichandran L.V., Sun Y., Koo S., Perera R.J., Jain R., et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Green H., Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kasashima K., Nakamura Y., Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- Kim J., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus S. Adipose tissue as a regulator of energy balance. Curr. Drug Targets. 2004;5:241–250. doi: 10.2174/1389450043490523. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu C.G., Calin G.A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C.D., Shimizu M., Zupo S., Dono M., et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa Y., Funahashi T., Kihara S., Shimomura I. Adiponectin and metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 2004;24:29–33. doi: 10.1161/01.ATV.0000099786.99623.EF. [DOI] [PubMed] [Google Scholar]
- Morrison R.F., Farmer S.R. Insights into the transcriptional control of adipocyte differentiation. J. Cell. Biochem. (Suppl.) 1999a;75:59–67. doi: 10.1002/(sici)1097-4644(1999)75:32+<59::aid-jcb8>3.3.co;2-t. [DOI] [PubMed] [Google Scholar]
- Morrison R.F., Farmer S.R. Role of PPARγ in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 1999b;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Sokol N.S., Dubrovsky E.B., Berger E.M., Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolar M.W. Insulin resistance, diabetes, and the adipocyte. Am. J. Health Syst. Pharm. 2002;59:S3–S8. doi: 10.1093/ajhp/59.suppl_9.S3. [DOI] [PubMed] [Google Scholar]
- Xu P., Vernooy S.Y., Guo M., Hay B.A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]