Abstract
The food-borne pathogen, Escherichia coli O157:H7, has been associated with gastrointestinal disease and the life-threatening sequela hemolytic uremic syndrome. The genes for the virulence factor, Shiga toxin 2 (Stx2), in E. coli O157:H7 are encoded on a temperate bacteriophage under the regulation of the late gene promoter. Induction of the phage lytic cycle is required for toxin synthesis and release. We investigated the hypothesis that nonpathogenic E. coli could amplify Stx2 production if infected with the toxin-encoding phage. Toxin-encoding phage were incubated with E. coli that were either susceptible or resistant to the phage. The addition of phage to phage-susceptible bacteria resulted in up to 40-fold more toxin than a pure culture of lysogens, whereas the addition of phage to phage-resistant bacteria resulted in significantly reduced levels of toxin. Intestinal E. coli isolates incubated with Shiga toxin-encoding phage produced variable amounts of toxin. Of 37 isolates, 3 produced significantly more toxin than was present in the inoculum, and 1 fecal isolate appeared to inactivate the toxin. Toxin production in the intestine was assessed in a murine model. Fecal toxin recovery was significantly reduced when phage-resistant E. coli was present. These results suggest that the susceptibility of the intestinal flora to the Shiga toxin phage could exert either a protective or an antagonistic influence on the severity of disease by pathogens with phage-encoded Shiga toxin. Toxin production by intestinal flora may represent a novel strategy of pathogenesis.
The food-borne pathogen Escherichia coli serotype O157:H7 causes an estimated 73,000 cases of disease per year in the United States (14). It is the causative agent of hemorrhagic colitis (25) and hemolytic-uremic syndrome (HUS) (9), a potentially fatal disease that affects mostly children under 10 years of age. The development of HUS is associated with Shiga toxin 2 (Stx2) production by E. coli O157:H7. Antibiotic treatment has been shown to increase the production of Stx2 (10, 11, 41) and has been associated with progression to HUS in some studies (39) but not in others (26).
Stx2 is a member of the AB5 family of bacterial toxins, which also includes pertussis toxin, cholera toxin, and the E. coli heat-labile enterotoxin. The five B-subunits form a ring crowned by the A-subunit (6, 30). The B-pentamer promotes internalization of the A-subunit into the host cell cytoplasm. The A-subunit of Stx2 cleaves out a single adenine residue from the host cell rRNA, halting protein synthesis and leading to cell death.
AB5 toxins have only been found in gram-negative bacteria, and conditions unique to the bacterial periplasm appear to be required for the toxin subunits to efficiently fold and assemble (31, 40). Once assembled, the outer bacterial membrane has proven to be a barrier to toxin secretion. Bordetella pertussis (38) and Vibrio cholerae (27) have dedicated secretion systems to promote release of toxin from viable bacteria. No dedicated secretion system has been described for Shiga toxin, and secretion appears to be achieved by bacterial lysis. Shiga toxin genes have been found in association with the late gene region of lambdoid phage (8, 18, 23, 28). The late genes are silent during lysogeny but are highly expressed when the phage enter the lytic cycle, resulting in phage production and bacterial lysis. Recent reports have demonstrated that expression of phage-encoded Shiga toxin is dependent on late gene transcription (16, 19). Toxin is not produced during latency but, after phage induction, the bacteria produce both viral particles and toxin. Toxin production is greatly diminished in phage defective for late gene expression (36). Although the coupling of toxin production to events that lead to bacterial lysis abolishes the need for a dedicated toxin secretion system, linking toxin secretion to cell death seems paradoxical unless this benefits the bacteria in another way.
The Shiga toxin phage are efficient vectors for lateral transfer of the toxin genes, a process that is thought to play an important role in the evolution of new pathogens (15, 21, 37). Acheson et al. (1) have demonstrated in vivo transduction of Shiga-toxin encoding phage to E. coli in the murine intestine. However, lysogeny is a rare event, and infection of susceptible bacteria often results in a lytic infection, ending with lysis of the bacterium, release of new phage particles and, in this case, production of Stx2. Stx2 production by intestinal E. coli could confer an advantage to having toxin production linked to phage production. It would eliminate the need for an energy-intensive toxin secretion mechanism and, furthermore, normal intestinal E. coli infected with the toxin-encoding phage would produce both phage and toxin, resulting in toxin levels exceeding what could be produced by the pathogenic strain by itself. We tested this hypothesis both in vitro and in the murine intestine and report here that Stx2 production is greatly increased in the presence of susceptible E. coli. In contrast, in the presence of resistant E. coli, toxin production is limited to that produced by the infecting E. coli. Human intestinal E. coli isolates were found to vary in susceptibility to the Shiga toxin phage. The presence of phage-susceptible intestinal E. coli could be a risk factor for development of severe disease after infection by E. coli O157:H7.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains and plasmids used in the present study are described in Table 1. E. coli strains C600 and C600::933W, which is C600 lysogenized with the 933W bacteriophage that encodes for Stx2, were obtained from A. D. O'Brien (20). The clinical E. coli O157:H7 strain, PT-32, was obtained from the culture collection of Cincinnati Children's Hospital Medical Center microbiology laboratory. Human fecal E. coli were isolated on MacConkey Agar (Difco, Detroit, Mich.) and confirmed by using BBL Enterotube II identification tubes (Becton Dickinson Microbiology Systems, Sparks, Md.). Informed consent and assent was obtained from all subjects and their parents or guardians. The present study was approved by the Institutional Review Board of the University of Cincinnati and conducted according to the experimental guidelines of the U.S. Department of Health and Human Services.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequencea | Reference or source |
|---|---|---|
| E. coli strains | ||
| EC One Shot | High-efficiency transformation | Invitrogen |
| DH5α | High-efficiency transformation; Nalr | Invitrogen |
| C600 | E. coli K-12 | 20 |
| C600::933W | C600 lysogenized with Stx2-converting phage 933W | 20 |
| C600::Δtox | C600 lysogenized with phage Δtox; Cmr; GFP | This study |
| C600-S | Spontaneous Strr C600 | This study |
| C600K | C600-S(pBBR1MCS-2); Strr Kanr | This study |
| C600A::933W | C600-S(pBBR1MCS-4) lysogenized with 933W; Strr Ampr | This study |
| C600G::Δtox | C600-S(pBBR1MCS-5) lysogenized with Δtox; Cmr Strr Genr | This study |
| PT-32 | E. coli O157:H7; stx1, stx2; isolated from a patient | This study |
| FI-4 | Fecal E. coli isolated from a healthy person | This study |
| FI-15 | Fecal E. coli isolated from a healthy person | This study |
| FI-31 | Fecal E. coli isolated from a healthy person | This study |
| FI-29 | Fecal E. coli isolated from a healthy person | This study |
| FI-37 | Fecal E. coli isolated from a healthy person | This study |
| Plasmids | ||
| pBluescript KS(+) | Cloning vector; Ampr | Stratagene |
| pCR2.1 | TA cloning vector; Ampr Kanr | Invitrogen |
| pGFPuv | GFPuv; Ampr | BD Biosciences Clontech |
| pST76-C | Cloning vector; Cmr | 24 |
| pPIR-K | Temperature sensitive suicide vector; Kanr | 24 |
| pSG039 | Upstream and downstream regions of stx2A, GFP, Cmr; in vector pPIR-K | This study |
| pBBR1MCS-2 | Cloning vector; Kanr | 12 |
| pBBR1MCS-4 | Cloning vector; Ampr | 12 |
| pBBR1MCS-5 | Cloning vector; Genr | 12 |
| Primers | ||
| STX-UP (forward) | 5′-GCGGCCGCACGTGTTGGGGGAGACGTTC (NotI) | This study |
| STX-UP (reverse) | 5′-GAATTCAAGCGAGCCTCCTAAATAAATATGG (EcoRI) | This study |
| STX-DOWN (forward) | 5′-ATCGATAATTCAGTCAGTTGACACTTGCCTG (ClaI) | This study |
| STX-DOWN (reverse) | 5′-GGGCCCGCGGCCGCACGGAACCAGCAGAATGCC (NotI, ApaI) | This study |
| GFP (forward) | 5′-GAATTCAAGCGAGCCTGGTAAATAAATATGG (EcoRI) | This study |
| GFP (reverse) | 5′-ATCGATTTATTTGTAGAGCTCATCCATGCC (ClaI) | This study |
| Cmr (forward) | 5′-ATCGATCAGCATCACCCGACGCACTTTG (ClaI) | This study |
| Cmr (reverse) | 5′-ATCGATCAGTGAGGCACCAATAACTGCC (ClaI) | This study |
| Verification of Δtox (forward) | 5′-CGAAATCGAATCGAACCATAACCG (upstream of stx2A) | This study |
| Verification of Δtox (reverse) | 5′-CCGAAGAAAAACCCAGTAACAGG (in stx2A) | This study |
Nalr, naladixic acid resistance; Ampr, ampicillin resistance; Genr, gentamicin resistance. Restriction sites introduced for cloning are underlined in the primer sequence and identified in parentheses.
E. coli strains were propagated at 37°C in Luria-Bertani (LB) broth or LB agar (Difco) or in modified LB containing 10 mM CaCl2 (LB modified). Where indicated, antibiotics were added to the media at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 15 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; and streptomycin, 30 μg/ml.
Construction of Δtox.
The genes for Stx2 in 933W were replaced by allelic exchange with sequences for green fluorescent protein (GFP) expression and chloramphenicol resistance (Cmr), as shown in Fig. 1, by using sequences upstream of the stx2 genes (STX2-UP, bp 20467 to 21393) and downstream of the stx2 genes (STX2-DOWN, bp 22752 to 23744) in the 933W genome (23). Primers used for the PCR are listed in Table 1. Restriction sites were added to the primers to facilitate cloning into pBluescript SK(+) (Stratagene, La Jolla, Calif.). The construct was cut with NotI, inserted into the temperature-sensitive, kanamycin-resistant (Kanr) suicide vector pPIR-K (24), and transformed into C600::933W at room temperature. Cmr and Kanr transformants were selected and grown at 37°C, with shaking, in LB broth to counterselect against the suicide vector. Phage were prepared by induction with ciprofloxacin (see below), and used to infect C600. A Cmr, kanamycin-sensitive colony was selected. Loss of stx2 was verified by lack of a PCR product by using verification primers (Table 1). The absence of Stx2 production by C600::Δtox was confirmed by enzyme-linked immunosorbent assay (ELISA; Premier EHEC kit, Meridian Bioscience, Inc., Cincinnati, Ohio).
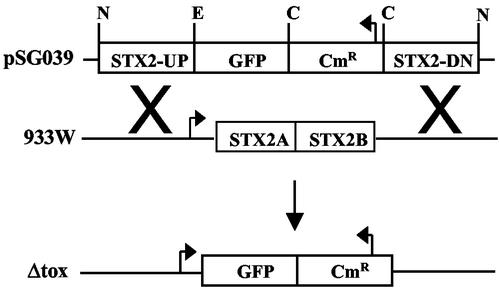
FIG. 1.
Construction of Δtox. The Stx2 genes in phage 933W were replaced with GFP and Cmr genes. Expression of GFP is under the control of the phage late gene promoter, and Cmr expression is under the control of its own promoter. N, NotI; E, EcoRI; C, ClaI.
Western analysis of GFP production.
Supernatants from C600::Δtox cultures were filter sterilized, mixed with an equal volume of sample buffer, boiled for 10 min, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 12% Tris-glycine precast gel (BioWhittaker, Rockland, Maine), and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) as previously described (2). The primary anti-GFP antibody (BD Biosciences Clontech, Palo Alto, Calif.) was used at a 1:100 dilution, followed by goat anti-rabbit secondary antibody (Cappel, West Chester, Pa.) used at a dilution of 1:37,500. Bands were visualized by using the Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, Mass.). GFP protein produced from E. coli DH5α (Invitrogen Life Technologies, Gaithersburg, Md.) containing pGFPuv (Clontech) was used as a positive control.
Phage induction.
Phage production was induced with ciprofloxacin (Serologicals Proteins, Inc., Kankakee, Ill.) (41). Lysogenic strains were grown in LB broth overnight at 37°C, shaking, diluted to an optical density at 600 nm (OD600) of 0.08 in LB-modified broth, and ciprofloxacin was added to a final concentration of 30 ng/ml. The cultures were incubated at 37°C on a roller wheel for approximately 10 h. Phage-mediated bacterial lysis was monitored by a decrease in OD600. For phage preparations, the culture was centrifuged (5,000 × g), and the supernatant was filter sterilized (0.22-μm pore size) and used immediately or stored at 4°C overnight. Phage were prepared fresh for each infection experiment, and phage titers were determined on the day of the infection.
Phage titers were determined by spotting 5 μl of dilutions of the phage preparations onto LB-modified agar overlaid with LB-modified soft agar containing the indicator strain C600. The plates were incubated at 37°C overnight, and the number of PFU per milliliter was determined.
Phage infection and coincubation experiments.
C600 and C600::Δtox were grown overnight at 37°C, with shaking, in LB broth, and adjusted to an OD600 of 1; 7-ml aliquots were then distributed into test tubes. Dilutions (0 to 105 PFU/ml) of 933W phage were added to the cultures, overlaid onto LB-modified agar in a deep petri dish, and incubated at 37°C as a static culture. After overnight incubation, the liquid cultures were collected from the petri dishes and centrifuged (5,000 × g), and the supernatants were filter sterilized. Phage titers and Stx2 production were determined. For coincubation experiments, overnight cultures of C600::933W lysogens were adjusted to an OD600 of 1 (approximately 109 CFU/ml), dilutions were added to C600 or C600::Δtox and incubated as described above.
These experiments were repeated with E. coli O157:H7 strain PT-32 as the source of Shiga toxin-encoding phage. PT-32 was induced with 30 ng of ciprofloxacin/ml to produce phage as described above. The induced culture had approximately 106 PFU/ml, and the filter-sterilized supernatant was added to 109 CFU of C600 or C600::Δtox/ml to a final dilution of 100 phage. After overnight incubation at 37°C as a static culture, supernatants were filter sterilized and characterized for phage and toxin production.
Screening of intestinal E. coli isolates.
E. coli isolated from healthy volunteers was grown overnight in LB broth and adjusted to an OD600 of 1. Approximately 100 phage from an induced culture of E. coli O157:H7 PT-32 were added to each strain of E. coli, followed by incubation at 37°C as a static culture. After overnight incubation, the supernatants were collected, filter sterilized, and characterized for toxin production.
Determination of toxin concentrations.
ELISA (Premier EHEC kit; Meridian Bioscience, Inc.) was used to determine the concentration of Stx2 for coincubation and phage infection experiments with C600::933W as the source of Shiga toxin-encoding phage. Dilutions of the samples were tested for Stx2 as described by the manufacturer, and results were compared to a standard curve by using purified Stx2 obtained from D. Acheson.
The Vero cell assay was used to measure Stx2 concentration for phage infection and coincubation experiments with PT-32 as the source of Shiga toxin-encoding phage. Briefly, twofold dilutions of filter-sterilized supernatants were incubated with 4 × 104 Vero cells/ml at 37°C in 5% CO2 for 3 days. Stx2 values were determined by comparison to Stx2 standards by using purified toxin from Toxin Technology, Inc. (Sarasota, Fla.).
Mouse challenge studies.
A spontaneous streptomycin-resistant (Strr) mutant of C600, C600-S, was obtained after selection on LB agar containing 30 μg of streptomycin/ml. To distinguish the strains, C600-S and its lysogenic derivatives were each transformed with a pBBR1MCS plasmid (12) encoding a different antibiotic resistance marker (Table 1).
All mouse experiments were conducted in a facility approved by the American Association for the Accreditation of Laboratory Animal Care. Six-week-old CD-1 male mice were obtained from Charles River Laboratories (Wilmington, Mass.). Each mouse was housed separately. Streptomycin (2 g/liter) was added to the drinking water 2 days prior to inoculation, and this treatement was maintained for the duration of the study. Fewer than 100 CFU were recovered on MacConkey agar from the feces of the mice before challenge, suggesting that the intestinal E. coli had been essentially eliminated. Food was removed the day before inoculation and returned postinoculation. Each bacterial strain was suspended in a 20% sucrose solution and delivered by intragastric inoculation. Singly infected mice received 0.1 ml of bacteria for a final dose of 109 C600K, 109 C600G::Δtox, or 107 C600A::933W. Doubly infected mice received either 0.05 ml each of 109 C600K and 107 C600A::933W or 109 C600G::Δtox and 107 C600A::933W. For mice receiving two strains, each strain was given separately. After 24 h, all of the feces were collected, weighed, and analyzed for bacterial and toxin recovery. Fecal bacterial recovery was determined by plating serial dilutions on McConkey agar containing streptomycin and a differentiating antibiotic.
RESULTS
Generation of Δtox, phage-resistant E. coli.
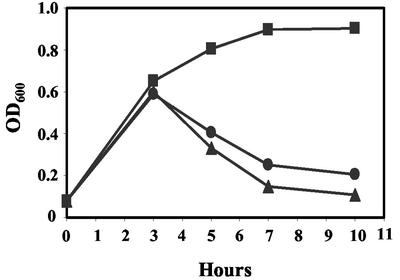
To construct Δtox, the Stx2 genes of 933W were replaced with GFP driven by the phage late gene promoter, and Cmr driven by its own promoter (Fig. 1). Spontaneous phage production by both C600::933W and C600::Δtox was approximately 104 PFU/ml. Ciprofloxacin has been shown to induce lysogenic phage to enter the lytic cycle (41). Treatment with ciprofloxacin resulted in induction and lysis of C600::933W and C600::Δtox but not the C600 control (Fig. 2); phage titers from either lysogen were similar, approximately 106 to 107 PFU/ml after induction. Induced cultures of C600::933W produced 23,700 ng of Stx2/ml and, as expected, no toxin was produced by C600::Δtox.
FIG. 2.
Phage-induced lysis after treatment with ciprofloxacin. Cultures of C600 and C600 lysogenized with either wild-type 933W or mutant Δtox were treated with 30 ng of ciprofloxacin/ml for 10 h. The OD600 was measured as an indication of bacterial lysis due to phage production. Symbols: ▪, C600; •, C600::933W; ▴, C600::Δtox.
933W and Δtox phage were unable to plaque on either lysogen, suggesting the lysogens were resistant to superinfection. In addition, GFP production by C600::Δtox was examined by Western blots. GFP was detected in pGFPuv (positive control, Fig. 3, lane 1) and when C600::Δtox was induced with ciprofloxacin (Fig. 3, lane 2). However, GFP production was below the limit of detection in uninduced C600::Δtox (Fig. 3, lane 4) or when C600::Δtox was superinfected with C600::933W (Fig. 3, lanes 6 to 10). These results suggest that, like Stx2 expression in 933W, GFP expression in Δtox is under the control of late gene expression and that Δtox confers resistance to 933W.
FIG. 3.
Western analysis of GFP production by C600::Δtox. GFP in culture supernatants was examined. Lane 1, GFP protein from pGFPuv; lane 2, C600::Δtox induced with ciprofloxacin; lane 3, C600::933W induced with ciprofloxacin; lane 4, uninduced C600::Δtox. Lanes 5 to 11: C600::Δtox incubated alone or with 933W phage as indicated: no phage added (lane 5), 105 phage added (lane 6), 104 phage added (lane 7), 103 phage added (lane 8), 102 phage added (lane 9), 101 phage added (lane 10), or 100 phage added (lane 11).
Phage and toxin production are amplified in the presence of susceptible E. coli.
The phage-susceptible E. coli strain, C600, and the phage-resistant strain, C600::Δtox, were incubated with various doses of 933W phage (Fig. 4). Infection of C600 resulted in lytic infection, as evidenced by amplified phage recovery at all doses (Fig. 4A). In contrast, the amount of phage recovered from the C600::Δtox cultures infected with 933W phage did not differ significantly from the approximately 104 PFU/ml produced spontaneously by lysogens.
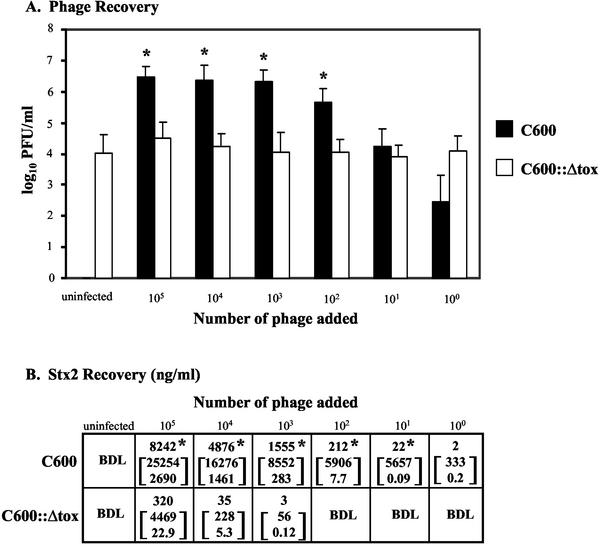
FIG. 4.
Phage (A) and toxin (B) recovery after infection with 933W phage. The results are the average of four trials. (A) Phage recovery (log10 PFU/milliliter + the SEM). An asterisk denotes statistically different values (P < 0.05 [Student t test]) of log10 phage counts from C600 compared to log10 counts from C600::Δtox for each challenge dose. (B) Toxin recovery. The top number is the geometric mean Stx2; the number in brackets represents the 95% confidence level of the mean. An asterisk indicates statistical significance (P < 0.05 [Student t test of log toxin values]) of Stx2 from C600 compared to C600::Δtox for each challenge dose. BDL, below detection limit of 0.15 ng/ml.
Stx2 recovery from C600 and C600::Δtox after incubation with 933W phage was also determined (Fig. 4B). Toxin production from C600 was significantly greater than toxin production from C600::Δtox at all challenge doses greater than 10 933W phage (Fig. 4B). At high doses of 933W phage, toxin production in the presence of C600 was 20 to 40 times greater than the amount of toxin recovered from an overnight culture of 933W lysogens, typically 35 to 700 ng/ml. In contrast, all of the toxin detected in C600::Δtox cultures superinfected with 933W phage could be attributed to the toxin present in the culture supernatant used as a source of phage, suggesting no new toxin synthesis had occurred. These studies suggest that susceptible bacteria infected with the toxin-encoding phage undergo lytic infection and amplify phage and toxin production, whereas phage-resistant bacteria do not undergo lytic infection.
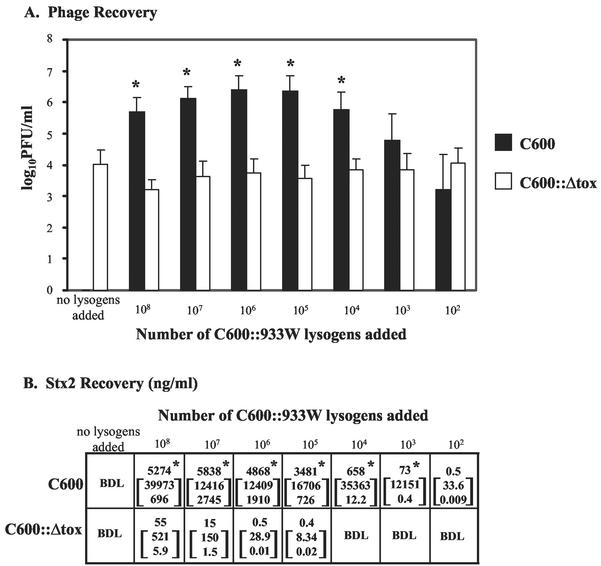
In diarrheal disease, a few toxin-producing pathogens will encounter large numbers of intestinal E. coli. To mimic these events, dilutions of the lysogen C600::933W were coincubated with approximately 109 susceptible E. coli (C600) or 109 resistant E. coli (lysogenic C600::Δtox). Phage and toxin production after coincubation with lysogens (Fig. 5) was similar to that observed with cells directly infected with phage. Coincubation of C600 with C600::933W resulted in phage production at all doses, which was significantly higher (P < 0.05) than that observed for C600::Δtox when ≥104 C600::933W were added (Fig. 5A). Similarly, more Stx2 was recovered after coincubation of C600::933W with C600 than with C600::Δtox, and these values were statistically significant (P < 0.05), except at the lowest coinfection dose (Fig. 5B).
FIG. 5.
Phage (A) and toxin (B) recovery after coincubation with C600::933W. The results are the average of four trials. (A) Phage recovery (log10 PFU/milliliter + the SEM). An asterisk denotes statistically different values (P < 0.05 [Student t test]) from C600 compared to C600::Δtox for each challenge dose. (B) Toxin recovery. The top number is the geometric mean Stx2; the number in brackets represents the 95% confidence levels of the mean. An asterisk indicates statistical significance (P < 0.05 [Student t test of log toxin values]) of Stx2 from C600 compared to C600::Δtox for each challenge dose. BDL, below detection limit of 0.15 ng/ml.
Toxin production by E. coli O157:H7 phage is amplified in the presence of susceptible E. coli.
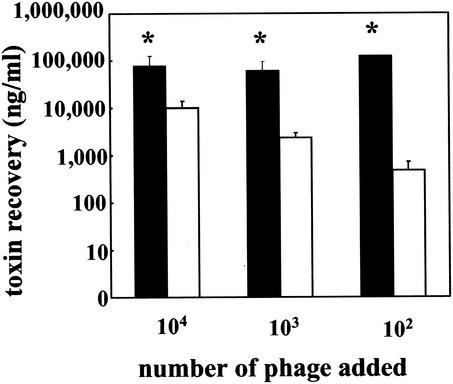
The coincubation studies were repeated with a human isolate of E. coli O157:H7, PT-32. Addition of phage from PT-32 to C600 resulted in significantly more (P < 0.05) toxin production than the phage-resistant C600::Δtox at all doses tested (Fig. 6), and the toxin recovered from the C600::Δtox cultures was largely due to the toxin present in the culture supernatant used as a source of phage. In similar experiments, coincubation of 109 CFU of C600/ml with 105 PT-32 lysogens resulted in significantly more (P < 0.05) toxin production than coincubation of 109 CFU of C600::Δtox/ml with 105 PT-32 (data not shown).
FIG. 6.
Toxin recovery after incubation with phage from E. coli O157:H7. A total of 109 CFU of either C600 (▪) or C600::Δtox (□)/ml was incubated overnight with various dilutions of Shiga toxin-encoding phage from E. coli O157:H7 strain PT-32. The toxin concentration in the supernatant (nanograms/milliliter + the SEM) was determined by Vero cell assay. An asterisk indicates statistically different values (P < 0.05 [Student t test]) from C600 compared to C600::Δtox for each challenge dose. The results are the average of three trials.
Stx2 production in the intestine.
To determine whether phage susceptibility influences toxin production in the intestinal environment, we used an established murine model (17, 34). In this model, facultative intestinal bacteria are eliminated by treatment with streptomycin, allowing the intestinal tract to be repopulated with Strr derivatives of the strains of interest. A spontaneous Strr derivative of C600, C600-S, was lysogenized with 933W or Δtox, and each strain was transformed with a plasmid containing a different antibiotic resistance marker. Since intestinal flora are present in greater numbers than invading pathogenic bacteria, non-toxin-producing strains (C600K and C600G::Δtox) were introduced at a dose of 109, whereas toxin-producing strain C600A::933W was introduced at a dose of 107. Five groups of mice were used. Three groups received only a single strain. The last two groups of mice received two strains: either 109 C600K or 109 C600G::Δtox to mimic normal flora and 107 toxin-producing C600A::933W to mimic infection. To avoid phage infection in vitro, the two strains were given in separate doses about 10 min apart. Feces were recovered 24 h later and used to evaluate bacterial colonization and Stx2 production.
The intestine is a dynamic environment, and both bacteria and phage are susceptible to intestinal immune defenses. Viable bacteria were recovered from all of the mice, but the numbers varied: (i) 106 to 1010 for the strains given at a dose of 109 and (ii) 104 to 109 for C600A::933W given at a dose of 107. These results indicate that bacterial replication had occurred in some mice, whereas in others the bacterial numbers were reduced. Mean bacterial recovery (in log10 CFU/ml ± the standard error of the mean [SEM]) for toxin-producing C600A::933W was 7.4 ± 0.3 when added alone, 6.9 ± 0.2 when added with C600K, and 7.2 ± 0.2 when added with C600G::Δtox; however, none of these values were statistically different.
Stx2 does not cause intestinal damage in this mouse model of infection (34). This is also true for cattle, which asymptomatically carry E. coli O157:H7 in the intestine (13). Although we were not able to evaluate toxin damage to infected mice, the amount of Stx2 was determined by ELISA. Like bacterial recovery, Stx2 production was also highly variable in the mice. Toxin recovery was below the limit of detection (5 ng/ml) for many of the mice (Table 2). Toxin was detected in the feces of 28% of the mice receiving only C600A::933W, in 19% of the mice receiving both C600A::933W and C600K, and in only 3% of the mice receiving both C600A::933W and C600G::Δtox. The highest amount recovered from a singly infected mouse was 100 ng/ml, but in the presence of susceptible E. coli, 10 to 20 times more toxin was recovered from two of the mice. Statistical analysis by using the Student t test of log toxin values was performed on the pooled data (toxin-positive and -negative samples) for each group. The amount of toxin recovered from the group challenged with the phage-resistant strain C600G::Δtox and C600A::933W was significantly less than the amount of toxin recovered from the mice receiving only C600A::933W, as well as from mice receiving both C600A::933W and C600K (P < 0.05). These results suggest that the presence of phage-resistant E. coli was protective, since fecal toxin levels were significantly lower for mice challenged with the toxin-producing lysogen when phage-resistant E. coli were present. This protective effect was not due to competition between the bacterial strains resulting in reduced numbers of toxin-producing lysogens, since the mean bacterial recovery was very similar in all cases.
TABLE 2.
Toxin recovery from mouse feces after bacterial challenge
| Bacterial challengea | No. of toxin-positive miceb/total mice (positive values [ng/ml]) |
|---|---|
| Single-infection groups | |
| 109 C600K | 0/12 |
| 109 C600G::Δtox | 0/12 |
| 107 C600A::933W | 5/18 (106, 42, 39, 37, 13) |
| Coinfection groups | |
| 109 C600K + 107 C600A::933W | 7/37 (2140, 1350, 100, 70, 16, 15, 13)c |
| 109 C600G::Δtox + 107 C600A::933W | 1/37 (17)d |
Mice were inoculated intragastrically with one or two strains. After 24 h, all of the feces from each mouse were collected and assayed for bacterial and toxin recovery.
The limit of detection for fecal Stx2 was 5 ng/ml.
Statistically different toxin recovery (P < 0.05 [Student t test of log toxin values]) compared to toxin from the doubly infected C600G::Δtox and C600A::933W group, determined by using the values for all of the mice in each group.
Statistically different toxin recovery (P < 0.05, Student t test of log toxin values) compared to toxin from the C600A::933W group, determined by using the values for all of the mice in each group.
Previous studies have shown phage transduction can occur in vivo (1). We examined C600K and C600G::Δtox isolates recovered from the mice coinfected with C600A::933W for the presence of stx2 genes by PCR. Stx2 DNA was detected in C600K isolates from 4 of the 37 mice that received both C600K and C600A::933W, suggesting lysogeny had occurred. However, no Stx2 DNA was detected in the C600G::Δtox isolates that were coinoculated with C600A::933W.
Characterization of human intestinal E. coli.
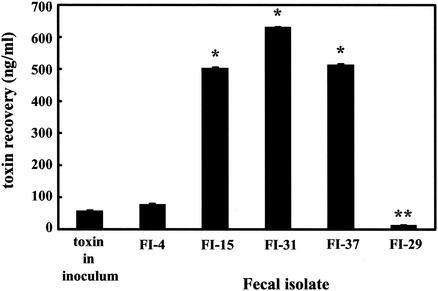
We examined the ability of normal intestinal E. coli to amplify Stx2. E. coli isolated from 37 healthy individuals were screened for toxin amplification after coincubation with 100 phage from PT-32. Thirty-three of the human isolates, represented by FI-4 (Fig. 7), were resistant to toxin amplification, and toxin recovery after infection was similar to the amount of toxin added with the phage inoculum. Three isolates—FI-15, FI-31, and FI-37—produced significantly (P < 0.01) more Stx2 after incubation with phage (Fig. 7). For one isolate, FI-29, the toxin recovery after coincubation was significantly (P < 0.01) reduced compared to the amount of toxin in the inoculum (Fig. 7).
FIG. 7.
Toxin recovery after incubation of human intestinal E. coli with E. coli O157:H7. A total of 109 CFU of fecal E. coli/ml was incubated overnight with 100 phage from E. coli O157:H7 PT-32. The values represent toxin recovery (nanograms/milliliter) + the SEM. An asterisk denotes significantly (P < 0.01) greater toxin production compared to toxin present in the initial phage inoculum. A double asterisk denotes significantly (P < 0.01) less toxin production compared to toxin in the initial phage inoculum. The results are the average of three trials.
DISCUSSION
Bacteriophage have long been recognized as vectors for the horizontal transfer of virulence factors (21). Transduction of Shiga toxin-encoding phage to other E. coli strains has been demonstrated in vitro (29) and in vivo in the murine intestine (1), and recently transduction to Shigella sonnei has been reported (32). However, lysogeny is a rare event, and infection of susceptible E. coli usually results in a lytic infection. Furthermore, unlike other phage-encoded virulence factors, such as diphtheria toxin, which is regulated by bacterial chromosomal genes (5, 33), phage-encoded Shiga toxin expression is under the regulatory control of the phage late genes (16, 19) and is produced during the phage lytic cycle. Factors that increase phage production, such as antibiotic treatment (10, 11, 41) or peroxide released from activated neutrophils (35), can induce Stx2 production.
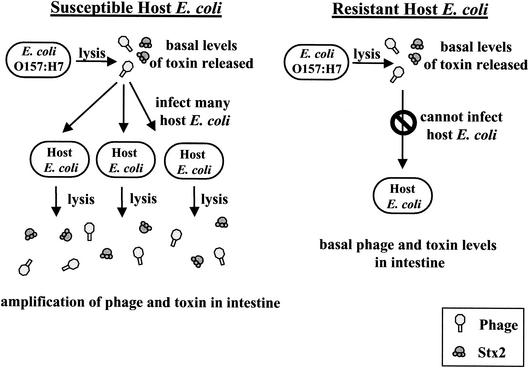
Unlike other AB5 toxins, which use energy-dependent secretion systems to promote toxin secretion from viable bacteria, phage-encoded Stx2 represents a novel strategy for secretion, coupling toxin production to bacterial lysis. As shown in Fig. 8, our results suggest that this strategy may confer an additional advantage by allowing the pathogenic bacteria to pass the burden of toxin production onto the harmless intestinal flora. To our knowledge, this represents the first evidence that nontoxinogenic E. coli can influence the amount of Stx2 produced by Stx2-encoding E. coli.
FIG. 8.
Model of the effect of normal intestinal E. coli on the production of Stx2 by E. coli O157:H7. During an E. coli O157:H7 infection, lysis of the pathogen occurs in the intestine, releasing a basal level of Stx2-encoding phage and toxin. If the normal E. coli in the intestine is susceptible to infection by the Shiga toxin-encoding phage, repeated cycles of infection and lysis will occur, resulting in an amplification of toxin production. If the normal intestinal E. coli are resistant to infection by the Shiga toxin-encoding phage, toxin production will be limited to the E. coli O157:H7, and only basal levels of toxin will be produced.
The murine model for E. coli O157:H7 infection is often used to examine pathogenic mechanisms in an environment that includes intestinal and mucosal factors. We used this model to substantiate our in vitro data that indicate that the presence of susceptible bacteria amplifies toxin when incubation occurs with the toxin-producing strain. A statistically significant increase in toxin production was observed in the presence of phage-susceptible intestinal flora compared to when the intestinal flora were phage resistant. A trend toward increased toxin production in the presence of phage-susceptible intestinal flora compared to the absence of intestinal flora was also observed, with two mice producing 10 and 20 times more toxin than was observed in the absence of susceptible flora, but statistical significance was not achieved. Toxin production was highly variable, and fewer mice were used in this group, and thus statistical significance may be achieved if more mice were studied. However, this experimental condition, a mouse lacking intestinal flora, is not a good model for human disease. All healthy humans possess intestinal flora, and a role for toxin amplification by susceptible intestinal flora compared to resistant flora was clearly demonstrated in these studies.
Stx2 is believed to play a role in the development of the hemorrhagic lesions (4, 7, 13) and HUS (13) in human disease. The course of disease in humans infected with Stx2-producing E. coli O157:H7 can be highly variable, with some individuals displaying few or no symptoms, whereas others succumb to lethal infection (13, 22). The variability observed in human disease could be related to variability in the amount of Stx2 produced in the gut. Antibiotic treatment can induce the lytic cycle, can increase the production of Stx2 (41), and may promote the development of HUS (39). We have shown that the presence of phage-susceptible flora can also influence the amount of Stx2 produced and could influence the severity of disease.
Characterization of a small sample of intestinal E. coli suggests that normal human flora are often resistant to the phage and subsequent toxin amplification. However, about 10% of the isolates were able to amplify toxin production. Severe disease is rare in patients with an E. coli O157:H7 infection (3, 13), with about 10% of patients with bloody diarrhea going on to develop HUS (13). This number agrees remarkably well with our observation that 10% of the intestinal E. coli are susceptible to the phage. It remains to be determined if intestinal bacteria play a role in the disease progression for the few patients who develop severe disease. Interestingly, one intestinal isolate was found to reduce the amount of Stx2 below the original amount present in the phage inoculum, suggesting that this strain can inactivate the toxin. Studies are ongoing to characterize the nature of this activity. Altering the susceptibility of the intestinal flora to the phage could represent a new approach to controlling the severity of disease caused by Shiga toxin-producing bacteria such as enterohemorrhagic E. coli O157:H7.
Acknowledgments
We thank David Acheson for purified Stx2, Joel Mortensen and the Cincinnati Children's Hospital Medical Center culture collection for E. coli O157:H7 strain PT-32, Linda Levin and Paul Succop for assistance with the statistical analysis, Angela Patton and Scott Millen for assistance with the mouse model studies, and Phillip Tarr and Ralph Giannella for insightful discussions.
This work was supported by grant RO1 AI23695 and grant R21 AI54762 to A.A.W. and by a University Research Council Summer Fellowship to S.D.G.
Editor: A. D. O'Brien
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, M. G., and A. A. Weiss. 2001. BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition. Infect. Immun. 69:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser, R. E., P. M. Griffin, and L. Slutsker. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355-367. [DOI] [PubMed] [Google Scholar]
- 4.Dykstra, S. A., R. A. Moxley, B. H. Janke, E. A. Nelson, and D. H. Francis. 1993. Clinical signs and lesions in gnotobiotic pigs inoculated with Shiga-like toxin I from Escherichia coli. Vet. Pathol. 30:410-417. [DOI] [PubMed] [Google Scholar]
- 5.Fourel, G., A. Phalipon, and M. Kaczorek. 1989. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect. Immun. 57:3221-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser, M. E., M. M. Chernaia, Y. V. Kozlov, and M. N. James. 1994. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 Å resolution. Nat. Struct. Biol. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 7.Griffin, P. M., L. C. Olmstead, and R. E. Petras. 1990. Escherichia coli O157:H7-associated colitis: a clinical and histological study of 11 cases. Gastroenterology 99:142-149. [DOI] [PubMed] [Google Scholar]
- 8.Karch, H., H. Schmidt, C. Janetzki-Mittmann, J. Scheef, and M. Kroger. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262:600-607. [DOI] [PubMed] [Google Scholar]
- 9.Karmali, M. A., B. T. Steele, M. Petric, and C. Lim. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619-620. [DOI] [PubMed]
- 10.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 1999. Induction of type 2 Shiga toxin synthesis in Escherichia coli O157 by 4-quinolones. Lancet 353:1588-1589. [DOI] [PubMed] [Google Scholar]
- 11.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 13.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 352:1207-1212. [DOI] [PubMed] [Google Scholar]
- 14.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhldorfer, I., J. Hacker, G. T. Keusch, D. W. Acheson, H. Tschape, A. V. Kane, A. Ritter, T. Olschlager, and A. Donohue-Rolfe. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect. Immun. 64:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myhal, M. L., D. C. Laux, and P. S. Cohen. 1982. Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur. J. Clin. Microbiol. 1:186-192. [DOI] [PubMed] [Google Scholar]
- 18.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 19.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255-1267. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 22.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plunkett, G., 3rd, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 26.Safdar, N., A. Said, R. E. Gangnon, and D. G. Maki. 2002. Risk of hemolytic uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 enteritis: a meta-analysis. JAMA 288:996-1001. [DOI] [PubMed] [Google Scholar]
- 27.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, and V. J. DiRita. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, H. 2001. Shiga-toxin-converting bacteriophages. Res. Microbiol. 152:687-695. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein, P. E., A. Boodhoo, G. J. Tyrrell, J. L. Brunton, and R. J. Read. 1992. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 355:748-750. [DOI] [PubMed] [Google Scholar]
- 31.Stenson, T. H., and A. A. Weiss. 2002. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infect. Immun. 70:2297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao, X., N. Schiering, H. Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 34.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 2001. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69:1934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 38.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, J., H. Webb, and T. R. Hirst. 1992. A homologue of the Escherichia coli DsbA protein involved in disulphide bond formation is required for enterotoxin biogenesis in Vibrio cholerae. Mol. Microbiol. 6:1949-1958. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]