Abstract
Salivary histatin 5 (Hst 5), a potent toxin for the human fungal pathogen Candida albicans, induces noncytolytic efflux of cellular ATP, potassium, and magnesium in the absence of cytolysis, implicating these ion movements in the toxin's fungicidal activity. Hst 5 action on Candida resembles, in many respects, the action of the K1 killer toxin on Saccharomyces cerevisiae, and in that system the yeast plasma membrane potassium channel, Tok1p, has recently been reported to be a primary target of toxin action. The question of whether the Candida homologue of Saccharomyces Tok1p might be a primary target of Hst 5 action has now been investigated by disruption of the C. albicans TOK1 gene. The resultant strains (TOK1/tok1) and (tok1/tok1) were compared with wild-type Candida (TOK1/TOK1) for relative ATP leakage and killing in response to Hst 5. Patch-clamp measurements on Candida protoplasts were used to verify the functional deletion of Tok1p and to provide its first description in Candida. Tok1p is an outwardly rectifying, noisily gated, 40-pS channel, very similar to that described in Saccharomyces. Knockout of CaTOK1 (tok1/tok1) completely abolishes the currents and gating events characteristic of Tok1p. Also, knockout (tok1/tok1) increases residual viability of Candida after Hst 5 treatment to 27%, from 7% in the wild type, while the single allele deletion (TOK1/tok1) increases viability to 18%. Comparable results were obtained for Hst-induced ATP efflux, but quantitative features of ATP loss suggest that wild-type TOK1 genes function cooperatively. Overall, very substantial killing and ATP efflux are produced by Hst 5 treatment after complete knockout of wild-type TOK1, making clear that Tok1p channels are not the primary site of Hst 5 action, even though they do play a modulating role.
Antimicrobial defense mechanisms in the oral cavity are provided by innate nonimmune proteins and peptides of salivary gland origin, which include lactoperoxidase, lysozyme, lactoferrin, cathelicidins, and histatins (Hst's). These agents have overlapping antimicrobial activities and are key initial host defense molecules in limiting oral infections (15, 26). The Hst's form a family of small, histidine-rich, cationic peptides secreted by human parotid and submandibular glands (27). Of this family, Hst 5 has the highest antifungal activity for Candida albicans and related yeasts (38). Oral candidiasis (thrush) is a common mucosal infection caused by C. albicans in patients undergoing chemotherapy or organ transplantation and in persons with immunodeficiency virus infections, underlying systemic disease states, or medications which reduce salivary flow.
The spectrum of innate eukaryotic antifungal molecules includes a large array of peptides and proteins, varying widely in both structure and molecular mechanism of action. Many of these agents, but not all, directly target the plasma membrane and/or organellar membranes for disruption; others kill fungi via multitarget, multistep pathways. Chromofungin is an antifungal peptide derived by natural processing of vasostatin-I and is secreted from human polymorphonuclear neutrophils (24). This protein acts primarily by penetrating and destabilizing fungal cell plasma membranes (24). Yeast K1 killer toxin, by contrast, kills sensitive Saccharomyces cerevisiae cells via a receptor-mediated multistage process. K1 toxin initially binds a receptor in the yeast cell wall, Kre1p (9), whence action is transferred to the plasma membrane, disrupting cell function by causing unregulated release of potassium and ATP (1, 33). Patch-clamp evidence has been presented (32) for K1 action on the TOK1 potassium channel in Saccharomyces, but killing via this target alone is very incomplete.
Superficially, the pathway of Hst 5 fungicidal activity now appears similar to that of K1 toxin and does not involve direct pore formation in the yeast plasmalemma (17, 18, 21, 36). Hst 5 effects on Candida plasma membranes include depolarization (30), loss of cellular ATP and potassium (22, 39), and uptake of the DNA marker propidium iodide (18) but not the passage of larger dyes, such as calcein (14). Hst 5 also appears to act, like K1 toxin, via a specific surface receptor distinct from the targets for permeabilization (14). For Hst 5, that receptor appears to be a 70-kDa binding protein in cell envelopes of both C. albicans and S. cerevisiae whose deletion makes cells less sensitive to Hst 5 (M. Edgerton, unpublished data). K1 toxin killing is blocked in 2,4-dinitrophenol (DNP)- and carbonyl cyanide m-chlorophenylhydrazone (CCCP)-energy-depleted cultures. Similarly, the cytotoxic effects of Hst 5 on C. albicans are dependent upon energy metabolism (21, 39). Hst 5 killing is inhibited by the ionophores DNP and CCCP (21), whose uncoupling action on mitochondria causes rapid deenergization of the cells (13). Saccharomyces cell death from K1 toxin was recently attributed, principally by one laboratory (1), to potassium efflux through Tok1p, since high external potassium seemed to protect yeast against K1 toxin and since deletion of the TOK1 gene also conferred resistance. In related experiments, we found Hst 5 fungicidal activity to be inhibited by increased extracellular potassium concentration, as well as by pretreatment of cells with ion-transport inhibitors, such as 5-nitro-2-(3-phenylpropylamino)benzoic acid (NPPB), 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS), and niflumic acid (2).
The total evidence to date on the candidacidal action of Hst 5 points toward a multistep process involving the C. albicans homolog of the S. cerevisiae TOK1 channel as a potential effector target. The experiments below identify the sole C. albicans TOK1 gene and describe construction of several TOK1-disruption strains. These studies provide the first direct functional assessment of Tok1p involvement in Hst 5 action.
MATERIALS AND METHODS
Yeast strains and media.
Escherichia coli strain DH5α (Invitrogen) was used as a host for plasmids and was cultivated in Luria-Bertani medium (Difco). Ampicillin was added at a final concentration of 100 μg ml−1. The C. albicans strains used and generated in this work are shown in Fig. 1. A rich medium, yeast extract-peptone-dextrose (YPD; Difco), was used to grow strain CAI4 for transformation. Yeast nitrogen base (YNB) medium (Qbiogene) without uracil or uridine and supplemented with 2% glucose as carbon source was used for selection of the URA+ transformants. Solid media were prepared by addition of 1.5% Difco agar. YCB-BSA medium was used for induction of FLP-mediated excision of the URA3 flipper cassette (UFC) (25). For selection of ura3 auxotrophic derivatives, cells were plated on solid YNB containing 50 μg of uridine ml−1 and 1 mg of 5-fluoroorotic acid (FOA; Sigma) ml−1. The FOA-resistant colonies were picked up after 3 days of incubation at room temperature.
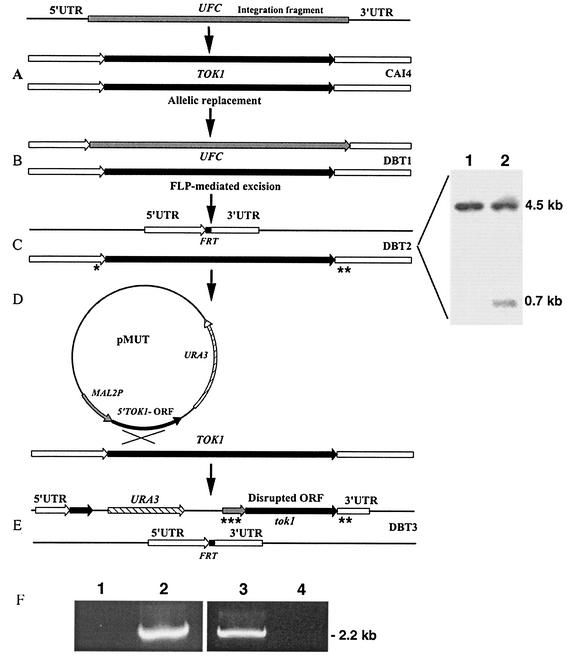
FIG. 1.
Construction of C. albicans TOK1-disrupted strains. Construction of C. albicans TOK1 knockout strains is described in Materials and Methods. (A) The integration fragment incorporating the UFC was introduced into the genome of the parental strain CAI4 (Ura−). (B) Integration of the UFC and replacement of the first TOK1 allele generates strain DBT1 (Ura+). (C) FLP-mediated excision of the UFC recycles the selectable marker and generates the single-allele deletion TOK1 strain DBT2 (Ura−). The small black box denotes the remaining single FRT site after UFC excision. Shown are the results of Southern hybridization analysis of BglII-digested genomic DNA from strains CAI4 (lane 1) and DBT2 (lane 2) with a 0.7-kb 5′ untranslated region (UTR) TOK1-specific probe. The sizes of the hybridizing fragments are as indicated: 4.5 kb, which corresponds to the intact TOK1 alleles in CAI4 (TOK1/TOK1) (lane 1), and 0.7 kb, which corresponds to the inactivated allele in strain DBT2 (TOK1/tok1) as well as the 4.5-kb fragment of the remaining wild-type allele (lane 2). (D) Integration of pMUT disrupts the second TOK1 allele. (E) Genomic structure of the null TOK1 strain DBT3 (Ura+). (F) PCR analysis of genomic DNA isolated from the homozygous (tok1/tok1) DBT3 (lanes 1 and 3) and the heterozygous (TOK1/tok1) DBT2 (lanes 2 and 4) strains. Lanes 1 and 2 were analyzed with the control primer combination TOK1Pctrl-f (*) and TOK1Uctrl-r (**) (Table 1), which amplify the remaining wild-type TOK1 allele of strain DBT2, while lanes 3 and 4 were analyzed with the primer combination MAL2Pctrl-f (***) and TOK1Uctrl-r (**) (Table 1), which amplify only the disrupted tok1 second allele. Asterisks on panels C and E indicate specific primer binding sites.
For growth testing of constructed strains, cells from each strain were grown overnight (∼15 h) in YNB-complete medium at 37°C, until they reached an optical density at 600 nm of 1.2. Cells were counted and serially diluted 1:10, 1:100, and 1:1,000 with water, after which 7 μl of each strain and concentration was pipetted onto YNB plates supplemented with 10 or 100 mM KCl or NaCl. Plates were incubated at 37°C for 48 h until visible colonies formed.
DNA manipulations.
Standard conditions for molecular cloning, hybridization, transformation, and electrophoresis were used (31). Restriction endonucleases were purchased from Fermentas. The Fast-Link DNA ligation kit was from Epicentre Technologies. Plasmid DNA was isolated from E. coli by the alkaline lysis method, employing a QIAprep Spin miniprep kit from Qiagen. Purification of restriction digest mixes, PCR mixes, and DNA fragment isolation from agarose gels were done by using QIAquick PCR purification and QIAquick gel extraction kits, respectively, both from Qiagen. All synthetic oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Synthetic oligonucleotides used in this study
| Purpose and primera | Sequenceb | Restriction sitec |
|---|---|---|
| Construction of pTOK-UFC-1 and pTOK-UFC-2 | ||
| TOKf-I | AAGgtcgacAGAAAAGCTTTTGCTATTGCCG | SalI |
| XhTOKr-I | AACctcgagTGTGGAGTATGGTCAGGTATG | XhoI |
| BgTOKf-I | GGAagatctACACTAAGATACAACCCAATATC | BglII |
| TOKr-I | AAGgaattcGACCCACCATCACTGCATTG | EcoRI |
| TOKf-II | AAGgtcgacTTTCATACCTGACCATACTCCA | SalI |
| XhTOKr-II | AACctcgagGCGGCAATCAATGGAAAATAAC | XhoI |
| BgTOKf-II | GGAagatctTGAGATTGCCTTTAAAAGAGCC | BglII |
| TOKr-II | AAGgaattcTCAGATATTGGGTTGTATCTTAG | EcoRI |
| Construction of pMUT | ||
| hTOK1-f | GCAaagcttATGCAAATAGTAAACAAAGCAAAAG | HindIII |
| bTOK1-r | AAGggatccTTTCTTCCAGGGTTAAATGTCTG | BamHI |
| Construction of pDBT | ||
| (S)TOK1L-f | AAGgtcgacCGTATTGAAAGTATACCAAAGATC | SalI |
| (B)TOK1L-r | AAGggatccCAGCAACAGCTTGACCCACC | BamHI |
| RP10-f | TTTCACTTCTGACAAATTAAGATC | |
| Rp10-r | GAAGTTGGTCAACAAGTTTTTAC | |
| PCR analysis of strain DBT3 | ||
| TOK1Pctrl-f | CATCTTTTGATTCTGCATCTGTC | |
| TOK1Uctrl-r | TGTAAATAAATGCATTCTGTGG | |
| MAL2Pctrl-f | TTTTGCAGTGGTTTAACTAATAATC | |
| RT-PCR analysis of TOK1 strains | ||
| TOK1-f | TGCAAATAGTAAACAAAGCAAAAG | |
| TOK1-r | TTATCATCATCAAACTTTATTGTTC |
Lowercase letters f and r indicate that the respective primer is forward or reverse.
All sequences are given in the conventional 5′-to-3′ direction.
Restriction sites introduced into primer sequences for cloning purposes are in lowercase letters.
Construction of C. albicans TOK1 mutant strains.
To study the possible role of the CaTOK1 gene product in the action of salivary Hst 5, knockout strains were constructed via the URA3-flipping method (25). C. albicans strain CAI4 was first transformed with linearized pTOK-UFC-1 in order to inactivate the first TOK1 allele (Fig. 1A). Correct allelic replacement was confirmed by PCR and Southern analysis for four randomly selected URA+ transformants (data not shown). One of them was grown on YCB-BSA medium for induction of FLP-mediated excision of the UFC (Fig. 1B). Two randomly selected Ura− colonies were PCR and Southern blot analyzed for the loss of the UFC (Fig. 1C). Thus, the heterozygous TOK1/tok1 strain DBT2 was obtained.
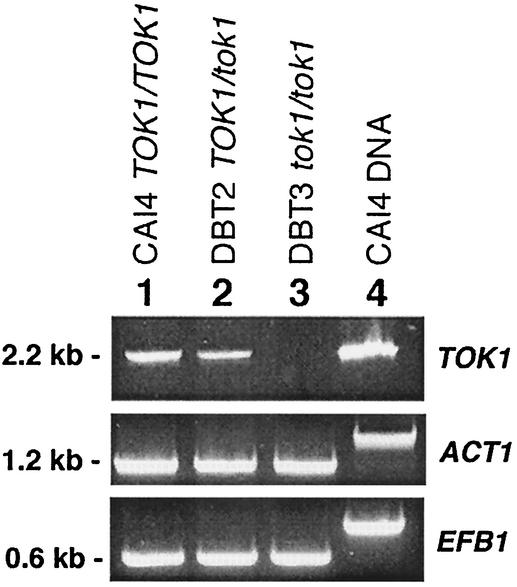
The heterozygous TOK1/tok1 strain was subjected to a second round of transformation with both pTOK-UFC-1 and/or pTOK-UFC-2 in order to obtain homozygote disruptants. Twelve URA+ transformants of each experiment were analyzed by PCR and Southern blotting, and all transformants maintained the wild-type allele, a phenomenon that has been recently described for C. albicans (10). Therefore, a second allele knockout strategy with plasmid pMUT was used (Fig. 1D). Spheroplasts of strain DBT2 were transformed with intact pMUT plasmid, and a C. albicans tok1 null mutant strain DBT3 with the insertion in the remaining TOK1 allele was obtained (Fig. 1E). PCR analysis of genomic DNA isolated from homozygous (tok1/tok1) DBT3 (Fig. 1F, lanes 1 and 3) and the heterozygous (TOK1/tok1) DBT2 (Fig. 1F, lanes 2 and 4) strains was done. Specific primers used are listed in Table 1, and their binding sites are indicated in Fig. 1C and E. The gene status of these strains was further verified by reverse transcription-PCR (RT-PCR) detection of the CaTOK1 transcript as described below and is shown in Fig. 2.
FIG. 2.
RT-PCR analysis of C. albicans strains for TOK1 transcription. RT-PCR products of TOK1 transcripts in total RNA samples are displayed for three C. albicans strains: lane 1, CAI4 (TOK1/TOK1); lane 2, DBT2 (TOK1/tok1); and lane 3, DBT3 (tok1/tok1). Lane 4 resulted from using CAI4 genomic DNA as template. Fragments obtained by amplification with oligos specific for ACT1 (1.2 kb) and EFB1 (0.6 kb) (3) served as internal mRNA controls and demonstrated the cDNA origin.
For complementation experiments with the single-allele deletion strain DBT2 (TOK1/tok1), the supplemental strain DBT4 was generated. The wild-type copy of TOK1 was cloned and introduced into the RP10 locus of DBT2, using the plasmid construct pDBT as described below to generate strain DBT4.
Construction of pTOK-UFC-1 and pTOK-UFC-2.
The UFC was isolated as a XhoI-BglII fragment from plasmid pSFUC2, kindly supplied by J. Morschhäuser. Two different constructs were generated. pTOK-UFC-1 was prepared as follows: using the primers TOKf-I and XhTOKr-I, a 697-bp fragment spanning positions −830 to −134 upstream of the putative translational start codon of the CaTOK1 open reading frame (ORF) was PCR amplified from strain CAI4 genomic DNA. Likewise, employing the BgTOKf-I and TOKr-I primers, a 609-bp PCR fragment spanning positions 266 to 875 downstream of the putative translational stop codon of the TOK1 ORF was generated. After purification, these PCR fragments were digested with SalI-XhoI and BglII-EcoRI restriction enzymes, respectively, and in a quadruple reaction they were ligated to a XhoI-BglII fragment bearing the UFC and the SalI-EcoRI-digested pUC18, to yield pTOK-UFC-1. Thus, this construct was designed to delete 2,541 bp of the CaTOK1 locus. In a similar way, pTOK-UFC-2 was prepared in order to delete the region of 1,646 bp within the TOK1 ORF. Using the primers TOKf-II and XhTOKr-II, a 455-bp PCR fragment was amplified, spanning the positions −157 upstream to 298 downstream of the putative translational start; by employing BgTOKf-II and TOKr-II primers, a 489-bp PCR fragment was generated, spanning the positions 200 upstream to 289 downstream of the putative translational stop signal. Like the previous construct, these two PCR fragments were digested with SalI-XhoI and BglII-EcoRI restriction enzymes, respectively, and in a quadruple reaction were ligated to an XhoI-BglII fragment bearing a UFC and SalI-EcoRI-digested pUC18 (40), to yield pTOK-UFC-2. These constructs were Eco31I linearized before transformation by using unique restriction enzyme cuts within the vector sequence.
Construction of pMUT.
The fragment bearing the first 1,078 bp of the TOK1 ORF, including the putative translational start signal, was PCR amplified using the primers hTOK1-f and bTOK1-r. After digestion with HindIII-BamHI, this fragment was ligated via the same enzymes used to digest pMAL/EFB3′UTR/URA3 (3), in order to generate pMUT.
Construction of pDBT.
A 3,895-bp fragment bearing the CaTOK1 locus was PCR amplified with the primers (S)TOK1L-f and (B)TOK1L-r. After SalI-BamHI digestion, this fragment was ligated to the same enzyme-digested vector pUC18 to yield pTOK1L. Following BamHI-SacI digestion, the resulting linearized construct was ligated to the BamHI-KpnI fragment bearing the CaRP10 locus and the KpnI-SacI-digested PCR fragment bearing a C. albicans URA3 gene as a selectable marker, to generate plasmid pDBT. This plasmid was used as template in a PCR amplification reaction with the RP10-f and RP10-r primers before transformation of strain DBT2, since the 3,895-bp DNA fragment bearing the CaTOK1 gene contains four NcoI restriction sites. NcoI digestion is usually employed to cut the C. albicans RP10 locus, in order to linearize a given construct before transformation, when the C. albicans RP10 locus is the target for integration. Therefore, divergent (long-distance) PCR was used in order to obtain pBBT in linear form.
RNA isolation, cDNA synthesis, and RT-PCR detection of the CaTOK1 transcript.
Isolation of total RNA from C. albicans strains CAI4, DBT2, and DBT3 grown in YNB (glucose) medium, control of DNA contamination, assessment of cDNA synthesis and efficiency, and RT-PCR analysis were performed essentially as reported previously (3). The TOK1-f/TOK1-r synthetic oligonucleotides used are listed in Table 1. PCR cycling conditions were as follows: initial denaturation was carried out for 3 min at 94°C followed by three-step cycling of 15-s denaturation at 94°C, 15-s annealing at 57°C, and 2-min extension at 72°C for 30 cycles, followed by a 5-min final extension at 72°C. All cDNA synthesis and RT-PCR experiments were performed at least three times and were highly reproducible.
Electrophysiological methods.
Protoplasts of C. albicans, required for patch recording from the plasma membrane, were prepared similarly to the published preparation of Saccharomyces protoplasts (6). Shaking cultures were grown to late exponential phase in YPD at 30°C, after which 5 to 8 ml of cell suspension (depending on the actual end optical density) was spun down and washed twice in 50 mM KH2PO4 buffer (brought to pH 7.2 with KOH). The resulting pellet was resuspended in the same buffer, with 0.2% β-mercaptoethanol added, and incubated for 30 min at 30°C. Partial digestion of the cell wall was accomplished by incubating the cells in a solution containing 50 mM KH2PO4 (pH 7.2), 1.2 M sorbitol, 0.2% β-mercaptoethanol, and 0.6 U of zymolyase-20T (catalog no. 320921; ICN) ml−1 at 30°C for 45 min. The resulting spheroplasts were centrifuged and gently resuspended in a solution of 220 mM KCl, 10 mM CaCl2, 5 mM MgCl2, 5 mM morpholineethanesulfonic acid (MES)-Tris (pH 7.2), 0.2% glucose, and adjusted to 800 mOsm with sorbitol. The resultant suspension was kept at room temperature (∼23°C) before use. For each test run, 1 μl was injected gently into the recording chamber flooded with sealing buffer (0.7 ml; see below) so that spheroplasts could adhere lightly to the glass bottom of the chamber.
Recording conditions.
Gigaseal formation and both the whole-cell and the isolated-patch configuration were achieved as described elsewhere for S. cerevisiae (6). Heat-polished, blunt-tip pipettes (see below) were attached to individual protoplasts by light suction, such that seals in excess of 10 GΩ formed within ∼5 min. Thereafter, the whole-cell recording mode was attained by breaking the patch with simultaneously applied suction and a high-voltage pulse (about 800 mV, 100 μs). Isolated (outside-out) patches were subsequently obtained by gently pulsing the sealing solution to and fro in the chamber, so as to draw the protoplast away from the pipette tip.
Patch pipettes were fabricated from borosilicate glass (Kimax-51 Kimble/Kontes 38500) by using a two-stage puller (model PC-10; Narishige) and were formed with an inside tip diameter of ∼1 μm. After heat polishing, pipettes were filled with pipette buffer (intracellular components, 175 mM KCl, 1 mM EGTA, 0.15 mM CaCl2, 4 mM MgCl2, 4 mM K2ATP; brought to pH 7.0 with KOH). When dipped into the sealing buffer (extracellular, 150 mM KCl, 20 mM CaCl2, 5 mM MgCl2, 1 mM MES-Tris [pH 7.5]; brought to 600 mOsm with sorbitol), pipette tip resistances were 3 to 5 MΩ. An Ag-AgCl reference electrode was connected to the bath by means of a 1 M KCl-agar bridge.
All recordings were made in the sealing buffer, at room temperature. Square voltage pulses lasting 2.5 or 3 s were delivered from a holding potential of −40 mV, in −20-mV increments, between +100 mV and −40 mV (see the voltage protocol in Fig. 4A), and the required membrane currents were recorded via an EPC9 amplifier under control of the PULSE software (HEKA Elektronik). Signals were filtered at 667 Hz and sampled at 2 kHz. Reported whole-cell currents have not been corrected for the small linear leak conductance.
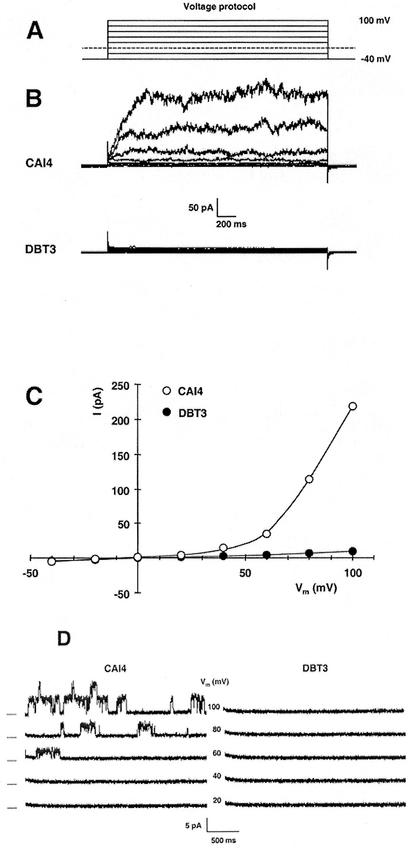
FIG. 4.
Disruption of the TOK1 gene in Candida removes outward rectifier currents that are characteristic of TOK1 proteins, as shown by patch-clamp records from C. albicans protoplasts. (A) Superimposed traces for the voltage-clamp protocol used in whole-cell recording. Membrane voltage was clamped, in 2.5-s pulses, from a holding potential of −40 mV to values of +100 mV to −40 mV in steps of −20 mV. (B) Superimposed current traces in response to the voltage-clamp protocol (panel A) for the wild-type strain CAI4 (TOK1/TOK1, upper record set) and for the fully disrupted strain DBT3 (tok1/tok1, lower record set). The outward currents, which were very similar to Tok1p-associated currents in Saccharomyces, were completely absent from DBT3. (C) Summary plot of the steady-state currents in panel B: for each pulse, current was averaged over the interval 1.2 to 2.2 s and plotted against the clamped membrane voltage. Values plotted are representative (near average) for 10 cells from each strain. (D) Current traces from an outside-out patch isolated from the wild-type strain (CAI4, left panel) and from the tok1-disrupted strain (DBT3, right panel). The voltage protocol was similar to that used for whole-cell recording, but with 3-s pulses. Dashes at the left indicate zero current for each pair of traces, and numbers in the middle are the clamped membrane voltages. The TOK1 patch contained at least two channels, each behaving similarly to Saccharomyces Tok1p, with a noisy open state previously shown to represent high-frequency close-open gating. Such single-channel events were completely absent from DBT3 patches. Data are representative of five patches for each strain.
Candidacidal assay.
Antifungal activity of Hst 5 was examined by microdilution plate assay (14). Briefly, C. albicans strains were grown in YNB medium, washed twice with 10 mM sodium phosphate buffer (Na2HPO4/NaH2PO4; pH 7.4), and resuspended in phosphate buffer at a cell concentration of 2.5 × 105 cells/ml. Hst 5 was added at serial dilutions from an initial concentration of 31 μM and incubated for an additional 1 h. Our standard test, at 31 μM, kills more than 90% of wild-type cells. Cell suspensions were diluted with phosphate buffer, and 50 μl (about 500 cells) was removed and plated onto Sabouraud dextrose agar. Cell viability was assessed by counting formed colonies after incubation for 24 h at 37°C. Candidacidal assays were performed in triplicate. Percent loss of viability was calculated as [1 − (number of colonies recovered from Hst 5-treated cells/number of colonies from control cells)] × 100. Statistical comparison of the loss of viability for each strain was done by using Student's t test for each applied Hst 5 concentration.
Bioluminescence assay for extracellular ATP.
Measurement of extracellular ATP levels was as previously described (21) with the following modifications. C. albicans cells (106) were mixed with 31 μM Hst 5, and extracellular ATP was measured at 5, 10, 20, 45, and 60 min after addition of Hst 5 by luminometry using an ATP assay kit (FL-AA; Sigma) at each indicated time point. Extracellular ATP concentrations were determined from ATP standard curves for each experiment, and results were expressed as nanomoles of ATP per 106 cells. Statistical comparisons of ATP release between the strains for each time point were done by using Student's t test.
RESULTS
Identification of the C. albicans TOK1 gene.
BLAST analysis of the C. albicans genome database, available at http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi?organism=euk, with the S. cerevisiae TOK1 protein sequence as query (ScTOK1; Swiss-Prot accession number P40310), revealed a homologous sequence in contig 6-2442. Further analysis of the 3,896-bp nucleotide sequence identified an ORF of 2,142 bp potentially encoding a protein of 714 amino acid residues. The deduced primary structure of the encoded protein showed 33% overall identity with the 691 amino acid residues of the ScTOK1 protein (Fig. 3). DIALIGN analysis (http://bibiserv.techfak.uni-bielefeld.de/cgi-bin/dialign) also identified, in the predicted C. albicans protein, the eight hydrophobic segments S1 to S8 (Fig. 3) and the two pore domains (P1 and P2) characteristic of the ScTOK1 potassium channel (Fig. 3) (19, 23, 29, 41). Sequence identity between the predicted C. albicans protein and Saccharomyces Tok1p was found to be 46% in the S1 to S8 spanners and 71% in the pore domains. In addition, TMpred hydropathy-profile analysis (http://www.ch.embnet.org/cgi-bin/TMPRED) yielded very similar plots for these two proteins (data not shown). These findings, plus additional analysis of the flanking regions for the identified ORF, demonstrated that contig 6-2442 contains the C. albicans TOK1 gene (CaTOK1) encoding the Tok1 potassium channel.
FIG. 3.
Alignment of the deduced primary structure of the C. albicans TOK1 protein with that of S. cerevisiae Tok1p. Both proteins are characterized by eight hydrophobic segments, presumed to be α-helical and membrane spanning, that are designated by boldface type and are labeled S1 to S8. Two pore domains in each protein are designated by bold underlining and are labeled P1 and P2; within the pore domains, the diagnostic K+ channel selectivity sequence, GYG, and its congeners (VYG, GLG) are marked by double underlining. Sequence identity between the C. albicans protein and its S. cerevisiae counterpart was found to be 33% overall, 46% in the S1 to S8 spanners, and 71% in the pore domains.
Extensive search of the C. albicans genome data bank using the S. cerevisiae TOK1 gene and/or different parts of its sequence as query failed to detect any other related sequences. Similarly, further searches using CaTOK1 itself and/or parts of its sequence did not reveal any homologous sequences. Therefore, like S. cerevisiae, in which the complete annotated genome does not contain any sequences that can be regarded as TOK1 related, C. albicans appears to harbor only one gene homologue sequence that can be referred to as CaTOK1.
Disruption of CaTOK1 abolishes the characteristic TOK1-associated currents.
Typical whole-cell currents in wild-type C. albicans protoplasts are demonstrated in Fig. 4B (upper panel) for voltage pulses positive to −40 mV. These outward currents (positive, upward in the plots) closely resemble Tok1p-associated currents in S. cerevisiae (5, 7) in time course and general noise characteristics, though they proved smaller than currents often reported in Saccharomyces, due in part to the smaller size of Candida protoplasts (∼4-μm diameter, cf. 8 μm). In Candida, as in Saccharomyces, the channels are potassium selective (data not shown), and the outward currents represent the outflow of K+ from cytoplasm to bath, made conspicuous at positive (reversed) membrane voltages. Disruption of both TOK1 alleles in strain DBT3 abolished the outward currents, leaving residual leakage currents—the baseline “smear” in Fig. 4B (lower panel)—amounting to ∼5% of the TOK1-associated currents.
Figure 4C summarizes the steady-state values of measured currents, averaged over the last half of each pulse and plotted against the corresponding clamped membrane voltage. The curve shape for CAI4, with both wild-type alleles of TOK1 functional, is typical for a simple voltage-gated channel having a gating voltage of approximately +60 mV. The plot for DBT3, with both alleles disrupted, represents a small and purely ohmic leak (linear, and passing through the origin), with no hint of gated channels.
The functioning of individual channels, recorded in isolated outside-out patches, is shown in Fig. 4D. Traces in the left panel, for the wild-type strain CAI4, demonstrate the presence of at least two TOK1 channels in the patch and a gating pattern which closely resembles that of Saccharomyces Tok1p, with long quiet intervals between noisy open-channel bursts. The noisy character of the open bursts represents high-frequency closing and reopening of the channel, which is too fast to be accurately recorded by the amplifier (5). Channel-gating events were completely absent from all membrane patches obtained on knockout DBT3 cells, as is demonstrated in the right panel of Fig. 4D.
Disruption of TOK1 reduces sensitivity of C. albicans cells to Hst 5 killing.
Since we were concerned that salt sensitivity of TOK1 mutants would be altered as an indirect consequence of gene disruption, we examined growth phenotypes of the constructed strains by standard drop tests on YPD-agar medium supplemented with 10 and 100 mM KCl or NaCl. No growth differences were observed among the three test strains, CAI4, DBT2, and DBT3, i.e., among the wild-type, the TOK1 single-deletion strain, and the knockout strain, nor among the different salt regimens imposed (data not shown). Growth curves for strains were also measured for qualitative comparison. Generation times for CAI4 and DBT2 were equivalent, and DBT3 was only slightly retarded (by 15%). Thus, the mutant strains could be compared directly with the wild-type parent under the conditions of our standard candidacidal assay.
Although data from viability tests at low concentrations of Hst 5 (<10 μM), were too variable, alone, to define differences between Candida strains, when taken together with data at higher Hst 5 concentrations (≥15 μM) they permitted a systematic analysis of the effects of TOK1 genes and proteins on the susceptibility of C. albicans to Hst 5 killing. A quantitative empirical treatment is provided in Fig. 5. The curves have been drawn by fitting a simple saturation function to the three plots, finding a single half-saturating concentration (14.2 μM).
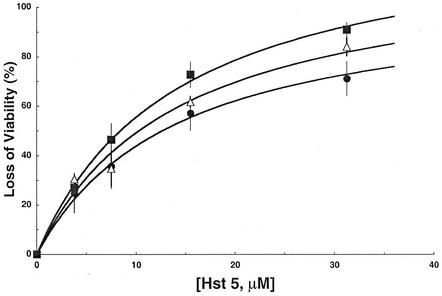
FIG. 5.
Disruption of TOK1 in C. albicans reduces sensitivity of cells to Hst 5 killing. C. allbicans strains were incubated at several concentrations of Hst 5, in 10 mM sodium phosphate buffer (pH 7.4) for 1.5 h at 37°C. Percent viability was calculated as the ratio of colonies recovered from Hst 5-treated cells/colonies recovered from control cells, multiplied by 100. Closed squares, CAI4 (TOK1/TOK1); open triangles, DBT2 (TOK1/tok1); closed circles, DBT3 (tok1/tok1). Each data point represents the mean ± standard deviation of at least three independent experiments. For a quantitative description, each of the plots was initially fitted with a simple saturation curve (rectangular hyperbola), but since all three half-saturation constants (K0.5) were close together, the smooth curves shown were fitted using a common K0.5, found to be 14.2 μM Hst 5.
Loss of viability in the knockout strain (DBT3) following Hst 5 treatment (31 μM) was only 73%, compared with 93% in the wild-type strain (CAI4). Viewed alternatively, survival of the wild type following 31 μM Hst 5 was only 7%, whereas it rose to 18% with one functional TOK1 allele (DBT2) and to 27% with no functional TOK1 alleles (DBT3). Thus, two TOK1 disruptions approximately doubled the viability increment produced by a single disruption. Finally, the concentration of Hst 5 required for 50% killing (LD50) was 12.6 μM with both alleles disrupted, 10.2 μM with one disrupted, and 8.4 μM with both intact, so that the incremental effect of two TOK1 genes was approximately twice that of one, as measured by LD50 values. Statistical analysis of the data was done, since differences in reduction of killing by Hst 5 upon deletion of the TOK1 channels were small. By Student's t test, the P values were <0.05 for the wild type (CAI4) versus the single-deletion strain (DBT2) or for DBT2 versus the full knockout strain (DBT3); P was <0.001 for CAI4 versus DBT3. Thus, the two smaller differences were weakly significant, but the larger one was strongly significant. The same conclusion was obtained from comparison of the Hst 5 concentrations required for 50% killing, with corresponding P values of <0.08 and <0.002.
The quantitative stability of TOK1 effects was verified by comparison of the wild-type strain (CAI4) with the DBT4 strain, in which a plasmid-borne copy of TOK1 had been inserted into the DBT2 strain. DBT4 and CAI4 displayed the same sensitivity to Hst 5 (data not shown), demonstrating full complementation of the TOK1/tok1 phenotype by the reintroduced copy of TOK1. Thus, the observed effects caused by the CaTOK1 disruption are specific to TOK1 deletion and are not a result of the genetic manipulation itself.
These data clearly demonstrate two important points. First, Hst 5 is strongly candidacidal in the complete absence of TOK1 function; indeed, killing extrapolates to 100% at high Hst 5 concentrations (Fig. 5, lower curve). Second, a functional presence of TOK1 genes and proteins systematically enhances the candidacidal action of Hst 5. Thus, the TOK1 protein is not the critical target for Hst 5 action, but it does augment killing.
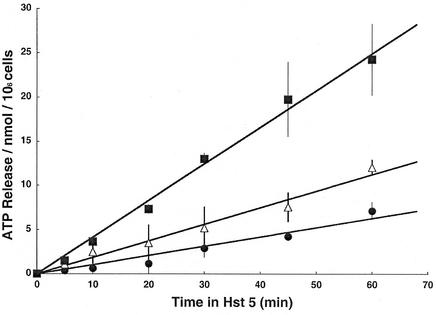
TOK1 disruption also reduces the characteristic ATP efflux associated with Hst 5 action.
Since loss of ATP commences rapidly after Hst 5 treatment of C. albicans (21) and that loss is strongly correlated with cell death, we also determined the effects of TOK1 disruption on ATP efflux from the cells, as shown in Fig. 6. All the data plots for ATP release during the first hour could be reasonably fitted by straight lines, which revealed the following quantitative relationships. Standard Hst 5 treatment (31 μM) released ATP from wild-type cells (CAI4) at the rate of 24.9 nmol/h · 106 cells (0.415 nmol/min · 106 cells), which represents approximately 40% of total cellular ATP during the initial hour. The rate of efflux was reduced by 55% (to 11.2 nmol/h · 106 cells) upon disruption of a single TOK1 allele and reduced by another 30% (to 6.3 nmol/h · 106 cells) upon loss of the second TOK1 allele. Statistical comparison of the fitted slopes in Fig. 6 using Student's t test found all three P values to be <0.001 at each time, so these differences were highly significant.
FIG. 6.
Strains with TOK1 disruption and killing-tolerance to Hst 5 have reduced ATP efflux. C. albicans strains were grown to late log phase in YNB, and then 106 cells from each strain were mixed with Hst 5 (31 μM) and incubated for the indicated times to establish ATP efflux. Closed squares, CAI4 (TOK1/TOK1); open triangles, DBT2 (TOK1/tok1); closed circles, DBT3(tok1/tok1). Extracellular ATP was measured by luminometry of cell supernatants and is expressed as nanomoles of ATP per 106 cells. Each data point represents the mean ± standard deviation of at least three independent experiments. The fitted regression lines have slopes of 6.3, 11.2, and 24.9 nmol of ATP/h · 106 cells, respectively, for zero, one, and two active TOK1 genes present.
Thus, again, TOK1 channels proved unnecessary for Hst 5 action, but their presence substantially enhanced that action. There was, however, an unexplained quantitative disparity between the TOK1 influence on Hst 5-evoked ATP efflux (Fig. 6) and its influence on Hst 5-evoked killing (or residual survival [Fig. 5]). The incremental effect of two functional copies of TOK1 on ATP efflux was nearly fourfold that of a single copy; but for residual survival, two functional copies had only twice the decrementing effect of a single copy.
DISCUSSION
The evidence presented here makes clear that the TOK1 channel, an outward rectifier for potassium, plays a supporting role rather than a central role in Hst 5-induced cell death of C. albicans, and even that supporting role appears to be limited by other factors. The cytotoxic effects of Hst 5 are likely to be partially offset by increased energy turnover and metabolite synthesis and, in that circumstance, cell killing might well be proportional to the steady-state accumulated loss of metabolites like ATP, as found by Koshlukova et al. (21, 22), but not necessarily be proportional to their rate of loss. However, the rate of loss of ATP and other metabolites is far more likely to illuminate the role of membrane transport proteins (such as TOK1 channels) in Hst action than is the steady-state loss of metabolites.
Given the fact that well-known anion channel inhibitors, such as DIDS, NPPB, and niflumic acid, all block Hst 5-induced ATP loss and killing in C. albicans (2), the simplest model for the primary membrane action of Hst 5 is that it somehow creates an anion channel-like conductance in one or more native membrane proteins. Under normal growth conditions, however, channel conductances in the plasma membranes of microorganisms must be very spare, lest they overwhelm the cells' total energy supply and run down the viable gradients of small molecules. The latter point is clear from studies on another ascomycete fungus, Neurospora crassa, where ion and metabolite transport at the plasma membrane consumes 25 to 30% of total cellular ATP turnover (35) and funnels the energy through the plasma membrane proton ATPase (Pma1p) into an electrochemical gradient for protons, which can exceed 350 mV. The resting membrane voltage (Vm) of Neurospora that is bathed in normal laboratory saline buffers approximates −200 mV (cell interior negative) and acts as a very large driving force tending to push out most intracellular (metabolic) anions and to push in most extracellular cations. All indirect evidence, including the apparent universal presence (in terrestrial fungi) of Pma1p-type membrane ATPases, suggests similar arrangements in yeasts, esp. Saccharomyces and Candida. Since ion channels typically have 103- to 106-fold-higher conductances than active transporters, bona fide channels opened other than very briefly and intermittently would cause both rapid depolarization and rapid rundown of the normal ion and metabolite gradients. Thus are produced the stark lethal effects of channel-forming antibiotics such as nystatin and proteins like complement, perforin, and pneumolysin.
If Hst 5 does indeed create anion channel-like pathways through C. albicans membrane proteins, TOK1 potassium channels could augment the performance of those anion channels either directly or indirectly. Indirect augmentation is easiest to envision, because strictly anion-selective channels themselves cannot run down metabolite gradients. Instead, opening of many such channels would clamp Vm to the anion equilibrium (presumably a large reversed [+] voltage), without much change in macroscopic gradients. But Tok1p could provide the parallel cation conductance needed for rapid rundown. Simultaneously open anion and cation channels would clamp Vm near zero and place Pma1p into overdrive, destroying any trapped ATP, while coincidently letting anion and cation gradients run down. Thus, potassium-ATP, and perhaps other metabolically important ion pairs, would rapidly drain out of the cells. The potential cogency of this kind of mechanism is increased by the fact that Tok1p functions as an outward rectifier: its open probability is very low at normal resting Vm (near −200 mV) but increases steeply as Vm approaches zero or positive values (in Saccharomyces [5]).
An obvious caveat of the above argument is that there is nothing special about Tok1p other than its appearance as the only well-defined, abundant, medium-conductance cation channel in yeast plasma membranes. Calculated from Fig. 4 above, there are ∼40 TOK1 channels per C. albicans cell, with an average single-channel conductance of ∼40 pS, similar to the numbers in S. cerevisiae (5). But almost any other revealed cation channel would serve the same purpose, as would nonselectivity (strictly among anions) in the postulated Hst 5-created anion channel, under the condition of high extracellular salt. For other possible defined channels in the Candida plasma membrane, we must wait on full annotation of the Candida genome sequence.
However, low extracellular concentrations of divalent cations, especially Ca2+, reveal an additional type of cation channel in both Saccharomyces (8, 28) and Candida (A. Rivetta, unpublished data) membranes. In S. cerevisiae, reducing extracellular Ca2+ and Mg2+ concentrations ([Ca2+]o, [Mg2+]o) below about 0.1 mM reveals a nonselective cation channel (NSC1) whose total conductance, at nanomolar Ca2+, approaches that of the sum of all TOK1 channels in the same membrane (8). This is equally true in wild-type yeast and in TOK1-null strains, and the molecular identity of the NSC1 channel is still unknown. In Saccharomyces, NSC1 mediates rapid K+ loss in exchange for almost any monovalent cation which is abundant in the medium: Na+, NH4+, Rb+, Cs+, or Li+ (11, 12). Under normal conditions, NSC1 is open at a low level (Ki for Ca2+ is about 0.1 mM), which implies both that lowered [Ca2+]o should enhance Candida's susceptibility to Hst 5-induced ATP loss (and killing) and that elevated [Ca2+]o should protect Candida cells against Hst 5 damage. Both of these conditions already have been experimentally verified (38).
However, one observation in the present experiments is inconsistent with the notion that Tok1p indirectly augments Hst 5 action. That is the disproportionately high rate of ATP loss permitted by two functional TOK1 alleles (Fig. 6), compared with that of a single functional allele. Assuming that two alleles contribute twice as much TOK1 protein to the membrane as does one allele, then two should also permit twice-as-rapid K+ efflux to accompany ATP loss rather than the fourfold loss measured. The actual result, then, implies cooperativity, either between the two functional genes or among their translational products. At least three distinct models for cooperativity are possible. Doubling the membrane density of TOK1 protein (i) could increase the single-channel conductance of Tok1p molecules via homologous physical interactions (e.g., homodimerization); (ii) it could increase the conductance of Hst 5-induced anion channels by binding of Tok1p to those anion channels (e.g., heterodimerization); or, alternatively, (iii) it could enhance formation of the postulated anion channels by itself binding to Hst 5 and acting as a conduit to the target protein. Further experiments to differentiate among these possibilities are needed.
It is also relevant to consider the well-demonstrated fact that metabolic inhibitors such as azide and DNP protect Candida against Hst 5 action (17, 21, 22). This phenomenon could be mediated by depletion of ATP, for example via shut-down of Tok1p, whose activation is ATP dependent (7); or it could result from the reduced driving force of Vm diminished by slowing of the plasma membrane H+-ATPase (Pma1p). More likely, however, the conspicuous protection afforded by cellular deenergization is complex and is due to global down regulation of membrane permeability in response to metabolic insults. This phenomenon, although not widely appreciated and certainly not understood, has been demonstrated in a variety of microorganisms, including E. coli, N. crassa, Chlorella vulgaris, and S. cerevisiae (4, 20, 34). A variety of metabolic attacks that trigger such down regulation are anoxia, respiratory blockade, respiratory uncoupling, glucose starvation, and treatment with lipid-soluble ions (including so-called membrane voltage probes). In all of these phenomena, however, the time interval between the metabolic attack and the test treatment is crucial. This latter fact affords another way to probe events surrounding Hst 5 action in C. albicans.
Because of similarities between the physiological effects of Hst 5 upon Candida and the effects of K1 toxin upon Saccharomyces, this demonstration that the TOK1 channel plays only a minor role in Hst 5 action reflects favorably upon the conclusion reached by many workers, most recently Breinig et al. (9), that the TOK1 channel in Saccharomyces plays only a minor role in K1 action.
Acknowledgments
This work was supported by U.S. PHS grants DE10641 and DE00406 from the National Institute of Dental and Craniofacial Research (to M.E.) and GM 60696 from the National Institute of General Medical Sciences (to C.L.S.).
Editor: T. R. Kozel
REFERENCES
- 1.Ahmed, A. F. Sesti, N. Ilan, T. M. Shih, S. L. Sturley, and S. A. N. Goldstein. 1999. A molecular target for viral killer toxin: TOK1 potassium channels. Cell 99:283-291. [DOI] [PubMed] [Google Scholar]
- 2.Baev, D., X. Li, J. Dong, P. Keng, and M. Edgerton. 2002. Human salivary histatin 5 causes disordered volume regulation and cell cycle arrest in Candida albicans. Infect. Immun. 70:4777-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baev, D., X. Li, and M. Edgerton. 2001. Genetically engineered human salivary histatin genes are functional in Candida albicans: development of a new system for studying histatin candidacidal activity. Microbiology 147:3323-3334. [DOI] [PubMed] [Google Scholar]
- 4.Ballarin-Denti, A., C. L. Slayman, and H. Kuroda. 1994. Small lipid-soluble cations are not membrane voltage probes for Neurospora or Saccharomyces. Biochim. Biophys. Acta 1190:43-56. [DOI] [PubMed] [Google Scholar]
- 5.Bertl, A., C. L. Slayman, and D. Gradmann. 1993. Gating and conductance in an outward-rectifying K+ channel from the plasma membrane of Saccharomyces cerevisiae. J. Membr. Biol. 132:183-199. [DOI] [PubMed] [Google Scholar]
- 6.Bertl, A., H. Bihler, C. Kettner, and C. L. Slayman. 1998. Electrophysiology in the eukaryotic model cell, Saccharomyces cerevisiae. Pflügers Arch. Eur. J. Physiol. 436:999-1013. [DOI] [PubMed] [Google Scholar]
- 7.Bertl, A., H. Bihler, J. D. Reid, C. Kettner, and C. L. Slayman. 1998. Physiological characterization of the yeast plasma membrane outward rectifying K+ channel, DUK1 (TOK1), in situ. J. Membr. Biol. 162:67-80. [DOI] [PubMed] [Google Scholar]
- 8.Bihler, H., C. L. Slayman, and A. Bertl. 1998. NSC1: a novel high-current inward rectifier for cations in the plasma membrane of Saccharomyces cerevisiae. FEBS Lett. 432:59-64. [DOI] [PubMed] [Google Scholar]
- 9.Breinig, F., D. J. Tipper, and M. J. Schmitt. 2002. Kre1p, the plasma membrane receptor for the yeast K1 viral toxin. Cell 108:395-405. [DOI] [PubMed] [Google Scholar]
- 10.Cognetti, D., D. Davis, and J. Sturtevant. 2002. The Candida albicans 14-3-3 gene, BMH1, is essential for growth. Yeast 19:55-67. [DOI] [PubMed] [Google Scholar]
- 11.Conway, E. J., and J. Breen. 1945. An “ammonia” yeast and some of its properties. Biochem. J. 39:368-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conway, E. J., and H. M. Gaffney. 1966. The further preparation of inorganic cationic yeasts and some of their chief properties. Biochem. J. 101:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Pena, P., F. Barros, S. Gascon, P. S. Lazo, and S. Ramos. 1981. Effect of yeast killer toxin on sensitive cells of Saccharomyces cerevisiae. J. Biol. Chem. 256:10420-10425. [PubMed] [Google Scholar]
- 14.Edgerton, M., S. E. Koshlukova, T. E. Lo, B. G. Chrzan, R. M. Straubinger, and P. A. Raj. 1998. Candidacidal activity of salivary Histatins: identification of a Histatin 5-binding protein on Candida albicans. J. Biol. Chem. 273:20438-20447. [DOI] [PubMed] [Google Scholar]
- 15.Edgerton, M., and S. E. Koshlukova. 2000. Salivary non-immune antimicrobial proteins. Adv. Dent. Res. 14:16-21. [DOI] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmerhorst, E. J., W. E. Van't Hof, P. Breeuwer, E. C. I. Veerman, T. Abee, R. F. Troxler, A. V. N. Amerongen, and F. G. Oppenheim. 2001. Charaterization of histatin 5 with respect to amphipathicity, hydrophobicity, and effects on cell and mitochondrial membrane integrity excludes a candidacidal mechanism of pore formation. J. Biol. Chem. 276:5643-5649. [DOI] [PubMed] [Google Scholar]
- 18.Helmerhorst, E. J., P. Breeuwer, W. E. Van't Hof, E. Walgreen-Weterings, L. C. Oomen, E. C. Veerman, A. V. Amerongen, and T. Abee. 1999. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274:7286-7291. [DOI] [PubMed] [Google Scholar]
- 19.Ketchum, K. A., W. J. Joiner, A. J. Sellers, L. K. Kaczmarek, and S. A. N. Goldstein. 1995. A new family of outwardly rectifying potassium channel proteins with two pore domains in tandem. Nature 376:690-695. [DOI] [PubMed] [Google Scholar]
- 20.Komor, E., H. Weber, and W. Tanner. 1979. Greatly decreased susceptibility of nonmetabolizing cells towards detergents. Proc. Natl. Acad. Sci. USA 76:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshlukova, S. E., T. L. Lloyd, M. W. B. Araujo, and M. Edgerton. 1999. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274:18872-18879. [DOI] [PubMed] [Google Scholar]
- 22.Koshlukova, S. E., M. W. B. Araujo, D. Baev, and M. Edgerton. 2000. Released ATP is an extracellular cytotoxic mediator in salivary histatin 5-induced killing of Candida albicans. Infect. Immun. 68:6848-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesage, F., E. Guillemare, M. Fink, F. Duprat, M. Lazdunski, G. Romen, and J. Barhanin. 1996. A pH-sensitive outward rectifier K+ channel with two pore domains and novel gating properties. J. Biol. Chem. 271:4183-4187. [DOI] [PubMed] [Google Scholar]
- 24.Lugardon, K., S. Chasserot-Golaz, A.-E. Kieffer, M.-D. Regine, G. Nullans, B. Kieffer, D. Aunis, and M.-H. Metz-Boutigue. 2001. Structural and biological characterization of chromofungin, the antifungal chromogranin A-(47-66)-derived peptide. J. Biol. Chem. 276:35875-35882. [DOI] [PubMed] [Google Scholar]
- 25.Morschhäuser, J., S. Michel, and P. Staib. 1999. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 32:547-556. [DOI] [PubMed] [Google Scholar]
- 26.Murakami, M., T. Ohtake, R. A. Dorschner, and R. L. Gallo. 2002. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J. Dent. Res. 81:845-850. [DOI] [PubMed] [Google Scholar]
- 27.Oppenheim, F. G., T. Xu, F. M. McMillian, S. M. Levitz, R. D. Diamond, and G. D. Offner. 1988. Histatins, a novel family of histidine-rich proteins in human parotid secretion. J. Biol. Chem. 263:7472-7477. [PubMed] [Google Scholar]
- 28.Roberts, S. K., M. Fischer, G. K. Dixon, and D. Sanders. 1999. Divalent cation block of inward currents and low-affinity K+ uptake in Saccharomyces cerevisiae. J. Bacteriol. 181:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid, J. D., W. Lukas, R. Shafaatian, A. Bertl, C. Scheurmann-Kettner, H. R. Guy, and R. A. North. 1996. The Saccharomyces cerevisiae outwardly rectifying potassium channel (DUK1) identifies a new family of channels with duplicated pore domains. Receptors Channels 4:51-62. [PubMed] [Google Scholar]
- 30.Ruissen, A. L. A., J. Groenink, E. J. Helmerhorst, E. Walgreen-Weterings, W. Van't Hof, E. C. I. Veerman, and A. V. N. Amerongen. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356: 361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sesti, F., T. M. Shih, N. Nikolaeva, and S. A. N. Goldstein. 2001. Immunity to K1 killer toxin: internal TOK1 blockade. Cell 105:637-644. [DOI] [PubMed] [Google Scholar]
- 33.Skipper, N., and H. Bussey. 1977. Mode of action of yeast toxins: energy requirement for Saccharomyces cerevesiae killer toxin. J. Bacteriol. 129:668-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slayman, C. L. 1980. Transport control phenomena in Neurospora, p. 179-190. In R. M. Spanswick, W. J. Lucas, and J. Dainty (ed.), Plant membrane transport: current conceptual issues. Elsevier, Amsterdam, The Netherlands.
- 35.Slayman, C. L., W. S. Long, and C. Y.-H. Lu. 1973. The relationship between ATP and an electrogenic pump in the plasma membrane of Neurospora crassa. J. Membr. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 36.Tsai, H., P. A. Raj, and L. A. Bobek. 1996. Candidacidal activity of recombinant salivary histatin-5 and variants. Infect. Immun. 64:5000-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Belle, D., and B. Andre. 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13:389-398. [DOI] [PubMed] [Google Scholar]
- 38.Xu, T., M. Levitz, R. Diamond, and F. Oppenheim. 1991. Anticandidal activity of major salivary histatins. Infect. Immun. 70:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, Y. Y., I. Ambudkar, H. Yamagishi, W. Swaim, T. J. Walsh, and B. C. O'Connell. 1999. Histatin 3-mediated killing of Candida albicans: effect of extracellular salt concentration on binding and internalization. Antimicrob. Agents Chemother. 43:2256-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Peron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, X.-L., B. Vaillant, S. H. Loukin, C. Kung, and Y. Saimi. 1995. YKC1 encodes the depolarization-activated K+ channel in the plasma membrane of yeast. FEBS Lett. 373:170-176. [DOI] [PubMed] [Google Scholar]