Abstract
Although many studies have been conducted to identify single nucleotide polymorphisms (SNPs) in humans, few studies have been conducted to identify alternative forms of natural genetic variation, such as insertion and deletion (INDEL) polymorphisms. In this report, we describe an initial map of human INDEL variation that contains 415,436 unique INDEL polymorphisms. These INDELs were identified with a computational approach using DNA re-sequencing traces that originally were generated for SNP discovery projects. They range from 1 bp to 9989 bp in length and are split almost equally between insertions and deletions, relative to the chimpanzee genome sequence. Five major classes of INDELs were identified, including (1) insertions and deletions of single-base pairs, (2) monomeric base pair expansions, (3) multi-base pair expansions of 2–15 bp repeat units, (4) transposon insertions, and (5) INDELs containing random DNA sequences. Our INDELs are distributed throughout the human genome with an average density of one INDEL per 7.2 kb of DNA. Variation hotspots were identified with up to 48-fold regional increases in INDEL and/or SNP variation compared with the chromosomal averages for the same chromosomes. Over 148,000 INDELs (35.7%) were identified within known genes, and 5542 of these INDELs were located in the promoters and exons of genes, where gene function would be expected to be influenced the greatest. All INDELs in this study have been deposited into dbSNP and have been integrated into maps of human genetic variation that are available to the research community.
A number of studies have been conducted to identify single nucleotide polymorphisms (SNPs) in the genomes of diverse humans. SNPs have been identified in specific genes (Nickerson et al. 1998; Rieder et al. 1999; Taillon-Miller et al. 1999; Taillon-Miller and Kwok 2000), on whole chromosomes (Mullikin et al. 2000; Dawson et al. 2001), and throughout the human genome (Altshuler 2000; The International SNP Map Working Group 2001). Most recently, the International HapMap project has completed an extensive SNP discovery effort to further increase the density of SNPs across the human genome (The International HapMap Consortium 2003). The human SNP map now has grown to over ten million (10,430,753) non-redundant polymorphisms due to these projects (www.ncbi.nlm.nih.gov/SNP; build 125). This collection of SNPs has been used to develop a comprehensive haplotype (HapMap) of the human genome, which will be useful for genetic linkage studies in humans (Daly et al. 2001; Patil et al. 2001; Stephens et al. 2001; Gabriel et al. 2002; The International HapMap Consortium 2003; The International HapMap Consortium 2005).
Some of these SNPs are directly responsible for phenotypic differences in humans, including differences in (1) physical traits, (2) susceptibility to diseases, and (3) physiological responses to the environment (Judson et al. 2002). Most of these functionally important SNPs are located within genes. For example, 64,549 non-synonomous human SNPs have been identified that cause amino acid substitutions in proteins (www.ncbi.nlm.nih.gov/SNP; build 125). SNPs also have been identified within other critical sites of genes, such as the promoters, 5′ and 3′ untranslated regions, and predicted splice sites (www.ncbi.nlm.nih.gov/SNP; build 125). Thus, SNPs can alter human genes (and phenotypes) through a variety of mechanisms.
In contrast to SNPs, which have been studied extensively, other forms of natural genetic variation in humans have received relatively little attention. At least some of these alternative forms of variation, such as insertion and deletion (INDEL) polymorphisms, are abundant in the genomes of model organisms and are expected to be abundant in humans as well. For example, studies of genetic variation in Drosophila melanogaster and Caenorhabditis elegans have shown that INDELs represent between 16% and 25% of all genetic polymorphisms in these species (Berger et al. 2001; Wicks et al. 2001). A study of genetic variation on human chromosome 22 has suggested that humans harbor similar levels of INDEL variation (INDELs represented 18% of the polymorphisms on this chromosome; Dawson et al. 2001). Based on these estimates, INDELs are likely to represent between 16% and 25% of all sequence polymorphisms in humans. Therefore, given that dbSNP currently contains ~10 million unique (rs) SNPs (www.ncbi.nlm.nih.gov/SNP; build 125), human populations are expected to collectively harbor at least 1.6–2.5 million INDEL polymorphisms. Unfortunately, INDEL discovery efforts have lagged significantly behind SNP discovery efforts, and relatively few INDELs have been identified.
In this report, we describe a new computational strategy to systematically identify INDEL polymorphisms in the genomes of diverse humans. We used this strategy to build an initial map of human INDEL variation that contains 415,436 novel INDELs distributed throughout the human genome. Approximately 36% of these INDELs are located within the promoters, introns, and exons of known genes. Thus, like SNPs, some of these INDELs are expected to have an impact on human gene function. All of these INDELs have been deposited into dbSNP and have been integrated into publically available maps of genetic variation. Fully integrated maps of natural genetic variation that include SNPs, INDELs, and other polymorphisms will be more useful than SNP maps alone for identifying polymorphisms that directly influence human phenotypes and diseases.
Results
Our INDEL discovery strategy involved mining insertion and deletion polymorphisms from DNA resequencing traces that originally were generated by genome centers for SNP discovery projects (The International SNP Map Working Group 2001; The International HapMap Consortium 2003). Because these traces were generated by shotgun sequencing the genomic DNA of 36 diverse humans, we expected them to harbor additional forms of natural genetic variation, including INDELs. Thus, we developed a new computational pipeline that utilized these traces to identify small INDELs in the human genome. The pipeline entailed (1) first mapping the traces to unique locations in the reference human genome, and (2) then aligning mappable traces to the equivalent genomic regions of the reference sequence to identify INDEL polymorphisms in the 1 bp to 10,000 bp size range (see Methods).
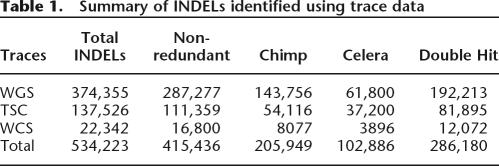
We used our computational pipeline to mine 534,223 INDELs from three independent sets of DNA sequencing traces (see Methods; Table 1; Supplemental Tables chr1–chrY). The majority of these INDELs (374,355) were identified from whole genome shotgun (WGS) traces that were generated by the HapMap Consortium using the genomic DNA of eight African Americans (Table 1; The International HapMap Consortium 2003). An additional 137,526 INDELs were identified from traces that were generated by The SNP Consortium (TSC) using the genomic DNA of 24 diverse humans (Table 1; The International SNP Map Working Group 2001). Finally, 17,217 INDELs were identified from whole chromosome 20 shotgun (WCS) traces that were generated by the Sanger Center from four diverse humans (Table 1; The International HapMap Consortium 2003). Redundant INDELs (118,787) were identified at least twice in our data sets and these were eliminated to create a non-redundant set. Thus, a total of 415,436 non-redundant INDELs were identified from the three diverse human populations, ranging from 1 bp to 9989 bp in length.
Table 1.
Summary of INDELs identified using trace data
We next mapped all INDEL candidates to the chimpanzee genome to further confirm these INDELs and to identify the ancestral allele, where possible. A total of 205,949 of our 415,436 non-redundant INDEL candidates (49.5%) were mapped successfully to unique positions in the chimpanzee genome (see Methods; Table 1; Supplemental Tables chr1–chrY). On the basis of these results, we determined that our INDELs were split almost equally between insertions and deletions in the human genome (47% of the INDELs were insertions in the human genome, and 53% were deletions, relative to the chimp genome). Thus, the underlying mechanisms that have led to DNA insertions and deletions in the human genome appear to have functioned at similar rates during the past ~6 million years (the time since the last common ancestor of these organisms). We also mapped our INDEL candidates to the Celera human genome assembly to further confirm our INDEL alleles. A total of 102,886 (24.7%) of our trace alleles were confirmed in the Celera assembly. By combining these results with the chimp data outlined above and with redundant trace data, we identified a total of 286,180 double-hit INDELs (Table 1; Supplemental Tables chr1–chrY). Thus, a large fraction of our INDELs (68.8%) were validated in the chimp genome or in other human genomes through these comparisons.
Five major INDEL classes were identified by analyzing the DNA sequences of our INDELs: (1) insertions and deletions of single-base pairs, (2) monomeric base pair expansions, (3) multibase pair expansions of 2–15 bp repeat units, (4) transposon insertions, and (5) INDELs containing apparently random DNA sequences (Table 2). Insertions and deletions of single-base pairs represented approximately one third (29.1%) of all INDELs in our collection. The majority of these INDELs were A:T and T:A base pairs, and these two classes together accounted for 84% of the single-base pair INDELs. Monomeric base pair expansions of various lengths and multi-base repeat expansions also represented almost one third (29.5%) of the INDELs in our collection (Table 2). This latter class includes the (CA)n repeat expansions that are commonly used as genetic markers and the trinucleotide repeat expansions that have been shown to cause human diseases (Warren et al. 1987). Although a variety of DNA sequences can participate in these repeat expansions, some sequences appear to undergo expansion much more readily than others. For example, six of the possible twelve dimeric expansions, (AC)n, (GT)n, (TG)n, (CA)n, (TA)n, and (AT)n, were much more abundant than the remaining dimeric expansions in our data sets (Table 2). Similar preferences were observed for larger repeat units (Table 2). Our repeat expansions were compared with maps of known micro-satellites, and 41% of our expansions mapped to such regions (Supplemental Tables chr1–chrY). In addition to repeat expansions, we also identified INDELs that were caused by de novo transposon insertions. Overall, transposons accounted for a small fraction of the INDELs (0.59%), and these polymorphisms have been described elsewhere in detail (Bennett et al. 2004). The remaining INDELs (40.8%) had a wide spectrum of apparently random DNA sequences ranging from 2 bp to 9989 bp in length (listed as “other” in Table 2). The complete size distribution of these “other” INDELs is provided in Supplemental Table 1. Greater than 99% of these “other” INDELs are <100 bp in length. Approximately 90% are repeated at least once in the genome (this is largely due to INDELs that are 2 bp to 20 bp, which may be repeated at least once by chance alone).
Table 2.
Classification of INDELs
Validation studies
We conducted several independent validation studies in which polymorphisms from our collections were subjected to PCR verification in human populations. In each case, a randomly selected INDEL candidate was amplified by PCR and the resulting PCR product was analyzed for the presence of the INDEL. In some cases, the INDEL affected a restriction endonuclease site and these INDELs were examined by digesting the PCR products with an appropriate restriction endonuclease. The remaining INDELs changed the sizes of the PCR products such that these changes could be observed directly with agarose gels (Fig. 1). Our validation studies were focused on polymorphisms that were identified from the TSC trace data, since genomic DNA samples were available for all 24 of the original individuals whose DNA was pooled and sequenced to generate these traces (Collins et al. 1999). Any polymorphism that was identified using the TSC trace data also should be found in the genome of at least one of these 24 individuals. Thus, the TSC trace data, together with the genomic DNA samples from these 24 individuals, represent an excellent system to validate polymorphism discovery pipelines (Collins et al. 1999).
Figure 1.
PCR validation study using the TSC panel. A PCR assay for INDEL detection is shown. (A) Arrows represent PCR primers. If a given segment of DNA is present (black box), a long PCR product (L) is produced. If it is absent, a short PCR product (S) is produced. (B) A PCR validation assay is shown for INDEL 647,421 located on chromosome 22 (also listed in Table 3 and Supplemental Table 2). The reference genome sequence indicated the presence of a 24-bp repeat of (AC)n at the coordinates given (Table 3). A trace from the TSC collection indicated that at least one individual from the Coriell panel lacked this DNA segment. Both alleles were identified in the Coriell panel of 24 individuals (Collins et al. 1999). Small (<10 bp) INDELs were located within predicted restriction sites and the PCR products were digested with an appropriate restriction enzyme to detect the INDEL (not shown).
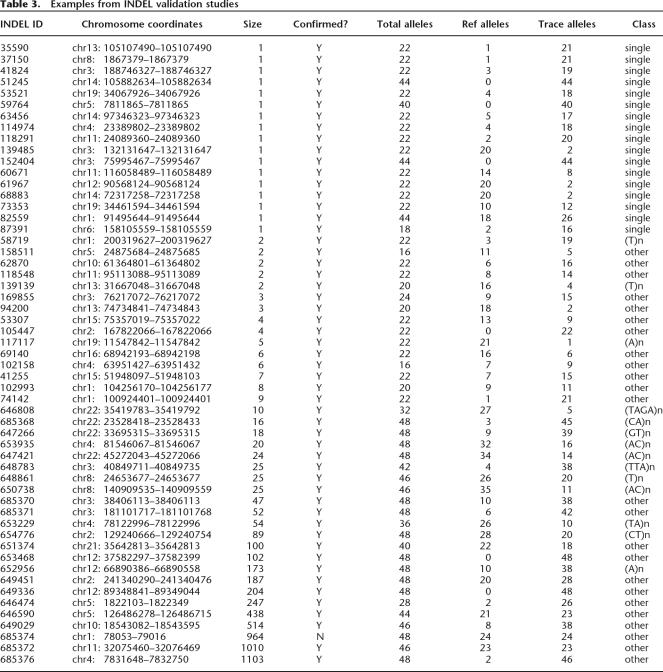
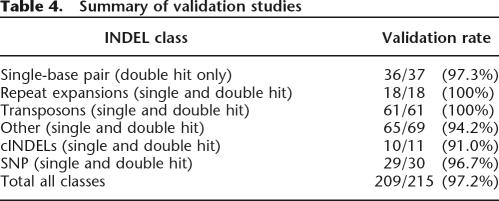
We systematically examined each of the INDEL classes listed in Table 2 in an effort to validate these classes in the TSC data set. Our initial validation efforts with single-base pair INDELs yielded relatively low validation rates (only about half of these INDELs were validated in our initial attempts). A previous study also reported relatively low validation rates for single-base pair INDELs (Weber et al. 2002). These results collectively indicate that single-base pair INDELs are particularly difficult to identify accurately from trace data, even when quality scores are used to guide INDEL discovery. We next examined a number of parameters in our pipeline to improve these results. Like the double-hit SNPs that have been described previously (The International HapMap Consortium 2003), we found that double-hit single-base INDELs had very high validation rates (36/37 or 97.3% were verified in the TSC population; Tables 3,4; Supplemental Table 2). On the basis of these results, we modified our INDEL discovery pipeline to include only double-hit single-base INDELs in our final collection of 415,436 INDELs (Table 1).
Table 3.
Examples from INDEL validation studies
Table 4.
Summary of validation studies
We also conducted additional validation studies with 18 INDELs from the “repeat expansion” class and 69 INDELs from the “other” class (Table 2). We observed validation rates of 18/18 (100%) and 65/69 (94.2%) for these studies, respectively (Fig. 1; Tables 3,4; Supplemental Table 2). We also examined 11 of our cINDELs (coding INDELs, see below) and confirmed 10/11 (91%) of these INDELs in the TSC population (Table 4; Supplemental Table 2). These 11 cINDELs belonged to the INDEL classes listed in Table 2 (single-base pair, repeat expansions, and “other”) and are likely to have identical validation rates as these classes. In a previous study, we found a validation rate of 61/61 (100%) for the transposon class of INDELs (Table 4; Bennett et al. 2004). In another previous study, we observed a validation rate of 29/30 (97%) for SNPs discovered with our pipeline (Table 4; Tsui et al. 2003). Therefore, with the exception of the single-base pair INDELs discussed above, our computational pipeline yields accurate predictions for the major polymorphism classes identified in our study. By including only double-hit single-base pair INDELs in our final collection, we also achieved high levels of accuracy for this class (Tables 3,4; Supplemental Table 2). Our pipeline has an overall accuracy level of 209/215 or 97.2% when combining all of the validation studies (Table 4).
We also found that a fraction of our INDELs had been independently discovered by other laboratories using completely different methods. In fact, 716 of the human INDELs in dbSNP (build 125) were discovered independently by our laboratory and other laboratories using a range of methods (www.ncbi.nlm.nih.gov/SNP; build 125; Supplemental Table 3). The allelic frequencies of these INDELs were measured by these other laboratories, and all of these INDELs were verified to exist in human populations (www.ncbi.nlm.nih.gov/SNP; build 125; Supplemental Table 3).
Finally, we also compared our INDELs with the 641 large genomic deletions that were identified in regions of the HapMap that had diminished (or absent) SNP genotyping signals (Conrad et al. 2006; McCarroll et al. 2006). Since the majority of these deletions occurred within or near segmental duplications, we did not expect to detect them efficiently with our pipeline (we eliminated traces that mapped to duplicated regions of the genome to avoid trace mapping errors). The HapMap deletions also generally exceeded the range of our INDEL discovery. Thus, we only detected a few of these HapMap deletions in our data sets (data not shown).
Genomic distribution of INDEL variation
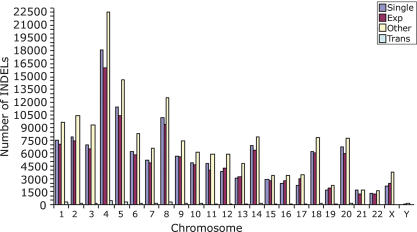
We next examined the genomic distribution of our INDELs and found that they were located throughout the genome at an average density of one INDEL per 7.2 kb of DNA (Fig. 2). INDELs generally were distributed according to the amount of DNA that was present on each chromosome (Fig. 2). However, in some cases (chromosomes 4, 5, 8, 14, and 18), the amount of INDEL variation was increased due to higher trace coverage on these chromosomes. Chromosome 20 also had higher levels of INDEL variation due to the inclusion of chromosome 20-specific WCS trace data in our experiments (Table 1). In contrast, chromosomes X and Y initially appeared to have lower levels of INDEL variation compared with the autosomes. Chromosome X had approximately half the level of INDEL variation found on the average autosome, whereas chromosome Y had only 5% of the INDEL variation on the average autosome (Fig. 2). Upon normalization to the number of trace bases that mapped to these chromosomes, however, the levels of INDEL variation on chromosomes X and Y were similar to the autosomes. Overall, the normalized averages for all chromosomes were similar and fell within the range of 1 INDEL for every 5.1–13.2 kb of genomic DNA (Supplemental Table 4).
Figure 2.
Genomic distribution of INDELs. The number of INDELs is plotted for each chromosome. The four major classes of INDELs shown in Table 2 are plotted (Single, single-base pair INDEL; Exp, single- and multi-base expansions; Other, other; Trans, transposons). Note that the INDELs generally are distributed throughout the genome according to the amount of DNA that is present on each chromosome. Chromosomes 4, 5, 8, 14, 18, 20, X, and Y are the exceptions. Chromosome 20 has more INDELs than average due to the inclusion of WCS traces from chromosome 20 in our experiments. The remaining exceptions had higher levels of trace coverage in the WGS and/or TSC trace sets.
To examine this variation on a finer scale, we next placed all of the polymorphisms that were discovered in this study (including both INDELs and SNPs) into 200 kb bins across each chromosome. The number of INDELs per 200 kb bin varied considerably across each chromosome and ranged from 0 to 4559 (Fig. 3; Supplemental Tables chr1–chrY). Similar results were observed for SNPs, which ranged from 0 to 59,042 SNPs per 200 kb bin. After normalizing these INDEL and SNP data to the number of trace bases that mapped to each bin, several major variation hotspots were identified on chromosomes 1, 2, 3, 4, 6, 7, 9, 10, 13, and 20, along with smaller hotspots throughout the genome (Supplemental Tables chr1–chrY). These hotspots contained up to 24-fold higher levels of INDEL variation and up to 48-fold higher levels SNP variation, compared with the averages for these chromosomes (Fig. 3). In most cases, both INDELs and SNPs were elevated at these sites, suggesting that these regions represent general hotspots for genetic variation. INDEL-only and SNP-only hotspots also were identified that contained statistically significant levels of only one type of variation (Supplemental Tables chr1–chrY; see Methods).
Figure 3.
Fine-scale maps of chromosomal INDEL and SNP variation. INDEL and SNP variation graphs are shown for 200 kb bins across chromosomes 1, 3, and 18. The number of traces mapping to each bin and the number of trace bases in each bin also are charted. Chromosome 1 has several large variation hotspots in which the INDEL and/or SNP variation levels are elevated. Chromosome 3 has one large variation hotspot along with smaller hotspots. Chromosome 18 lacks large hotspots. The color key is indicated in the top panel. Similar graphs were generated for all human chromosomes and are found in Supplemental Tables chr1–chrY.
Distribution of INDELs relative to human genes
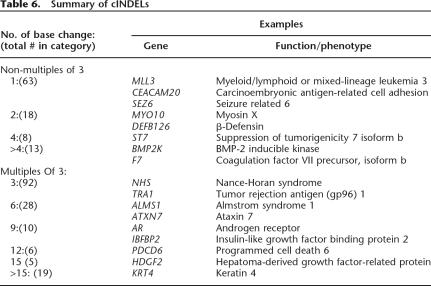
We next examined the distribution of our 415,436 INDELs relative to human genes and found that 148,335 INDELs (35.7%) were located within known genes (Tables 5,6; Supplemental Tables 5, 6). Moreover, 5542 of these INDELs were located in the promoters and exons of these genes, where gene function would be expected to be influenced the greatest. Interestingly, 262 INDELs were located within the coding regions of genes (Table 6; Supplemental Table 6). One hundred and two (38.9%) of these cINDELs were predicted to cause frameshifts that would lead to the premature termination of the encoded proteins (Table 6; Supplemental Table 6). Most of these cINDELs would be expected to diminish or altogether abolish gene function, and we propose that they represent recessive alleles of these genes that are carried by human populations. The remaining 160 cINDELs were multiples of three base pairs and thus resulted in the precise insertion or deletion of codons (Table 6; Supplemental Table 6). Overall, these data indicate that INDELs, like SNPs, are likely to have an impact on human gene function.
Table 5.
Summary of INDELs in known RefSeq genes
Table 6.
Summary of cINDELs
We also examined the INDEL densities within all annotated human genes (Supplemental Table 5). The average INDEL density within genes (one INDEL per 6.3 kb) was very similar to the average INDEL density observed for the whole genome (one INDEL per 7.2 kb). However, at least 84 genes had INDEL densities that were up to two orders of magnitude higher than this average (in the range of one INDEL per 25–500 bp; Supplemental Table 5). For example, the 5-kb RPS3A gene on chromosome 4 had 117 INDELs. With a density of one INDEL per 42 bp, this gene had one of the highest INDEL densities of all human genes. At least 83 other genes had similarly elevated levels of INDEL variation (Supplemental Table 5). In most cases, these sites also had elevated levels of SNP variation. Thus, like the macro hotspots described above (Fig. 3), many of these micro hotspots within genes also appear to represent general hotspots of genetic variation.
How many common INDELs in human populations?
We identified a total of 3.3 million SNPs and 534,223 INDELs from the three populations examined in this study (for a total of 3.8 million polymorphisms). On the basis of these results, we estimate that INDELs represent ~15.6% of the polymorphisms discovered. This estimate is in excellent agreement with a previous measurement of polymorphisms on human chromosome 22 in which 18% of the polymorphisms were INDELs (Dawson et al. 2001). Given that dbSNP has ~10,000,000 unique SNPs (www.ncbi.nlm.nih.gov/SNP; build 125), our data indicate that human populations should harbor a minimum of 1.56 million INDELs. Therefore, our 415,436 unique INDELs represent ~27% of the expected INDEL polymorphisms in human populations.
Discussion
Despite the fact that both SNPs and INDELs generally are abundant in genomes, INDELs (particularly in the 1 bp to 10,000 bp range) have received relatively little attention in humans. Several lines of evidence suggested that such INDELs may be abundant in humans, and we now confirm that this is the case. We have identified over 415,000 unique INDELs in the genomes of 36 diverse humans using DNA sequencing traces that initially were generated for SNP discovery projects. This study was conducted with only 16 million of the ~40 million DNA re-sequencing traces that were generated by TSC and the HapMap project. Thus, our methods now could be applied to the remaining traces (and to other resequencing data such as the Celera human genome assembly) to develop a more complete map of INDEL variation in humans. Comprehensive variation maps, which include both SNPs and INDELs, will be more effective than SNP maps alone for identifying variants that underlie specific human phenotypes and diseases.
INDELs and human diseases
Like other forms of natural genetic variation in humans such as SNPs, INDEL polymorphisms are of great interest because they can alter human phenotypes and also cause human diseases. One of the most common genetic diseases in humans, cystic fibrosis, is frequently caused by an allele of the CFTR gene that contains a three-base pair deletion. This deletion leads to the elimination of a single amino acid from the encoded protein, which, in turn, leads to the disease (Collins et al. 1987). Trinucleotide expansion diseases, such as Fragile X Syndrome, likewise are caused by DNA insertions that result from the expansion of short trinucleotide repeat units (Warren et al. 1987). Transposable genetic elements, by integrating into genomes, also produce DNA insertions that may cause human diseases (Ostertag and Kazazian 2001). In particular, Alu, L1, and SVA transposon insertions have been found to disrupt gene function and cause human diseases such as hemophilia, neurofibromatosis, muscular dystrophy, and cancer (Ostertag and Kazazian 2001). Our collection of 415,000 INDELs contains these very same classes of INDELs. For example, we identified a 63-bp (CAG)n repeat expansion allele within a region of the human ATXN3 gene that previously has been reported to undergo repeat expansion. We identified 262 cINDELs that were located within the coding regions of genes and up to 34 of these alleles had been identified previously in the Human Gene Mutation Database (Stenson et al. 2003).
INDEL hotspots
We identified hotspots of INDEL variation that contain up to 24-fold higher levels of INDEL variation compared with the remaining regions of the genome. Most of these sites also represented SNP hotspots (with up to 48-fold higher levels of SNP variation), indicating that these regions are general hotspots of natural genetic variation (although some INDEL-only and SNP-only hotspots also were identified). On a finer scale, we also identified at least 84 smaller hotspots that were located within genes (where the INDEL and SNP densities were up to two orders of magnitude higher than most genes). Several explanations have been put forth previously to explain sites of hypervariation in genomes (The International SNP Map Working Group 2001). In some cases, these unusual levels of genetic variation may reflect older evolutionary ages for the DNA segments involved, or unusual levels of homologous recombination in the region. In other cases, high levels of variation may indicate that balancing selection, in which diverse alleles are maintained under selective pressure, is occurring in the region for a biological purpose. Finally, it is also possible that such sites lack functionally important sequences and lack selective pressure altogether. Additional studies will be necessary to determine the underlying causes and possible biological roles of these hotspots.
Utility of our INDEL map
We envision that our initial INDEL map will be useful for a variety of purposes. As outlined above, we expect that some of these INDELs will affect human genes and, therefore, will alter human phenotypes or cause diseases. Thus, it would be useful to integrate our emerging map of human INDEL variation into the HapMap. This would provide a more complete description of the variation that is carried in each haplotype block and would facilitate efforts to identify the specific variation that influences human phenotypes and diseases. Our INDELs could be genotyped in the same DNA trios that were used to generate the HapMap in order to complete this goal. Since the majority of INDELs examined in our validation studies (147/185 or 80%) had minor allelic frequencies that were greater than 5%, a large fraction of our INDELs also could serve as genetic markers for the HapMap. Previous studies have indicated that small INDELs resembling those detected by our pipeline can be used effectively as genetic markers in humans (Weber et al. 2002; Bhangale et al. 2005).
Methods
General computational methods
All sequences, databases, and programs were generated or obtained as follows. The human genome sequence was obtained from the University of California Santa Cruz (UCSC) browser (Kent et al. 2002; www.genome.ucsc.edu). Trace data and accompanying quality files were obtained from The SNP Consortium (TSC; http://snp.cshl.org) or from the National Center for Bio-technology Information (NCBI; www.ncbi.nlm.nih.gov). BLAST and Vecscreen programs also were obtained from NCBI. Repeat-Masker was obtained from Arian Smit (Institute for Systems Biology, Seattle, WA). RepBase was obtained from Jerzy Jurka (Jurka 2000). Custom MySQL databases and PERL scripts were generated as necessary. All analysis was performed locally on SUN SunFire v40z or Dell Power Edge 2500 servers running Linux operating systems.
INDEL discovery pipeline
Our polymorphism discovery pipeline is described below. This pipeline has been used previously to identify SNPs (Tsui et al. 2003) and transposon insertions (Bennett et al. 2004). Traces obtained from TSC or NCBI first were trimmed to remove vector sequences using the VecScreen system from NCBI. Low quality regions containing at least five bases in a row with Phred scores below 25 (Ewing and Green 1998) then were trimmed using a custom PERL script. The longest high quality (LHQ) region from each trace was selected for further evaluation, and the remaining trimmed regions of the traces were set aside. The LHQ regions were further required to have average Phred scores of at least 25 and had to be longer than 100 bases in length. Repeats were identified and masked within the LHQ region of each trimmed trace using RepeatMasker and RepBase. The longest unmasked “anchor” region, which had to be at least 50 bases in length, then was used to assign each trace to a unique genomic location in build hg17 of the human genome using BLAST. Successfully mapped anchor sequences were required to have a single 100% match to a unique genomic location. Traces containing anchor sequences with more than one perfect match were set aside to avoid traces that mapped to segmental duplications (Bailey et al. 2002). The LHQ regions of successfully mapped traces then were unmasked and aligned to their assigned genomic locations using BLAST2seq (NCBI). Polymorphisms were mined from these alignments using custom PERL scripts. We required the five bases on each side of a polymorphism candidate to have Phred scores that were 25 or higher. For SNP discovery, the SNP base also was required to have a Phred score of 25 or higher. Single-base pair INDELs were screened to identify double-hit INDELs, and only these were included in our final collections. Since BLAST only allows for up to a 16-base gap in the alignments, a custom PERL script was developed to identify INDELs that were larger than 16 bp in length. Upon encountering a region in the alignment that no longer matched the query, this program split trace data into two blocks. The first block (which matched the query) was maintained at the original position, whereas the second block (which did not match the query) was moved over one base at a time until a perfect match was obtained, or a distance of 10,000 bases (the maximum distance allowed by the program) was reached.
Mapping INDELs to the chimpanzee genome and identification of double-hit INDELs
The pipeline outlined above also was used to map our INDELs to the chimp and Celera genomes. Traces that were found to harbor INDELs first were mapped to the chimp genome (build pantro1). BLAST2seq alignments were generated as outlined above and used to identify the status of the chimp sequence at the INDEL site. The INDEL was successfully mapped if the trace allele or the reference human genome allele was detected at the equivalent position. The same approach was used to screen the Celera human genome sequence to further confirm our INDELs. Redundant INDELs that were detected in at least two separate traces also were identified (Supplemental Table 7) and cross-referenced in our INDEL database. This information collectively was used to identify double-hit INDELs. In order to be assigned double-hit status, an INDEL was required to have been detected independently at least twice among the chimp, Celera, and/or trace data. Double-hit status is indicated in Supplemental Tables chr1–chrY for all INDELs.
PCR validation studies
Validation studies were carried out as described previously (Tsui et al. 2003; Bennett et al. 2004). In each case, a set of oligonucleotide primers was designed to amplify genomic segments of ~0.2–1.5 kb that contained the region carrying the polymorphism (Supplemental Table 1). For small (<10 bp) INDELs, the presence or absence of the INDEL was assessed by cutting the PCR product with a restriction enzyme. In each case, the INDEL was located within the restriction endonuclease site and differentially affected cleavage. For larger INDELs, the presence or absence of an INDEL was assessed by examining the sizes of PCR products by running the PCR reactions on 1%–2% agarose gels. Each PCR assay was performed on 10–24 of the genomic DNAs from the TSC diversity panel (Collins et al. 1999). Genomic DNA for the TSC diversity panel was obtained from the Coriell Cell Repository. An INDEL was confirmed if the trace allele was observed at least once in the TSC diversity panel.
Statistical analysis of variation hotspots
The genome was split into 200 kb bins and the variation levels were calculated relative to the number of bases of trace data that mapped to each bin. These values were plotted using curve fitting programs in the “R” package and followed linear curves with strong statistical significance (these curves are depicted for each chromosome in Supplemental Tables chr1–chrY). A mean and standard deviation was determined for INDEL and SNP variation levels for each chromosome (Supplemental Table 4) and one-sided z tests were performed for each bin. The resulting P-values were analyzed by FDR analysis, which controls for multiple-sampling error (Storey and Tibshirani 2003). A conservative FDR cutoff of 0.0001 was used, and the resulting (highly significant) hotspots are indicated in Supplemental Tables chr1–Y along with P-values.
Acknowledgments
This work was supported by an equipment grant from SUN Microsystems (W.S.P. and S.E.D.) and NIH grant 1R01HG002898 (S.E.D.). We thank Laura Coleman and Summer Goodson for help with PCR experiments and Andrew Hill for help with statistical analysis of the data. We thank Shari Corin for helpful discussions and critical review of the manuscript. We thank The SNP Consortium, the HapMap Consortium, dbSNP, and U.C. Santa Cruz for the use of their data. Finally, we thank the anonymous reviewers for helpful constructive feedback.
Footnotes
[Supplemental material is available online at www.genome.org. All INDELs described in this manuscript have been deposited into dbSNP under the “Devine_lab” handle.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4565806.
References
- Altshuler D., Pollara V.J., Cowles C.R., Van Etten W.J., Baldwin J., Linton L., Lander E.S., Pollara V.J., Cowles C.R., Van Etten W.J., Baldwin J., Linton L., Lander E.S., Cowles C.R., Van Etten W.J., Baldwin J., Linton L., Lander E.S., Van Etten W.J., Baldwin J., Linton L., Lander E.S., Baldwin J., Linton L., Lander E.S., Linton L., Lander E.S., Lander E.S. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bennett E.A., Coleman L.E., Tsui C., Pittard W.S., Devine S.E., Coleman L.E., Tsui C., Pittard W.S., Devine S.E., Tsui C., Pittard W.S., Devine S.E., Pittard W.S., Devine S.E., Devine S.E. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J., Suzuki T., Senti K.A., Stubbs J., Schaffner G., Dickson B.J., Suzuki T., Senti K.A., Stubbs J., Schaffner G., Dickson B.J., Senti K.A., Stubbs J., Schaffner G., Dickson B.J., Stubbs J., Schaffner G., Dickson B.J., Schaffner G., Dickson B.J., Dickson B.J. Genetic mapping with SNP markers in Drosophila . Nat. Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- Bhangale T.R., Rieder M.J., Livingston R.J., Nickerson D.A., Rieder M.J., Livingston R.J., Nickerson D.A., Livingston R.J., Nickerson D.A., Nickerson D.A. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 2005;14:59–69. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Drumm M.L., Cole J.L., Lockwood W.K., Vande Woude G.F., Iannuzzi M.C., Drumm M.L., Cole J.L., Lockwood W.K., Vande Woude G.F., Iannuzzi M.C., Cole J.L., Lockwood W.K., Vande Woude G.F., Iannuzzi M.C., Lockwood W.K., Vande Woude G.F., Iannuzzi M.C., Vande Woude G.F., Iannuzzi M.C., Iannuzzi M.C. Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science. 1987;235:1046–1049. doi: 10.1126/science.2950591. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Brooks L.D., Chakravarti A., Brooks L.D., Chakravarti A., Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1999;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Carter N.P., Hurles M.E., Pritchard J.K., Hurles M.E., Pritchard J.K., Pritchard J.K. A high resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Daly M.J., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S., Schaffner S.F., Hudson T.J., Lander E.S., Hudson T.J., Lander E.S., Lander E.S. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- Dawson E., Chen Y., Hunt S., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Chen Y., Hunt S., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Hunt S., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Livingston S., Bumpstead S., Bruskiewich R., Sham P., Bumpstead S., Bruskiewich R., Sham P., Bruskiewich R., Sham P., Sham P., et al. A SNP resource for human chromosome 22: Extracting dense clusters of SNPs from the genomic sequence. Genome Res. 2001;11:170–178. doi: 10.1101/gr.156901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Higgins J., DeFelice M., Lochner A., Faggart M., DeFelice M., Lochner A., Faggart M., Lochner A., Faggart M., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- ———2005A haplotype map of the human genome Nature 4371299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Judson R., Salisbury B., Schneider J., Windemuth A., Stephens J.C., Salisbury B., Schneider J., Windemuth A., Stephens J.C., Schneider J., Windemuth A., Stephens J.C., Windemuth A., Stephens J.C., Stephens J.C. How many SNPs does a genome-wide haplotype map require? Pharmacogenomics. 2002;3:379–391. doi: 10.1517/14622416.3.3.379. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Hausler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Hausler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Hausler D., Roskin K.M., Pringle T.H., Zahler A.M., Hausler D., Pringle T.H., Zahler A.M., Hausler D., Zahler A.M., Hausler D., Hausler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Gabriel S.B., Lee C., Daly M.J., Lee C., Daly M.J., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Mullikin J.C., Hunt S.E., Cole C.G., Mortimore B.J., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Hunt S.E., Cole C.G., Mortimore B.J., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Cole C.G., Mortimore B.J., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Mortimore B.J., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Burton J., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Matthews L.H., Pavitt R., Plumb R.W., Sims S.K., Pavitt R., Plumb R.W., Sims S.K., Plumb R.W., Sims S.K., Sims S.K., et al. An SNP map of human chromosome 22. Nature. 2000;407:516–520. doi: 10.1038/35035089. [DOI] [PubMed] [Google Scholar]
- Nickerson D.A., Taylor S.L., Weiss K.M., Clark A.G., Hutchinson R.G., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Taylor S.L., Weiss K.M., Clark A.G., Hutchinson R.G., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Weiss K.M., Clark A.G., Hutchinson R.G., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Clark A.G., Hutchinson R.G., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Hutchinson R.G., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Stengard J., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Saloma V., Vartianen E., Boerwinkle E., Sing C.F., Vartianen E., Boerwinkle E., Sing C.F., Boerwinkle E., Sing C.F., Sing C.F. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat. Genet. 1998;19:216–217. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H., Kazazian H.H. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Patil N., Berno A.J., Hinds D.A., Barrett W.A., Doshi J.M., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Berno A.J., Hinds D.A., Barrett W.A., Doshi J.M., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Hinds D.A., Barrett W.A., Doshi J.M., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Barrett W.A., Doshi J.M., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Doshi J.M., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Hacker C.R., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Kautzer C.R., Lee D.H., Marjoribanks C., McDonough D.P., Lee D.H., Marjoribanks C., McDonough D.P., Marjoribanks C., McDonough D.P., McDonough D.P., et al. Blocks of limited haplotype diversity revealed by high resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- Rieder M.J., Taylor S.L., Clark A.G., Nickerson D.A., Taylor S.L., Clark A.G., Nickerson D.A., Clark A.G., Nickerson D.A., Nickerson D.A. Sequence variation in the human angiotensin converting enzyme. Nat. Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Abeysinghe S., Krawczak M., Cooper D.N., Krawczak M., Cooper D.N., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Stephens J.C., Schneider J.A., Tanguay D.A., Choi J., Acharya T., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Schneider J.A., Tanguay D.A., Choi J., Acharya T., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Tanguay D.A., Choi J., Acharya T., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Choi J., Acharya T., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Acharya T., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Stanley S.E., Jiang R., Messer C.J., Chew A., Han J.H., Jiang R., Messer C.J., Chew A., Han J.H., Messer C.J., Chew A., Han J.H., Chew A., Han J.H., Han J.H., et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P., Kwok P.Y., Kwok P.Y. A high-density single-nucleotide polymorphism map of Xq25-q28. Genomics. 2000;65:195–202. doi: 10.1006/geno.2000.6165. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P., Piernot E.E., Kwok P.Y., Piernot E.E., Kwok P.Y., Kwok P.Y. Efficient approach to unique single-nucleotide polymorphism discovery. Genome Res. 1999;9:499–505. [PMC free article] [PubMed] [Google Scholar]
- Tsui C., Coleman L.E., Griffith J.L., Bennett E.A., Goodson S.G., Scott J.D., Pittard W.S., Devine S.E., Coleman L.E., Griffith J.L., Bennett E.A., Goodson S.G., Scott J.D., Pittard W.S., Devine S.E., Griffith J.L., Bennett E.A., Goodson S.G., Scott J.D., Pittard W.S., Devine S.E., Bennett E.A., Goodson S.G., Scott J.D., Pittard W.S., Devine S.E., Goodson S.G., Scott J.D., Pittard W.S., Devine S.E., Scott J.D., Pittard W.S., Devine S.E., Pittard W.S., Devine S.E., Devine S.E. Single nucleotide polymorphisms (SNPs) that map to gaps in the human SNP map. Nucleic Acids Res. 2003;31:4910–4916. doi: 10.1093/nar/gkg664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren S.T., Zhang F., Licameli G.R., Peters J.F., Zhang F., Licameli G.R., Peters J.F., Licameli G.R., Peters J.F., Peters J.F. The fragile X sites in somatic cell hybrids: An approach for molecular cloning of fragile sites. Science. 1987;237:420–423. doi: 10.1126/science.3603029. [DOI] [PubMed] [Google Scholar]
- Weber J.L., David D., Heil J., Fan Y., Zhao C., Marth G., David D., Heil J., Fan Y., Zhao C., Marth G., Heil J., Fan Y., Zhao C., Marth G., Fan Y., Zhao C., Marth G., Zhao C., Marth G., Marth G. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H.A., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H.A., Gish W.R., Waterston R.H., Plasterk R.H.A., Waterston R.H., Plasterk R.H.A., Plasterk R.H.A. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]