Abstract
Essential genes are required for an organism's viability, and the ability to identify these genes in pathogens is crucial to directed drug development. Predicting essential genes through computational methods is appealing because it circumvents expensive and difficult experimental screens. Most such prediction is based on homology mapping to experimentally verified essential genes in model organisms. We present here a different approach, one that relies exclusively on sequence features of a gene to estimate essentiality and offers a promising way to identify essential genes in unstudied or uncultured organisms. We identified 14 characteristic sequence features potentially associated with essentiality, such as localization signals, codon adaptation, GC content, and overall hydrophobicity. Using the well-characterized baker's yeast Saccharomyces cerevisiae, we employed a simple Bayesian framework to measure the correlation of each of these features with essentiality. We then employed the 14 features to learn the parameters of a machine learning classifier capable of predicting essential genes. We trained our classifier on known essential genes in S. cerevisiae and applied it to the closely related and relatively unstudied yeast Saccharomyces mikatae. We assessed predictive success in two ways: First, we compared all of our predictions with those generated by homology mapping between these two species. Second, we verified a subset of our predictions with eight in vivo knockouts in S. mikatae, and we present here the first experimentally confirmed essential genes in this species.
Essential genes are those that, when absent, confer a lethal phenotype upon an organism. Such genes make excellent potential drug targets (Cole 2002), and the ability to rapidly identify them has been described as “the most important task of genomics-based target validation” (Chalker and Lunsford 2002). However, experimentally screening for lethal gene disruptions is challenging and time consuming, even in well-studied species. We propose a machine-learning approach to predicting essential genes in sequenced but largely unstudied fungal species, one that depends not on homology comparison to known essential genes but instead on sequence features that correlate with essentiality at the gene level.
Essentiality can be defined as a lethal phenotype associated with the deletion or disablement of a given gene. Essential genes account for only a small subset of the genome (Reich 2000). In fungal species they account for roughly one fifth of the total genes: 17.8% in Saccharomyces cerevisiae (Winzeler et al. 1999; Giaever et al. 2002), and 17.5% of the small subset in Schizosaccharomyces pombe studied to date (Decottignies et al. 2003). Because phenotype is a product of both genotype and environment, it is only meaningful to discuss essentiality in relation to a given environmental condition; for yeast studies, the condition under which essentiality is assessed is typically laboratory growth on rich media.
Several techniques exist to identify essential genes. The most effective approach—also the most difficult—is large-scale experimental gene disruption. Such screens represent a massive investment of time and resources, and they are not always feasible. For instance, traditional essentiality screens are difficult in Candida albicans because of its partial-diploid nature, mating differences, and a lack of insertional mutagenesis methods (De Backer et al. 2001). Furthermore, recent shotgun sequencing of microbial communities has suggested that <1% of species are amenable to laboratory culture (Riesenfeld et al. 2004; Chen and Pachter 2005). Thus, to identify essential genes in the vast majority of pathogens, we must look beyond direct experimental methods.
Early comparative genomics approaches involved comparing multiple genomes to find a core conserved minimal genome and labeling its component genes essential (Mushegian and Koonin 1996; Arigoni et al. 1998; Bruccoleri et al. 1998). Bayesian statistical approaches have been used to predict microbial essential genes in tandem with transposon mutagenesis (Lamichhane et al. 2003). Another machine learning system has been trained to identify essential genes in S. cerevisiae by integrating genomic experimental data (Jeong et al. 2003). The latter approach demonstrates an ability to predict essential genes accurately via computational methods alone but does not address the applicability of such methods to novel genomes. A useful predictor must classify genes whose essentiality is unknown; thus, such a system must perform well outside the organism on which it was trained.
Because essential genes are thought to evolve more slowly than their non-essential counterparts (Wilson et al. 1977; Hurst and Smith 1999), researchers tasked with identifying them in newly sequenced organisms often rely on homology mapping to known essential genes in model organisms.
Such homology mapping depends on the accessibility of a closely related organism with experimentally determined essential genes. This approach is particularly limited when seeking to identify drug targets because it is inherently biased toward genes conserved outside the host organism. As drug targets, such broadly conserved genes are naturally less useful than genes unique to the parasite in question. To identify essential drug targets in pathogenic organisms, therefore, it is disadvantageous to rely on homology mapping.
We propose that gene essentiality can be predicted in sequenced but unstudied species using a weighted combination of certain hallmark features. Whereas homology mapping relies continually on model organisms—and their core reference set of known essential genes—our approach requires these only once: to initially identify relevant features and train the classifier. Thereafter, all information necessary to classify a gene can be drawn from the sequence of that gene itself, freeing us from the constant demand for closely related, well-studied organisms to chaperone our predictions.
Broadly, genomic features can be divided into three groups: those intrinsic to a gene's sequence (e.g., GC content, length), those derived from sequence (such as localization signals and codon adaptation measures), and experimental functional genomics data.
Certain features have already been shown to correlate with gene essentiality. For example, protein–protein interaction hubs are more likely to be essential than less-connected nodes (Dezso et al. 2003; Jeong et al. 2003; Yu et al. 2004). Although such functional genomics data as mRNA expression or protein interaction data might be useful in characterizing essential genes, it is not sensible to train a classifier to expect such input when no large-scale experiments have been done on the organisms of interest. A classifier that considers only sequence-derived features is best suited to predicting essential genes in unstudied organisms.
Our 14 features were selected for their accessibility from genomic sequence data and their proposed link to essentiality. We hypothesize that translational stalling is minimized in essential genes—if true, this may be manifest in preferential codon usage and a paucity of rare amino acids in the coding sequence. Further, we propose that essential genes are insulated against nonsense mutations and are therefore less likely to contain in-frame codons sequentially similar to stop codons. Because subcellular localization is relevant to essentiality, we consider localization signals and predicted transmembrane helices. We supplement these with easily accessible sequence features: GC content, gene length, and hydrophobicity. All features listed can be derived from sequence data (Drawid and Gerstein 2000; Lu et al. 2004; http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html) and are thus available for virtually any microbial genome.
To learn the relationship of these features to essentiality, we require a comprehensive set of known essential genes. The baker's yeast S. cerevisiae is extremely well studied; through comprehensive and sustained efforts, over 95% of genes in this species have been systematically deleted (Winzeler et al. 1999; Giaever et al. 2002), yielding a definitive mapping of the essential genes in this species.
We selected a related and relatively unstudied species on which to test our classifier. The yeast S. mikatae was sequenced in 2003 and shares a high level of sequence homology with S. cerevisiae (Kellis et al. 2003). Because of its phylogenetic proximity and the high conservation rate among essential genes in general (Jordan et al. 2002), we expect orthologs to most S. cerevisiae essential genes to exist—and also to be essential—in S. mikatae. Thus, homology mapping by BLAST (Altschul et al. 1990) should be an effective method for the identification of essential genes in S. mikatae and a strong standard against which to evaluate our predictive success. Eight of our predictions were experimentally tested with in vivo knockout mutagenesis, yielding the first experimental verification of heterogenomic essentiality prediction.
Results
S. cerevisiae and S. mikatae display similar essentiality profiles through BLAST comparison
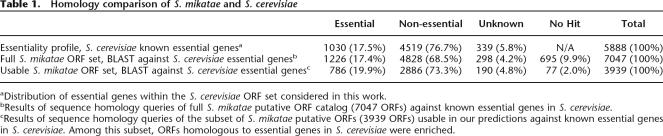
A data file of S. cerevisiae open reading frames (ORFs) was annotated to include known essential genes, as determined experimentally by the Saccharomyces Genome Deletion Consortium (Winzeler et al. 1999; Giaever et al. 2002). Dubious ORFs and pseudogenes were excluded from this list. Within the set of 5888 S. cerevisiae ORFs considered, 1030 (17.5%) were essential, 339 (5.8%) were not successfully deleted and are marked unknown, and the remaining 4519 (76.7%) were marked non-essential (Table 1).
Table 1.
Homology comparison of S. mikatae and S. cerevisiae
The S. mikatae genome contains 7047 putative ORFs, of which a subset of 3939 was ultimately usable by our predictive technique (see Methods). The complete set of S. mikatae ORF sequences was queried against the S. cerevisiae list described above, with an E-value reporting threshold of 0.0001 (1e-4). Using sequence homology criteria as a guide, S. mikatae exhibits a similar essentiality profile to S. cerevisiae, with 17.4% of ORFs homologous to known essential genes in S. cerevisiae. Unique S. mikatae ORFs (those with no BLAST hit to S. cerevisiae at this E-value threshold) number 695 (9.9%).
Within the subset of 3939 S. mikatae ORFs ultimately amenable to classification by our system, putative essential genes are enriched, with 19.9% of ORFs homologous to known essential genes in S. cerevisiae. The number of unique ORFs (no BLAST hit to S. cerevisiae at 1e-4) is sharply reduced in this subset (2.0%).
Identifying features related to essentiality
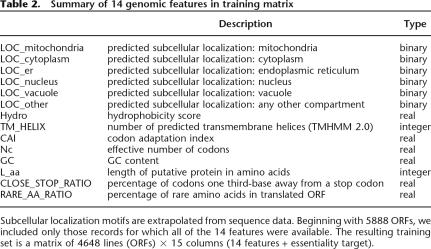
We compiled a list of 14 features available from gene sequence that we hypothesize to be related to essentiality (Table 2). We predicted subcellular localization motifs using the Proteome Analyst Specialized Subcellular Localization Server v2.5 (Lu et al. 2004) and transmembrane helices for each ORF using TMHMM v2.0 (Sonnhammer et al. 1998; Krogh et al. 2001). Using the CodonW program (http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html), we computed two measures of codon adaptation: the effective number of codons (Nc) (Wright 1990; Fuglsang 2004), and the codon adaptation index (CAI) (Sharp and Li 1987). We calculated the frequency of rare amino acids and the number of “close-to-stop” codons (those which are a single third-base substitution removed from a stop codon). Finally, we computed the GC content, the length of the translated protein product, and predicted protein hydrophobicity, again with CodonW.
Table 2.
Summary of 14 genomic features in training matrix
To train our classifier, these 14 features were compiled where available for each ORF in S. cerevisiae (all features available for 4648 ORFs total) and annotated with known essentiality values from the Saccharomyces Genome Database, thereby creating our training data set.
Individual genomic features contain information about essentiality
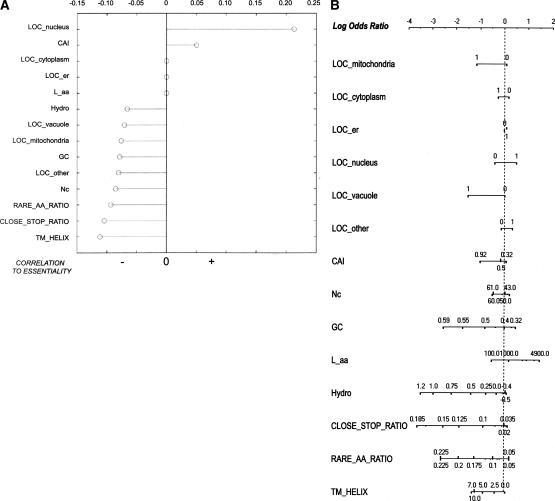
It is important to quantify how much information each genomic feature carries with respect to essentiality. To this end, we computed the correlation with essentiality for all features in the training matrix (Fig. 1). Three features (predicted localization: ER, predicted localization: cytoplasm, and length of predicted protein) had pairwise correlations with essentiality that were not significant (P > 0.05) and were therefore set to zero in our figure. Predicted nuclear localization showed the strongest positive correlation with essentiality, while CAI showed a weak positive correlation. The presence of predicted transmembrane helices, the fraction of close-stop codons, and the fraction of rare amino acids in the sequence exhibited the strongest inverse correlation to essentiality.
Figure 1.
(A) Stem plot of correlation coefficients of each feature with essentiality. Negative correlations are shown to the left of the vertical axis, positive correlations to the right. Correlation coefficients were filtered by P value: Coefficients with P > 0.05 (deemed not significant) were set to zero. (This occurred for features LOC_cytoplasm, LOC_er, and L_aa.) Higher-order correlations are not shown. (B) The nomogram for the Naive Bayes analysis, illustrating the relative contributions of each predictive feature to the target class (essentiality). For each feature, values are drawn along a line according to their influence on the target class: The longer the line, the more important the feature for final prediction. The top line gives the log-odds ratio. Given a set of feature states, a prediction can be obtained by performing a vertical lookup to the log-odds ratio for each feature state and summing the log-odds ratio contributions of all 14 features. The higher the sum, the more likely that gene is essential.
To assess the relative importance of each individual feature as a predictor of essentiality, we also employed the Naive Bayes technique (implemented using the Orange software package [http://www.ailab.si/orange]). The results are depicted in nomogram form in Figure 1B, which shows the relative importance of the features in this framework.
The classifier is trained and tested on S. cerevisiae
Several types of classifiers were trained and tested using the 14-feature data set on S. cerevisiae in order to build the best essentiality predictor for this organism. The best performance was obtained by a hybrid system that combined the output of diverse classification schemes including decision trees, Naive Bayes, and a logistic regression model (see Methods for exact implementation details). Given an ORF and its associated 14 input variables, each classification scheme generates a separate probability estimate of essentiality. These estimates are then combined in an unweighted average to generate the final essentiality prediction for that ORF.
Ten-fold cross-validation in S. cerevisiae (4648-gene training set), with average probability threshold set at 0.5, yielded 88 true positives (TP) and 875 false negatives (FN) from a total of 963 actual essential; 39 false positives (FP) and 3646 true negatives (TN) from a total of 3685 non-essential.
Positive predictive value (PPV) measures how many genes predicted as essential are indeed essential (i.e., precision). PPV is calculated as TP / [TP+FP] and, thus, PPV = 0.69 in this case. Recall (R) measures how many of the true essential genes are classified correctly (i.e., sensitivity). Using our values, R = 0.10 (given by TP / [TP+FN]).
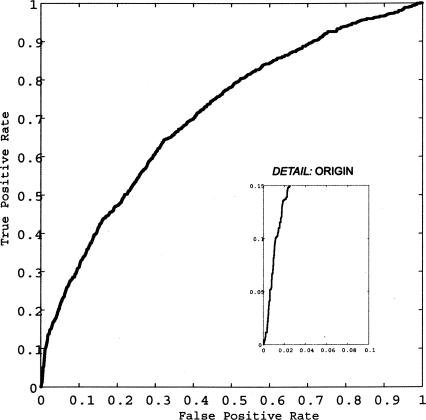
Because we are interested in making a small number of high-value predictions, we readily tolerate false negatives and focus instead on minimizing false-positive predictions among the top-scoring predictions. Thus, we demand a Receiver Operating Characteristic (ROC) curve that is steep near the origin. The ROC curve for the learned classifier is shown in Figure 2.
Figure 2.
Receiver Operating Characteristic curve for the classifier on the S. cerevisiae data set. This plots the true positive rate versus the false positive rate for different thresholds of classifier probability output.(Inset) The steep slope found near the origin signifies a high percentage of correct essentiality assignments among the top probability scores output by the classifier. Because we aim to predict a small subset of essential genes with high confidence, only the predictions found in this subset are relevant to our approach.
Predicting S. mikatae essential genes
To predict essential genes in S. mikatae, the same 14 features comprising the training data set were compiled for each ORF. As with S. cerevisiae, the subcellular localization server was unable to generate predictions for a subset of genes; in total, we gathered the full feature set for 3939 S. mikatae ORFs. The classifier trained on S. cerevisiae was then applied to this data set. (Full results of essentiality prediction for all 3939 S. mikatae ORFs considered can be found in the Supplemental material online.) Notice that the learner is not directly exposed to BLAST results between these species or other direct information about sequence homology. Instead, homology comparison is used along with in vivo knockouts to test our predictions.
Predicting essential genes in other species
In addition to S. mikatae, we also applied our classifier to two additional yeast species, generating essential gene predictions for S. bayanus and Schizosaccharomyces pombe. Although we were unable to confirm these predictions with knockouts, we report the results in the Supplemental material available online (http://www.gersteinlab.org/proj/predess).
Examining predictive success by comparison with BLAST predictions
Our predictive success was first assessed by BLAST comparison. Among the top 100 predicted essential ORFs in S. mikatae, 75 (75%) are homologous to known essential genes in S. cerevisiae, and among the top 30 predicted essential in S. mikatae, 25 (83%) match S. cerevisiae essential genes.
To ensure that our classifier learned traits actually associated with essentiality (as opposed to the quirks of a random subset of genes), we performed a permutation test (Edgington 1995). The “essential” label was randomly reassigned among S. cerevisiae ORFs; a new learner was trained with this spurious “essentiality” data, and applied to S. mikatae. Among the top 100 highest-scoring predicted essential ORFs in S. mikatae, only 45 (45%) are homologous to the correct “essential” genes in the S. cerevisiae training set. Among the top 30 predictions, 17 (57%) match S. cerevisiae “essential” genes. Thus, performance is diminished considerably when training on a random subset of genes.
Predictions verified with knockouts in S. mikatae
We pursued a PCR-directed knockout strategy to explore our essential gene predictions further.
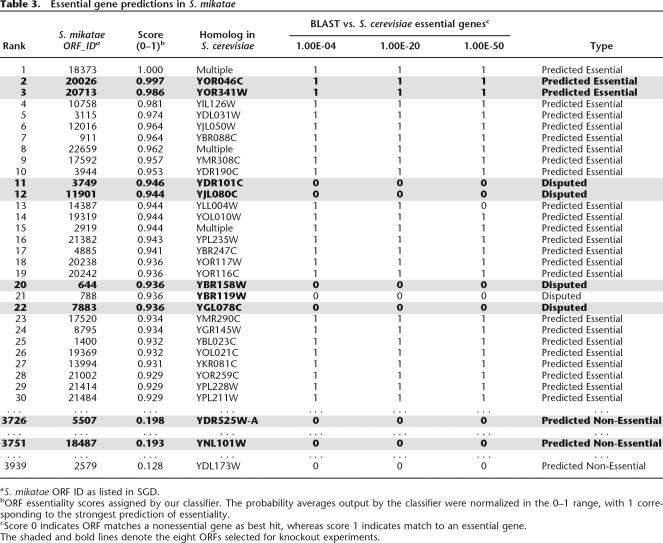
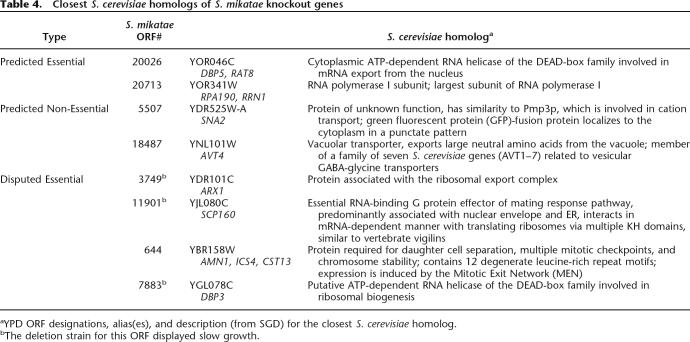
In total, eight ORFs were selected for further study (Table 3; Supplemental Table 1). This set contained two Predicted Essential ORFs (both homology prediction and machine learner agree: gene is essential), two Predicted Non-Essential ORFs (both homology and machine learner agree: gene is non-essential), and four Disputed Essential ORFs (machine learner predicts essential; homology predicts non-essential). Results of these S. mikatae knockouts are shown in Figures 3and4. Table 4 reports the Yeast Proteome Database (YPD) ORF designation, alias, and description of the closest S. cerevisiae homologs to these genes.
Table 3.
Essential gene predictions in S. mikatae
Figure 3.
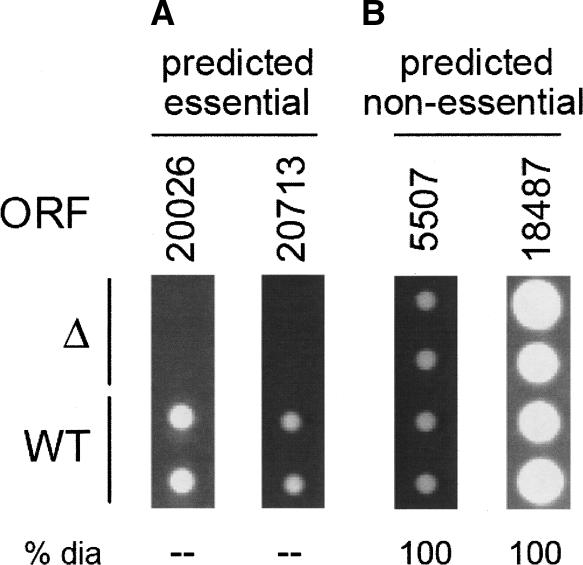
Observed colony sizes for knockout strains in S. mikatae, spotted in duplicate. ORFs were deleted with PCR and transformed into diploids. Sporulation was induced and tetrads dissected to recover haploid cells carrying the deletion of interest. Each column represents a different deletion strain; the top two spots represent colonies grown from deletion strains, while the bottom two spots are wild-type colonies. Four ORFs were selected for knockout mutagenesis: (A) two high-scoring ORFs representing our top essentiality predictions; deletions of predicted essential ORFs were non-viable; (B) two low-scoring ORFs likely to represent non-essential genes; deletions of predicted non-essential ORFs displayed wild-type growth. (ORF) S. mikatae ORF deleted in this strain. (Δ) Growth of knockout strain (deletion of ORF listed above). (WT) Growth of wild-type strain. (% dia) Percent of mutant colony diameter compared with WT for those with slow growth.
Figure 4.
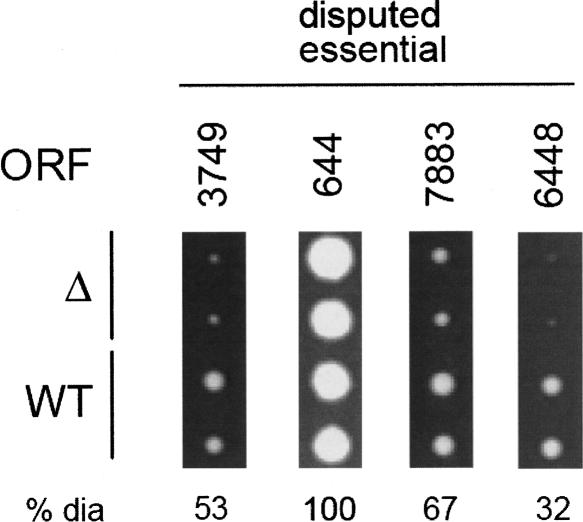
Four additional ORFs were selected for knockout mutagenesis to examine in detail the small subset of cases where predictions generated by our system disagreed with homology comparison to S. cerevisiae (Disputed Essential ORFs). All four deletions were viable; three showed significant degrees of growth arrest. Experimental details are the same as stated in Figure 3.
Table 4.
Closest S. cerevisiae homologs of S. mikatae knockout genes
The two Predicted Essential ORFs were taken from among the top three to which our classifier assigned the highest essentiality scores (ORF 20,026, score = 0.997 and ORF 20,713, score = 0.986) and that were also found to be homologous to known essential genes in S. cerevisiae. (A knockout strain could not be generated for ORF 18,373 because of sequence/contig constraints.) Classifier predictions for both Predicted Essential genes were correct: deletions Δ20026 and Δ20713 were non-viable (Fig. 3A)
Putative ORFs 5507 and 18,487 were chosen as Predicted Non-Essential, selected from the pool of lowest-scoring predictions overall that were also found to be homologous to non-essential genes in S. cerevisiae. Deletions of both these ORFs were viable, displaying wild-type growth. (Fig. 3B)
Four Disputed Essential genes were selected from the small subset of top-scoring putative essential genes for which BLAST comparison contradicted our classifier. There were five such ORFs among the top 30 predictions, and none among the top ten predictions. Knockouts were generated for four of these five Disputed Essential ORFs. Although none show true lethality, three display growth rates significantly slower than wild type (doubling time ~76 min). Specifically, Δ3749 (94 min), Δ7883 (83 min), and Δ6448 (135 min) all exhibit varying degrees of growth arrest. The growth curves on these S. mikatae clones agreed well with the observed colony sizes. The fourth deletion strain (Δ644) demonstrated near wild-type growth (Fig. 4).
Discussion
Our machine learning classifier is able to effectively identify genes necessary for growth in S. mikatae, considering as input only sequenced-derived features; our system is competitive with homology-based prediction, with 25 of the top 30 predictions in S. mikatae found to be homologous to essential genes in S. cerevisiae. Among such closely related species, homology sets a high standard. Conceptually, we expect that our approach is portable to many unstudied species, provided that the necessary features can be derived for the organism in question.
The genomic features and their correlations
Because our aim was to predict essential genes in largely unstudied organisms, we selected sequence-derived features available de novo in freshly sequenced genomes. (After all, it is not useful to train a classifier with experimental genomic data when such data do not yet exist for genes we actually care to classify.) Analysis of the correlations between the genomic features and essentiality suggests that no single feature in our set is sufficient to act alone as a classifier for essentiality. However, even those features that display no direct correlation to essentiality can prove useful. The correlation coefficients shown in Figure 1 were calculated pairwise, and higher-order correlations were not explicitly analyzed; our success in training a classifier demonstrates that a non-linear combination of these features—each having a small correlation to the target variable—is indeed predictive of essentiality. The nomogram (Fig. 2) illustrates the relative importance of these features to a simple Naive Bayes classifier. Here, features common to all ORFs (close-stop ratio, GC content, hydrophobicity, gene length, and rare amino acid ratio) tend to rank higher in importance than features found in only some ORFs (i.e., transmembrane helices or localization signals for a specific subcellular compartment).
Some general trends are visible among the features. Essential genes are likely to be nuclear-localized, unlikely to contain close-stop codons, predicted transmembrane helices or rare amino acids (see Methods), and tend to show preferential codon bias.
Our classifier: Caveats
Our results show that a relationship exists between our 14 genomic features and essentiality, and this relationship can be learned with machine learning techniques. However, we stress that our particular machine learning approach may not represent the optimal solution to this problem; to determine the best approach, other applicable machine learning algorithms should be assayed.
Because our learner correlates sequence features to a target variable for a subset of yeast genes—specifically, the one fifth of genes defined as essential—the risk exists that we are not learning features truly meaningful to essentiality per se, but have instead merely designed a potent system to identify quirks in any genomic subset to which it is exposed. Thus, as a control, we randomly shuffled the essential label among genes and retrained our classifier with this mock data. The resulting classification was considerably less effective than that of a classifier trained on true essential genes. This suggests that there is more to be learned from the true essential subset than an equally large set selected at random, and that our classifier is indeed learning traits characteristic to essential genes.
The S. pombe genome is predicted to contain 865 essential genes. A recent gene-deletion pilot study assayed 80 genes drawn from a single 253-kb region and identified 14 as essential (Decottignies et al. 2003). To explore our predictive success outside the Saccharomyces clade, we compared these 14 known essential genes to our predictions on S. pombe. Our classifier is designed to generate a small number of high-value positive predictions, at the expense of a large number of false-negative predictions. Thus, in assessing its performance, it only makes sense to evaluate its top ~30 predictions. Unfortunately, our top 30 essential gene predictions in S. pombe contained no overlap to the 80 genes assayed in the aforementioned study, and consequently we are unable to use these data to verify our predictions.
Success of our predictions—homology, knockouts
In drug discovery terms, a few promising targets are more valuable than hundreds of questionable leads. We elected to focus on effectively identifying a handful of promising essential genes with a high degree of confidence, and optimized our learning to penalize false-positive predictions. Our classifier is therefore intentionally skewed toward producing a small number of reliable, high-value predictions.
This approach proved successful. Our full top 10—and 25 of our top 30—S. mikatae predictions are corroborated by homology mapping to known essential genes in S. cerevisiae. Despite radically different approaches, the predictions generated by our system agree well with homology-based prediction overall.
We performed experimental gene deletion assays in S. mikatae to interrogate our predictions further. Two representative high-scoring predicted-essential genes were selected for assay by in vivo knockout mutagenesis; both of these deletion strains proved non-viable. Conversely, two low-scoring ORFs, predicted to be non-essential, displayed full wild-type growth when deleted in vivo.
Disputed Essential knockouts
We took particular interest in five genes among our top 30: those for which our classifier predicted essentiality but BLAST comparison to S. cerevisiae revealed homology with a non-essential gene. We examined four such Disputed Essential ORFs by knockout mutagenesis.
All four Disputed Essential ORF deletion strains were viable, although they displayed varying degrees of growth arrest. While the sample size of our knockout pool is insufficient to draw reliable statistical conclusions, these four strains are enriched in slow-growth phenotypes. According to SGD deletion data (Winzeler et al. 1999; Giaever et al. 2002), if we exclude non-viable deletions, only 16% of genes (755 of 4694 total) display slow-growth phenotypes. Thus, selecting viable knockouts at random, we expect roughly one in six (~17%) to exhibit growth arrest. Our results show three in four (75%) with severely impaired growth.
This suggests that our predictor, even when directly contradicted by homology comparison, is nonetheless identifying genes important to cellular function.
Table 4 reports the closest S. cerevisiae homolog to all eight genes knocked out in S. mikatae. Among these, our machine learner classified two DEAD-box helicases as essential (S. mikatae ORFs 20,026 and 7883), although only the former was supported by homology comparison. Looking at the four Disputed Essential genes, it is interesting to note that the words “essential” or “required” occur in two of the SGD descriptions (11901, 644) although neither deletion strain is actually inviable in S. mikatae. Anecdotally, our machine learner can be said to identify genes with a good likelihood of being essential, but when such prediction disagrees with experimental results from extremely close sequence homologs in S. cerevisiae, this homology mapping trumps our predictions.
Portability and unique genes
To assay the performance of our classifier on large numbers of unique genes—those for which no known homolog exists—we would need to apply it to distantly related species. As more distant species are considered, both homology mapping and our classifier are likely to suffer: BLAST will lose efficacy because more divergent sequences provide less sequence traction to identify essential orthologs, whereas for our classifier, what was learned in the training species may decay in distant relatives. Nonetheless, we believe that a system predicting essentiality from an amalgamation of secondary characteristics will prove more robust and portable than one relying strictly on comparison of nucleotide sequence.
Consider a hypothetical novel gene, unrelated to any known gene in sequence but displaying all the hallmark feature-states associated with essentiality. While a homology-based predictor is frustrated by the divergent nucleotide sequence of this gene, our system is capable of generating an educated guess based on secondary characteristics. This will hold true so long as these secondary features can be adequately determined from sequence cues and what has been learned about these features continues to make sense in the target organism.
Conclusions
The ability to consistently identify essential genes is of great value to drug discovery operations. Experimentally determining essential genes is challenging and requires that the organism in question be amenable to growth in culture and receptive to standard molecular biological techniques. To limit essential gene discovery to those few species cooperative to laboratory study is not ideal; indeed, the strongest demand exists in the area of microbial drug lead identification, an area rich in unstudied genomes and novel pathogens.
We demonstrate that it is possible to learn traits associated with essential genes in yeast species and to use these features in a predictive manner. Our approach therefore shows promise for the identification of drug targets in novel and pathogenic species. Future work will involve continued study of feature sets: For instance, if functional genomics information is available for a given species, does its inclusion in the training data noticeably improve predictive power? Broader applications of our system might include a bacterial classifier trained on an amalgam of bacterial essential genes, and retraining our system on higher eukaryotes with available essentiality data such as Caenorhabditis elegans and Drosophila melanogaster. Our system is inherently flexible, capable of integrating any available experimental or sequence-derived data. As a stand-alone approach, this machine learning classifier is an effective tool for predicting essential genes.
Methods
Preparing fungal genomes for study
We used the S. cerevisiae coding ORF list (orf_coding.fasta.gz, revision 12/16/2004, available at ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/genomic_sequence/orf_dna/).
This excludes 5′-UTR, 3′-UTR, intron sequences, and bases not translated because of translational frameshifting. Pseudogenes and dubious ORFs were excluded, yielding 5888 total ORFs.
We used the S. mikatae genomic ORF list (orf_genomic.fasta.gz, revision 12/15/2004, available at ftp://genome-ftp.stanford. edu/pub/yeast/data_download/sequence/fungal_genomes/S_mikatae/MIT/orf_dna/). This includes the protein-coding region of all ORFs identified previously (Kellis et al. 2003) and excludes UTR sequences. We then removed any putative ORFs under 150 nucleotides in length, as well as any ORF not beginning with a start codon (ATG) or terminating with a stop codon (TAA/TAG/TGA). This removed 2010 ORFs, yielding 7047 total ORFs.
Comparative genomics
We continued working with the S. cerevisiae ORF list detailed above. For convenience, the FASTA headers in this ORF list were annotated to include the essentiality values for each ORF, drawn from the Saccharomyces Genome Deletion project (Winzeler et al. 1999; Giaever et al. 2002). This ORF list was then formatted as a BLAST database. Performing a homology search of any sequence against this database thereby yields results with essentiality annotated directly in the BLAST output; this facilitates identification of those querying ORFs for which the top hit is an essential gene.
S. cerevisiae training data set
To train the classifier, a training data set was generated comprising 14 sequenced-derived features (Table 2). We compiled a record for every S. cerevisiae ORF for which these 14 features could be calculated. The targets for the learning were given by the SGD experimentally determined essentiality result in binary form (0 = non-essential, 1 = essential). Because we sought to train our classifier on ORFs where essentiality was clearly defined, we excluded the 283 ORFs for which essentiality was unknown, and we also excluded the 19 questionable ORFs (qORFs) clearly labeled in the ORF catalog. The CodonW program (http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html) was used to evaluate Kyte and Doolittle's grand average of hydropathicity (GRAVY) (Kyte and Doolittle 1982) which we call “Hydro”, protein length (amino acids), GC content, and two measures of codon usage: effective Nc (Wright 1990; Fuglsang 2004) and CAI (Sharp and Li 1987). (Nc could not be calculated for 4 ORFs.) Transmembrane helices were predicted by hidden Markov model techniques with the TMHMM v2.0 Web server (http://www.cbs.dtu.dk/services/TMHMM/) (Sonnhammer et al. 1998; Krogh et al. 2001).
We calculated the ratio of rare amino acids in the sequence by counting the occurrence of Cys, Trp, His, and Met codons and expressed the total as a ratio of total ORF length. Thus, if an ORF is 200 codons long, and the translated sequence contains 3 Met, 2 Trp, and 1 His, the total RARE_AA_RATIO would be (3 + 2 + 1 = 6) / 200 = 0.03.
We also calculated CLOSE STOP RATIO, the number of codons that are one third-base mutation removed from a stop codon. There are five such codons: TAC and TAT encoding Tyr are close to TAA and TAG; TGC and TGT encoding Cys, and TGG encoding Trp are one third-base mutation away from TGA. Such codons were counted and the ratio calculated as with RARE_AA_RATIO above.
Subcellular localization values were predicted from sequence data using the Proteome Analyst Specialized Subcellular Localization Server v2.5 (Lu et al. 2004). (The server was unable to generate localization predictions for 1240 ORFs; removing these reduced the final number of usable ORF records in this array to 4648.)
Compiling features for S. mikatae
The same 14 sequence-based features comprising the S. cerevisiae training data set were compiled for every S. mikatae ORF. As with S. cerevisiae, the Specialized Subcellular Localization Server (Lu et al. 2004) was unable to generate localization predictions for a subset of the ORF list: 3098 ORFs were thus excluded. Nc could not be calculated for an additional 10 ORFs. This reduced the final count of usable ORFs in S. mikatae to 3939.
Correlation of features to essentiality
To assess the relative importance of each individual feature as a predictor of essentiality, we employed three distinct measures of how each feature related to the target column, SGD essentiality: correlation coefficient, information gain, and information gain ratio. These three measures produced highly similar results, and here we report only the values of the correlation coefficients. Our Naive Bayes classifier, employed to visualize the relative contributions of the 14 features to a simple learner, was implemented with the Orange software package (http://www.ailab.si/orange), and the schema is available in the Supplemental material available online (http://www.gersteinlab.org/proj/predess/).
Classifier design
Our efforts were focused on obtaining a system that could make a few high-value essential gene predictions. The objective of our learning was therefore to reduce the number of false positives, rather than minimizing the total number of misclassifications on the test set. To achieve this, we assigned a higher cost to false positives than to false negatives.
Because the training (S. cerevisiae) and testing (S. mikatae) sets are not samples drawn from the same distribution, they have somewhat different characteristics. Thus, we elected to avoid being overly aggressive in learning the positives for S. cerevisiae and chose instead to impose a relatively small bias against false positives in the form of a slightly higher cost. False positives were assigned a cost of 1.1, while the cost for false negatives was set equal to 1.
As previously explained, our models were trained on S. cerevisiae data. To make our predictions in S. mikatae, we trained several models and chose that which gave the best results during testing in S. cerevisiae. To assess the performance of our models on S. cerevisiae data, we employed 10-fold cross-validation. All models were implemented using the WEKA software package (Witten and Frank 2005).
The model that provided the best performance combined the output of diverse classifiers using an unweighted average of probability estimates. Seven different learners were trained, and the output of each participated in the average. The component learners comprising our model are: (1) a decision stump boosted using the Adaboost algorithm for 10 iterations (Freund and Schapire 1996); (2) a random forest, constituted by 10 trees (Breiman 2001); (3) an alternating decision tree with 10 boosting iterations (Freund and Mason 1999); (4) a multinomial logistic regression model with a ridge estimator (le Cessie and van Houwelingen 1992), where the ridge regularization parameter was set to 10−8; (5) a zeroR rule; (6) a Naive Bayes classifier; and (7) a C4.5 decision tree (Quinlan 1993).
S. mikatae gene disruption, sporulation, and lethality scoring
Heterozygous gene deletions were constructed in S. mikatae strain IFO1815 (courtesy of M. Johnson, Washington University, St. Louis, MO) using a long-primer PCR approach developed for S. cerevisiae (Lorenz et al. 1995). Primers were designed to each ORF such that they contained 60 bp immediately flanking either the start or stop codon followed by a 20 bp sequence homologous to the plasmid template used for the deletion (see Supplemental Table 1 for the primer sequences). The plasmid pFA6a-KanMX, which contains the KanMX gene (G418R) as a selectable marker, was used as the PCR template (Longtine et al. 1998).
PCR products were transformed into S. mikatae using a lithium acetate (LiAc) technique (Gietz and Woods 2002). Transformants were selected by growth on YPD plates containing 200 mg/mL of G418. Strains were tested for correct integration by PCR using a primer internal to the KanMX cassette (KanB) (Longtine et al. 1998) in addition to a gene specific primer designed to be ~200 bp upstream of the start codon of that particular ORF.
Confirmed heterozygous deletion strains were then sporulated according to the standard S. cerevisiae procedures (Winzeler et al. 1999). Sixteen individual tetrads were dissected onto YPAD plates with growth scored and photographed after incubating for 40 h to 48 h at 30°C. These plates were then replica plated onto YPD plates containing 200 mg/mL of G418 to check for the correct 2:2 segregation of the deletion marker and to associate the slow growth phenotype with the presence of G418 resistance in those strains that showed growth of both the homozygous wild-type and deletion progeny or to confirm the absence of G418 resistance progeny for those deletion strains that displayed homozygous lethality.
For those strains that showed a decrease in growth rate, this was quantitated both by measuring the average (n = 8) colony size compared with wild type following the initial tetrad dissection and incubation and by performing growth tests on these strains in liquid YPAD. For this, strains were inoculated in YPAD with the optical density (OD600) recorded hourly. The final growth rate is expressed as the average doubling time (n = 4) of an exponentially growing culture of each of these strains.
Predictions in other species
The same 14 sequence-based features were compiled for every ORF in S. bayanus (all features available for 5447 ORFs) and S. pombe (all features available for 4267 ORFs). The same classifier detailed above was applied to these data files. The genome data files, feature sets, and predictions can all be found on our Supplemental material Web site (http://www.gersteinlab.org/proj/predess/).
ACKNOWLEDGMENTS
This work was supported by NIH grants to Mark Gerstein and Michael Snyder. The authors wish to thank Tom Royce, Tara Gianoulis, and Joel Rozowsky for helpful comments on the manuscript, and Andrea Sboner for his assistance with Naive Bayes implementation.
Footnotes
[Supplemental material is available online at www.genome.org. and http://www.gersteinlab.org/proj/predess/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5144106. Freely available online through the Genome Research Open Access option.
REFERENCES
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arigoni F., Talabot F., Peitsch M., Edgerton M.D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Talabot F., Peitsch M., Edgerton M.D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Peitsch M., Edgerton M.D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Edgerton M.D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H., Fish R., Jamotte T., Curchod M.L., Loferer H., Jamotte T., Curchod M.L., Loferer H., Curchod M.L., Loferer H., Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Bruccoleri R.E., Dougherty T.J., Davison D.B., Dougherty T.J., Davison D.B., Davison D.B. Concordance analysis of microbial genomes. Nucleic Acids Res. 1998;26:4482–4486. doi: 10.1093/nar/26.19.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker A., Lunsford R., Lunsford R. Rational identification of new antibacterial drug targets that are essential for viability using a genomics-based approach. Pharmacol. Ther. 2002;95 doi: 10.1016/s0163-7258(02)00222-x. [DOI] [PubMed] [Google Scholar]
- Chen K., Pachter L., Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.T. Comparative mycobacterial genomics as a tool for drug target and antigen discovery. Eur. Respir. J. Suppl. 2002;36:78s–86s. doi: 10.1183/09031936.02.00400202. [DOI] [PubMed] [Google Scholar]
- De Backer M.D., Nelissen B., Logghe M., Viaene J., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Nelissen B., Logghe M., Viaene J., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Logghe M., Viaene J., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Viaene J., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., Dewaele S., Simons F.A., Verhasselt P., Simons F.A., Verhasselt P., Verhasselt P., et al. An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans . Nat. Biotechnol. 2001;19:235–241. doi: 10.1038/85677. [DOI] [PubMed] [Google Scholar]
- Decottignies A., Sanchez-Perez I., Nurse P., Sanchez-Perez I., Nurse P., Nurse P. Schizosaccharomyces pombe essential genes: A pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z., Oltvai Z.N., Barabasi A.L., Oltvai Z.N., Barabasi A.L., Barabasi A.L. Bioinformatics analysis of experimentally determined protein complexes in the yeast Saccharomyces cerevisiae . Genome Res. 2003;13:2450–2454. doi: 10.1101/gr.1073603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawid A., Gerstein M., Gerstein M. A Bayesian system integrating expression data with sequence patterns for localizing proteins: Comprehensive application to the yeast genome. J. Mol. Biol. 2000;301:1059–1075. doi: 10.1006/jmbi.2000.3968. [DOI] [PubMed] [Google Scholar]
- Edgington E.S. Randomization tests. Marcel Dekker; New York: 1995. [Google Scholar]
- Freund Y., Mason L., Mason L. Proceeding of the Sixteenth International Conference on Machine Learning. Morgan Kaufmann; San Francisco: 1999. The alternating decision tree learning algorithm; pp. 124–133. [Google Scholar]
- Freund Y., Schapire R.E., Schapire R.E. Proceedings of the International Conference on Machine Learning. Morgan Kaufmann; San Francisco: 1996. Experiments with a new boosting algorithm; pp. 148–156. [Google Scholar]
- Fuglsang A. The ‘effective number of codons’ revisited. Biochem. Biophys. Res. Commun. 2004;317:957–964. doi: 10.1016/j.bbrc.2004.03.138. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Dow S., Lucau-Danila A., Anderson K., Andre B., Lucau-Danila A., Anderson K., Andre B., Anderson K., Andre B., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Hurst L.D., Smith N.G., Smith N.G. Do essential genes evolve slowly? Curr. Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Jeong H., Oltvai Z.N., Barabasi A.-L., Oltvai Z.N., Barabasi A.-L., Barabasi A.-L. Prediction of protein essentiality based on genomic data. ComPlexUs. 2003;1:19–28. [Google Scholar]
- Jordan I.K., Rogozin I.B., Wolf Y.I., Koonin E.V., Rogozin I.B., Wolf Y.I., Koonin E.V., Wolf Y.I., Koonin E.V., Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L., Larsson B., von Heijne G., Sonnhammer E.L., von Heijne G., Sonnhammer E.L., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lamichhane G., Zignol M., Blades N.J., Geiman D.E., Dougherty A., Grosset J., Broman K.W., Bishai W.R., Zignol M., Blades N.J., Geiman D.E., Dougherty A., Grosset J., Broman K.W., Bishai W.R., Blades N.J., Geiman D.E., Dougherty A., Grosset J., Broman K.W., Bishai W.R., Geiman D.E., Dougherty A., Grosset J., Broman K.W., Bishai W.R., Dougherty A., Grosset J., Broman K.W., Bishai W.R., Grosset J., Broman K.W., Bishai W.R., Broman K.W., Bishai W.R., Bishai W.R. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: Application to Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Cessie S., van Houwelingen J.C., van Houwelingen J.C. Ridge estimators in logistic regression. Appl. Stat. 1992;41:191–201. [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Wach A., Brachat A., Philippsen P., Pringle J.R., Brachat A., Philippsen P., Pringle J.R., Philippsen P., Pringle J.R., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Muir R.S., Lim E., McElver J., Weber S.C., Heitman J., Muir R.S., Lim E., McElver J., Weber S.C., Heitman J., Lim E., McElver J., Weber S.C., Heitman J., McElver J., Weber S.C., Heitman J., Weber S.C., Heitman J., Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae . Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Lu Z., Szafron D., Greiner R., Lu P., Wishart D.S., Poulin B., Anvik J., Macdonell C., Eisner R., Szafron D., Greiner R., Lu P., Wishart D.S., Poulin B., Anvik J., Macdonell C., Eisner R., Greiner R., Lu P., Wishart D.S., Poulin B., Anvik J., Macdonell C., Eisner R., Lu P., Wishart D.S., Poulin B., Anvik J., Macdonell C., Eisner R., Wishart D.S., Poulin B., Anvik J., Macdonell C., Eisner R., Poulin B., Anvik J., Macdonell C., Eisner R., Anvik J., Macdonell C., Eisner R., Macdonell C., Eisner R., Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Koonin E.V., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan J.R. Machine Learning. Morgan Kaufmann; San Mateo, CA: 1993. C4.5: Programs for machine learning. [Google Scholar]
- Reich K.A. The search for essential genes. Res. Microbiol. 2000;151:319–324. doi: 10.1016/s0923-2508(00)00153-4. [DOI] [PubMed] [Google Scholar]
- Riesenfeld C.S., Schloss P.D., Handelsman J., Schloss P.D., Handelsman J., Handelsman J. Metagenomics: Genomic analysis of microbial communities. Annu. Rev. Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H., Li W.H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A., von Heijne G., Krogh A., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Wilson A.C., Carlson S.S., White T.J., Carlson S.S., White T.J., White T.J. Biochemical evolution. Annu. Rev. Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Bangham R., Benito R., Boeke J.D., Bussey H., Benito R., Boeke J.D., Bussey H., Boeke J.D., Bussey H., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Witten I.H., Frank E., Frank E. Data mining: Practical machine learning tools and techniques. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Yu H., Greenbaum D., Xin Lu H., Zhu X., Gerstein M., Greenbaum D., Xin Lu H., Zhu X., Gerstein M., Xin Lu H., Zhu X., Gerstein M., Zhu X., Gerstein M., Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]