Abstract
Using 1128 protein-coding gene families from 11 completely sequenced cyanobacterial genomes, we attempt to quantify horizontal gene transfer events within cyanobacteria, as well as between cyanobacteria and other phyla. A novel method of detecting and enumerating potential horizontal gene transfer events within a group of organisms based on analyses of “embedded quartets” allows us to identify phylogenetic signal consistent with a plurality of gene families, as well as to delineate cases of conflict to the plurality signal, which include horizontally transferred genes. To infer horizontal gene transfer events between cyanobacteria and other phyla, we added homologs from 168 available genomes. We screened phylogenetic trees reconstructed for each of these extended gene families for highly supported monophyly of cyanobacteria (or lack of it). Cyanobacterial genomes reveal a complex evolutionary history, which cannot be represented by a single strictly bifurcating tree for all genes or even most genes, although a single completely resolved phylogeny was recovered from the quartets’ plurality signals. We find more conflicts within cyanobacteria than between cyanobacteria and other phyla. We also find that genes from all functional categories are subject to transfer. However, in interphylum as compared to intraphylum transfers, the proportion of metabolic (operational) gene transfers increases, while the proportion of informational gene transfers decreases.
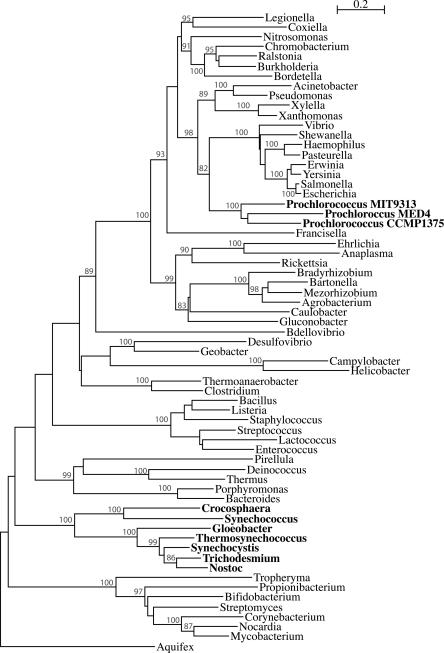
Cyanobacteria occupy a diverse range of habitats. The 11 genome sequences included in this study represent freshwater, marine, and hot spring species, including four closely related marine cyanobacteria from the Prochlorococcus/marine Synechococcus group. Based on the shared traits of oxygenic photosynthesis, several single gene analyses (e.g., Giovannoni et al. 1988), and analyses of shared indels (Gupta et al. 2003), all cyanobacteria form a monophyletic phylogenetic group. Indeed, the “coherence” of cyanobacteria—by which we mean monophyly for all or the vast majority of genes—is often considered self-evident, and asserted without elaboration or citation (e.g., Hagen and Meeks 2001; Otero and Vincenzini 2004).
In 1979, Rippka et al. (1979) divided cyanobacteria into five sections based on morphology. However, this classification does not correspond with molecular markers, including ribosomal RNAs. In 16S rRNA phylogenies (Turner 1997; Honda et al. 1999; Turner et al. 1999; Wilmotte and Herdman 2001), cyanobacteria form several statistically supported clusters different from the sections of Rippka et al. (1979), and with no clearly resolved relationships among the clusters. This poor resolution might be explained by rapid radiation or by recombination within 16S rRNA—enough to scramble phylogenetic signal (e.g., Yap et al. 1999; Boucher et al. 2004; Miller et al. 2005; Morandi et al. 2005).
Horizontal (or lateral) gene transfer (HGT), potentially followed by recombination with or replacement of resident homologs (orthologous replacement), is now recognized as a major force shaping evolutionary histories of prokaryotes (e.g., Koonin et al. 2001; Zhaxybayeva and Gogarten 2002; Boucher et al. 2003) and eukaryotes (e.g., Mitreva et al. 2005). Among methods for detecting instances of HGT are observations of unusual evolutionary patterns in gene phylogenies, patchy phylogenetic distribution, and atypical nucleotide composition (Ochman et al. 2000; Ragan 2001). These different methodologies produce varying estimates of HGT.
Atypical nucleotide composition methods indicate that individual cyanobacterial genomes have acquired between 9.5% and 16.6% of their genes through HGT (Ochman et al. 2000; Nakamura et al. 2004). Because acquired genes eventually “ameliorate” to have the compositional characteristics of their new environment, these are almost certainly serious underestimates. Individual instances of molecular markers in cyanobacteria with contradicting phylogenetic histories continue to accumulate as well (e.g., Rudi et al. 1998; Seo and Yokota 2003). In an earlier study, some of us (Zhaxybayeva et al. 2004) performed genome-wide bipartition analyses of 678 data sets of orthologous genes (or data sets, for short) present in 10 cyanobacterial genomes. The plurality consensus of these data sets was poorly resolved. Nevertheless, many individual gene families contradicted it, suggesting that gene families in cyanobacterial genomes have complex, frequently noncongruent, phylogenetic histories. Population studies of Microcoleus chthonoplastes and Nodularia sp. indicate very high rates of homologous recombination (Barker et al. 2000; Lodders et al. 2005). Cyanophages infecting marine cyanobacteria have been reported to contain genes important for photosynthesis (Mann et al. 2003; Lindell et al. 2004; Millard et al. 2004; Sullivan et al. 2005; Zeidner et al. 2005), and likely mediate transfer and recombination of these genes among marine cyanobacteria (Zeidner et al. 2005). Indeed, Zeidner et al. (2005) postulated that the diversity accumulated in phage psbA genes may serve as an evolutionary reservoir for hosts and increases the hosts’ chances of adapting to changing environments. Additional evidence for the occurrence of HGT in cyanobacteria comes from laboratory experiments with Synechocystis sp. PCC6803 and Thermosynechococcus elongatus BP1 in which mutants are made through the recombination with exogenous DNA (e.g., Ikeuchi and Tabata 2001; Iwai et al. 2004).
In spite of such evidence for HGT involving individual gene families in cyanobacteria and many other bacterial groups, coherence of the phyla is often assumed, and invoked as evidence that HGT is in the long run a weak force, and no serious challenge to the historical accuracy of the rRNA-based Tree of Life. There have been few systematic and exhaustive assessments of the extent to which bacterial phyla really are coherent (Beiko et al. 2005; Kunin et al. 2005). Furthermore, coherence, even if well documented, does not mean that HGT is unimportant or infrequent within phyla. There are many reasons to expect that within-phylum HGT will be more vigorous and more fruitful than between-phylum exchange. In some cases, members of a phylum are more likely to occupy similar environments, and encounter each others’ DNA. There will be phylum-specific constraints based on physiology: for instance, cyanobacteria could not profitably incorporate individual genes of the methanogenesis pathway, but there might be many circumstances in which variant photosynthetic genes could benefit near or distant relatives within the cyanobacteria. Finally, between-phylum differences in genome organization (Lawrence and Hendrickson 2005) and in the machinery of gene expression and its regulation may constrain effective HGT.
More frequent within-phylum orthologous replacement (and homologous recombination) would serve to maintain similarity between members, while allowing divergence between phyla. Thus, the preferential sharing of a common gene pool could be itself the principal cause of coherence (Gogarten et al. 2002; Olendzenski et al. 2002). To test this, comparative estimations of the extent of inter and intragroup transfers are needed. In this study, we analyze sets of orthologous genes from 11 available cyanobacterial genomes and their homologs in 168 other prokaryotes and attempt to quantify the number of transfers that occurred within cyanobacteria and between cyanobacteria and other phyla.
Results
Selection of sets of orthologous genes
Detection of orthologous genes is an important step in attempts to estimate HGT events. Poor selection of sets of orthologous genes leads to hidden paralogy, which is a serious problem in phylogenetic reconstruction. Several different approaches are used frequently to detect sets of orthologous genes (e.g., Zhaxybayeva and Gogarten 2002; Lerat et al. 2003; Tatusov et al. 2003; Harlow et al. 2004); none is perfect. Our very conservative method (see Zhaxybayeva and Gogarten 2002 for methodology), requiring a reciprocal top-scoring BLAST hit for each member of a set of orthologs, may miss many sets of legitimate orthologous genes, but it minimizes data sets contaminated with paralogs.
Many genome-wide analyses (including analyses of cyanobacterial genomes in Zhaxybayeva et al. 2004) are based on a core set of genes, that is, genes present in all analyzed genomes. This restricts such studies to a limited number of very conserved genes, and as more genomes are added to improve taxon sampling, the size of the core set of genes decreases (Charlebois and Doolittle 2004). Using our selection criterion, there are, for instance, only 663 genes present in all 11 of the cyanobacterial genomes, although there are 3804 found in at least four genomes, the minimum for any comparative phylogenetic analysis (see Table 1). Here we use a “relaxed core” of 1128 genes, those identified in at least nine of the 11 genomes, reasoning that such nearly ubiquitous genes probably determine many of the characters by which cyanobacteria are judged to be a “coherent” group.
Table 1.
Number of sets of orthologous genes detected in 11 cyanobacterial genomes
Embedded quartet decomposition analyses
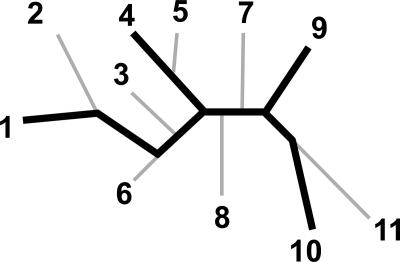
For this analysis, we developed a new tool that allows the inclusion of sets of orthologous genes with missing data. The tool is designed to examine all possible “embedded quartets” for each set of orthologous genes detected in a group of analyzed genomes, that is, all possible four-taxon trees that are consistent with (embedded within) a corresponding gene tree (Fig. 1). This method—“embedded quartet decomposition analyses” or “quartet decomposition,” for short—is conceptually similar to the spectral analysis method of Hendy and Penny (1993) and Lento et al. (1995). A data set of orthologous genes can be, in principle, as small as four taxa (containing one quartet) or as large as 11 taxa (containing 330 embedded quartets). The resulting embedded quartets can be summarized to depict the evolutionary relationships among the genomes that are supported by a plurality of orthologous sets, as well as to delineate genes that conflict with this plurality consensus. We applied this method to the relaxed core of cyanobacteria.
Figure 1.
Illustration of an embedded quartet in a gene tree. In each 11-taxon unrooted gene tree (shown in gray thin lines), we look at the relationship of any four taxa at a time (shown in thick black lines is an example of an embedded tree for taxa 1, 4, 9, and 10), which we call an embedded quartet. In one gene tree, an embedded quartet can support only one of three possible phylogenetic relationships among the four taxa. However, in other gene trees, an alternative phylogenetic relationship for this quartet can be observed. In each 11-taxon gene tree, all possible (4 11) four-taxon trees embedded within the gene tree are examined.
Screening quartets for minimization of false inferences
Quartets with a very short internal branch can produce misleading results because of the absence of sufficient phylogenetic information, thus we removed from our relaxed core data sets all quartets with fewer than three amino acid substitutions along the internal branch: 27 data sets had at least one embedded quartet with such a very short internal branch. To reduce long branch attraction artifacts (Felsenstein 1978), we also excluded embedded quartets with any unbroken external branch more than 10 times longer than the internal branch, and 798 data sets had at least one quartet excluded at this step.
Power of detection as assessed through simulations
Even in the absence of phylogenetic reconstruction artifacts, one expects some false positives at any bootstrap support cutoff (i.e., phylogenies that conflict with the plurality signal by chance), because of the finite amount of phylogenetic data used for phylogenetic reconstruction. False positives will be particularly frequent among quartets resolved only by a few gene families. We performed genome evolution simulations without any gene loss or gain, that is, genes in genomes following strictly vertical inheritance (see Methods for details), to estimate the frequency of such false positives and the likely efficacy of the screening methods for minimizing false inference described above.
We found that when >30% of the data sets resolve an embedded quartet (i.e., support one of the three possible tree topologies with at least 80% bootstrap support), the number of false positives is negligible (see Table 2 and Supplemental material). However, simulations introducing HGT events showed that the conservative approach of excluding quartets resolved by <30% of the data sets increases the number of false negatives (i.e., undetected transfer events). Simulations of either sort produced similar results whether analyzed with PhyML (Guindon and Gascuel 2003) or TREE-PUZZLE (Schmidt et al. 2002), thus we used the latter, faster, method in subsequent study of real data. We conclude that our screening methods likely result in underestimates of HGT, overall.
Table 2.
Simulation results: Number of data sets that conflict with the plurality signal at different cutoff levels, at 80% bootstrap support
Plurality signal and estimation of conflicts within the cyanobacterial group based on analyses of the relaxed core
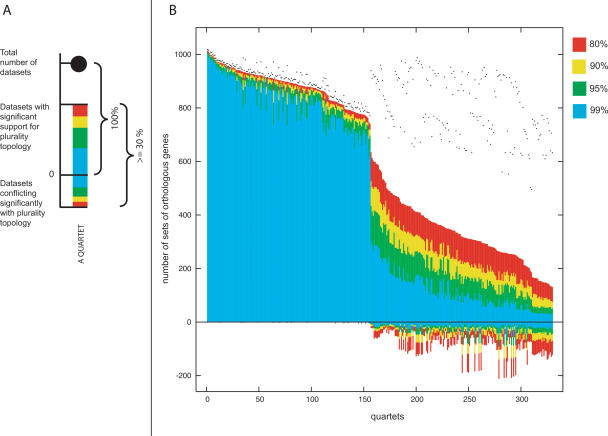
All embedded quartets retained after removal of those with short internal or long external branches were resolved by at least 30% of data sets. We summarized the plurality support for all embedded quartets across all data sets as well as conflicts with plurality in a diagram that we call a quartet spectrum (because we assess all possible combinations of four taxa, providing a full spectrum of possible relationships) (see Fig. 2). All quartet topologies supported by a plurality of data sets are compatible with each other, and therefore only one most parsimonious tree exists (a so-called perfect phylogeny, Felsenstein 2004). This tree, found using a supertree reconstruction algorithm (see Methods), is shown in Figure 3.
Figure 2.
Quartet decomposition analysis of cyanobacteria. Panel A illustrates a component of quartet decomposition analysis. Each embedded quartet is represented by a vertical bar and a black dot. The black dot indicates how many data sets contain this embedded quartet. The vertical bar shows the number of data sets having the topology of the quartet that is supported by a plurality of gene families (value above zero) and the number of data sets having one of the other two quartet topologies (value below zero). The bar is color-coded with respect to bootstrap support. Only quartets that resolve the quartet relationships in at least 30% of analyzed data sets are visualized. The value of 30% was chosen based on simulations (see text for details). Panel B shows the quartet spectrum of 1128 sets of orthologous genes from cyanobacteria. Columns are sorted according to the number of supporting data sets with at least 80% bootstrap support. Quartets with a very short internal branch or very long external branches were excluded from the analyses to minimize artifacts of phylogenetic reconstruction. Quartets above the x-axis are combined into a plurality signal (see Fig. 3 and text for more details). Quartets below the x-axis are embedded into 685 unique sets of orthologous genes. The appearance of discontinuities in the spectrogram corresponds to the uneven phylogenetic distances among 11 analyzed genomes. For example, quartets containing two very closely related taxa (such as the ones containing both Nostoc and Anabaena) will almost unanimously agree on one of three possible quartet topologies.
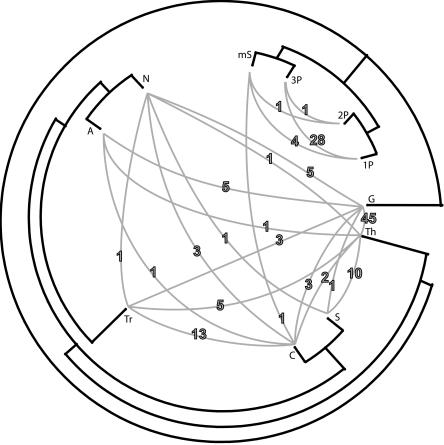
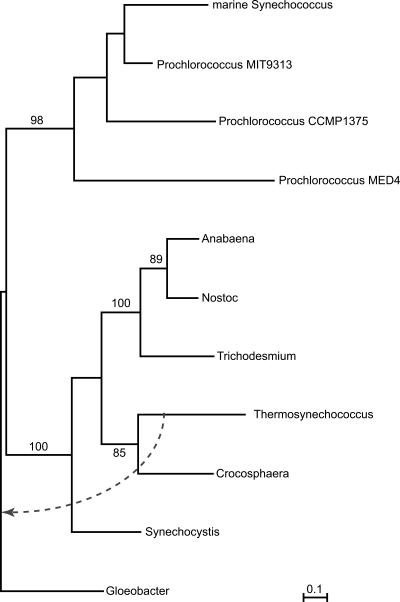
Figure 3.
Visualization of the evolutionary history of cyanobacteria as inferred from quartet decomposition analyses. The unrooted tree topology was calculated from the embedded quartets supported by the plurality of sets of orthologous genes, and it is shown in black. Conflicts to the plurality topology that involve two particular taxa grouping together with at least 80% bootstrap support are shown as gray lines connecting two taxa in conflict. The number of conflicting data sets that have the two connected taxa grouping together are shown on the corresponding gray line. Note that the number of conflicting sets shown represents only a small subset of observed conflicts to the plurality signal, since not all topological incongruencies are of this kind. The genes involved in conflicts, as well as corresponding topologies, are provided as Supplemental material. (A) Anabaena sp. PCC7120; (Tr) Trichodesmium erythraeum IMS101; (S) Synechocystis sp. PCC6803; (1P) Prochlorococcus marinus CCMP1375; (2P) Prochlorococcus marinus MED4; (3P) Prochlorococcus marinus MIT9313; (mS) marine Synechococcus WH8102; (Th) Thermosynechococcus elongatus BP-1; (G) Gloeobacter violaceus PCC7421; (N) Nostoc punctiforme ATCC29133; (C) Crocosphaera watsonii WH8501.
While the plurality signal supports one fully resolved tree topology, we found that a substantial proportion of data sets (685 data sets, or roughly 61% of analyzed data sets) exhibits conflict with the plurality signal in at least one embedded quartet. Some of these conflicts (those involving alternative sister relationships between terminal taxa and having at least 80% bootstrap support) are visualized in Figure 3. In the Supplemental material, we provide trees for the 131 data sets involved in conflicts indicated in Figure 3. One example is provided in Figure 4. Among genes conflicting with the plurality signal are genes involved in photosynthesis (see Table 3), including genes recently found in phages infecting Prochlorococcus (Lindell et al. 2004) and marine Synechococcus (Millard et al. 2004).
Figure 4.
Example of intraphylum transfer: a hemolysin-like protein. This example of horizontal gene transfer was extracted from the list of data sets exhibiting conflicts with the plurality signal. This gene family has detectable homologs in other phyla, but phylogenetic analysis of an extended data set shows that cyanobacteria form a coherent phylogenetic group. Interestingly, in cyanobacteria this protein seems to have acquired a different function (Nagai et al. 2001). This suggests that this protein is probably not being exchanged with the organisms outside of cyanobacteria, which makes it a good example of intraphylum transfer. In this phylogeny Thermosynechococcus and Crocosphaera are sister taxa, which contradicts the relationships observed in the plurality tree. The arrow indicates placement of the Thermosynechococcus sequences in the consensus tree. The tree topology and branch lengths were calculated in PhyML (Guindon and Gascuel 2003) under the JTT+Gamma model. The bootstrap support values are from TREE-PUZZLE + NEIGHBOR phylogenetic analyses as described in Methods. Only bootstrap support values above 80% are shown. Additional examples of putative intraphylum transfers are available in the Supplemental material.
Table 3.
Photosynthesis genes that are observed to conflict with the plurality signal
Incongruence of gene histories among Prochlorococcus/Synechococcus
Among the 11 genomes, four belong to the Prochlorococcus/ marine Synechococcus group. Members of the Prochlorococcus genus have only been recently discovered because of their anomalously low fluorescence and small size (Chisholm et al. 1988). Marine Synechococcus and Prochlorococcus are proposed to diverge from a common phycobilisome-containing ancestor (Ting et al. 2002). While marine Synechococcus still uses phycobilisomes as light-harvesting antennae, members of the Prochlorococcus genus lack phycobilisomes and use a different antenna complex (Pcb), as well as possessing derivatives of chlorophyll a and b that are unique to this genus (for a recent review, see Partensky et al. 1999). In addition, marine Synechococcus and Prochlorococcus are adapted to different ecological niches: Marine Synechococcus is prevalent in coastal waters, while Prochlorococcus is ubiquitous in open subtropical and tropical ocean. Within Prochlorococcus marinus, two “ecotypes” are differentiated: low-light-adapted and high-light-adapted types (Rocap et al. 2003). In the 16S rRNA tree, high-light-adapted Prochlorococcus spp. arise from within a low-light-adapted clade (Ting et al. 2002).
We find numerous conflicts between these four genomes, those involving highly supported apparent transfers between terminal taxa being shown in Figure 3. Similarly, in a recent study, Beiko et al. (2005) report >250 HGT events among these marine cyanobacteria. Although these genomes are reported to have accelerated rates of evolution (Dufresne et al. 2005), and hence could be more prone to the long branch attraction artifact, the quartets with long branches were excluded from our analyses (see above). Interestingly, the relationship among these four genomes captured by the plurality of gene families supports neither the relationship inferred from phylogenetic analyses of 16S rRNA (e.g., Ting et al. 2002; Dufresne et al. 2005), nor the grouping based on proposed ecotypes (Ting et al. 2002; Rocap et al. 2003). This can be explained by rampant gene flow among these genomes, with the plurality consensus no longer reflecting ecotype physiology. Notably, the majority of observed conflicts with the plurality occur between the two low-light-adapted ecotypes (see Fig. 3).
Transfers between cyanobacteria and other phyla
Quartet decomposition analysis only detects conflicts within the group of analyzed genomes. However, observed conflicts could be instances of incongruence produced by transfers from outside of cyanobacteria into only one or a few cyanobacterial lineages. To correct for this and to estimate the number of transfers that occurred between the cyanobacteria and organisms from other phyla, we added homologous genes from other completely sequenced genomes spanning Bacterial and Archaeal domains. Phylogenetic analysis of such “extended data sets” identifies putative instances of horizontal gene transfer from/to the cyanobacteria.
Out of 1128 data sets, 879 had detectable homologs in selected prokaryotic genomes, and the remaining 249 data sets were “cyanobacteria-specific” (and therefore not suitable for estimation of interphylum transfers). Seven hundred of these 879 data sets were “phylogenetically useful,” that is, were sufficiently resolved and either had cyanobacteria as a coherent group with 80% bootstrap support or had other taxa grouping within cyanobacteria at 80% bootstrap support. Of these 700 data sets, 540 support cyanobacteria as a coherent group (~77%), while 160 data sets (~23%) either have sequences from other taxa interspersed among cyanobacteria or some cyanobacterial sequences grouping somewhere else, suggesting possible transfer events to or from cyanobacteria (an example of such a data set is shown in Fig. 5, and additional examples are available as Supplemental material). Interestingly, 294 out of 540 data sets that support
Figure 5.
Example of horizontal gene transfer to cyanobacteria: threonyl tRNA synthetase. This is a phylogenetic tree reconstructed from a data set in which the Anabaena sp. genome did not have a detectable homolog in its annotation. In this tree, sequences of three Prochlorococcus spp. group within Gamma-proteobacteria with high bootstrap support. The presence of an ancestrally transferred gene constitutes a shared derived character for the descendents (Andersson et al. 2005; Huang et al. 2005), suggesting that the three Prochlorococcus spp. form a monophyletic group. Note that despite coherency of the remaining cyanobacteria as a group, the relationships within cyanobacteria do not follow the plurality topology. tRNA synthetases are known to have complex evolutionary histories involving multiple HGT events (e.g., Wolf et al. 1999). Cyano-bacteria are shown in bold. To obtain bootstrap support values, a consensus tree was generated from 100 bootstrap sampled trees (see Methods for more details). Only bootstrap support values above 80% are shown. Additional examples of interphylum transfers are available in the Supplemental material.
cyanobacteria as a monophyletic group (54%, or 42% of the 700 phylogenetically useful data sets) conflict with the plurality consensus based on the quartet decomposition analyses (see above). This estimation suggests that there are more conflicts observed within cyanobacteria than between cyanobacteria and other phyla.
Distribution of genes among functional categories
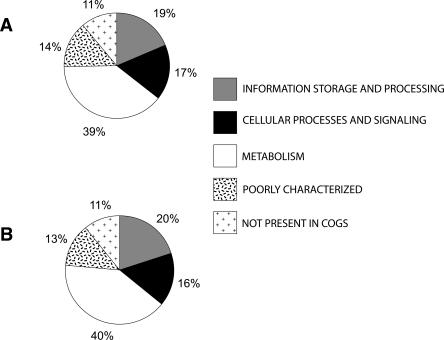
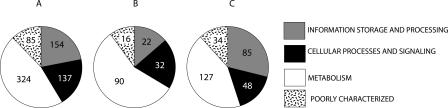
We looked at the distribution of all analyzed cyanobacterial gene families, as well as the distribution of gene families present in extended data sets, across functional categories as defined in the COG database (see Figs. 6 and 7). Functional category analysis of cyanobacterial data sets conflicting with the plurality signal shows that genes from all functional categories are among the conflicting genes (see Fig. 6), including genes from information storage and processing categories (categories J and K, according to the COG database abbreviations) (Tatusov et al. 2003). However, when we analyzed the distribution of extended data sets across functional categories (see Fig. 7), we found that in inter-phylum transfers, metabolic genes are overrepresented, and “information storage and processing” genes are underrepresented.
Figure 6.
Distribution of cyanobacterial sets of orthologous genes across functional categories. The functional categories are according to the COG database, March 2003 release (Tatusov et al. 2003). Panel A shows the distribution of all 1128 analyzed genes, while panel B shows the distribution of 685 genes that conflict with the plurality signal. Conflicts with the plurality signal are observed in sets of orthologs across all functional categories, including genes involved in translation and transcription, and no particular functional category shows bias toward conflicts.
Figure 7.
Distribution of phylogenetically useful extended genes across functional categories. Panel A shows the distribution of 700 phylogenetically useful extended data sets. Panel B shows the distribution of 160 sets where cyanobacteria do not form a monophyletic group (putative interphylum transfers). Panel C shows the distribution of 294 sets where cyanobacteria form a monophyletic group, but they conflict with the plurality signal (putative intraphylum transfers). Numbers inside the pie graphs refer to the number of sets of orthologous genes in each corresponding functional category. Across short phylogenetic distances, all types of genes appear to be equally affected by transfer, while across large phylogenetic distances, genes encoding metabolic functions are more frequently transferred, and genes in transcription and translation are transferred less frequently.
Discussion
Several recent studies attempt to estimate the number of HGT events at different taxonomic levels (e.g., Snel et al. 2002; Lerat et al. 2003; Mirkin et al. 2003; Beiko et al. 2005; Ge et al. 2005; Kunin et al. 2005). In some, investigators were only interested in the transfer of novel genes into genomes (Snel et al. 2002; Mirkin et al. 2003; Kunin et al. 2005), thus neglecting orthologous replacement, and underestimating the total number of transfer events among genomes of interest. In others, addressing orthologous replacement, investigators have limited themselves to analyses of very strictly defined (ubiquitous) core genes (Lerat et al. 2003; Ge et al. 2005), also underestimating the total number of horizontally transferred genes. In studies of either sort, consensus trees derived from bootstrap or posterior probability analyses, or trees based on concatenated data sets have been used as references against which to assess HGT. The former are often only partially resolved (e.g., as in Snel et al. 2002) and thus preclude detection of many phylogenetic conflicts, while the latter, as commonly used, entail the assumption that most genes do have a single history. For instance, Lerat et al. (2003) assume that individual data sets that do not statistically reject a tree based on their concatenated sequences have not experienced HGT when, in fact, their weak phylogenetic signals are compatible with many conflicting topologies (Bapteste et al. 2004).
To avoid such problems, we make an a priori assumption that individual sets of orthologous genes may not have to have the same evolutionary history, and therefore are not suitable for concatenation. Quartet decomposition analyses also avoid the “averaging” effect of consensus trees, since they partition trees inferred for each bootstrapped sample into sets of possible embedded quartets, and allow summarizing data sets with varying numbers of taxa in a single diagram (Fig. 2). In addition, the quartet decomposition method represents an improvement over methods that rely on analyses of bipartitions, since the support values for individual embedded quartets do not decay when the internal branches become shorter because of more sequences being included in the analysis.
While a majority of analyzed extended data sets (~77%) support coherence (monophyly) of cyanobacteria, some do not, and we find significant conflicting phylogenetic signals within cyanobacteria (~61% of analyzed data sets). Such conflicts could be caused by (1) instances of horizontally transferred genes; (2) differentially lost paralogs, which are impossible to discriminate from transfer events; (3) systematic artifacts of phylogenetic reconstruction (e.g., long branch attraction, compositional biases, or biases introduced through wrong models of phylogenetic reconstruction); and (4) false positives caused by insufficient phylogenetic signal. All analyses based on phylogenetic inference face these problems (Gogarten and Townsend 2005), and we took necessary precautions to minimize their impact on our genome-wide analyses (see Results for more details). Indeed, our simulation studies suggest that our screening approach is conservative, and likely to result in underestimating the extent of HGT. No doubt, among the observed conflicts are instances of real transfer events. For example, we observe conflicting signals in genes that are found in phages (see Table 3 and Lindell et al. 2004; Millard et al. 2004; Sullivan et al. 2005; Zeidner et al. 2005), and therefore are very probable candidates for HGT. (At the same time, our simulation study shows how frequently false positives arise, and casts a shadow on the reliability of phylogenetic reconstruction in general.)
False negatives are also inevitable. Transfers between sister taxa are undetectable, as will be many from unsequenced donors with no sequenced close relatives. Simulations confirm that many transfers escape detection (Table 2), probably because of the causes mentioned above. Thus, the number of detected transfers in cyanobacteria that we report here should, indeed, be considered an underestimate.
Although a majority of our data sets conflict with the plurality signal, all plurality quartets are compatible with a single fully resolved phylogenetic tree (see Fig. 3). Does the plurality topology reflect an “organismal phylogeny”? Gary Olsen has suggested a rope metaphor to illustrate the evolution of organisms and their genes (cited in Zhaxybayeva et al. 2004). The rope (rep-resenting an organismal lineage) has continuity despite the fact that no individual rope fiber (representing genes) persists throughout the entire rope. Using this metaphor, we might define the organismal lineage as that determined by the plurality of genes passed on over short time intervals. While this metaphor yields a theoretical definition of organismal lineage, it is not clear that the organismal lineage always can be reconstructed from the character of the individual fibers (genes).
Given this definition of organismal lineage, our plurality topology can be interpreted as a snapshot of relationships among extant cyanobacterial lineages. However, the picture is incomplete without providing information about observed conflicts to the plurality signal. Figure 3 depicts 135 conflicts observed between the tips of the plurality topology. This accounts only for a subset of all 685 observed conflicts, since the majority of transfers, deeper in the tree, affect the positions of multiple taxa.
The relationships recovered for the Prochlorococcus/ Synechococcus group point toward extensive gene flow between well-characterized groups of organisms.
The three Prochlorococcus marinus strains share many derived characteristics including cell shape, environment, type of antenna pigments (Partensky et al. 1999), and a distinctive threonyl tRNA synthetase likely acquired by HGT (Fig. 5). These synapomorphies notwithstanding, the plurality signal recovered from our analysis and the analysis by Beiko et al. (2005) group one of the P. marinus strains as sister taxon to a marine Synechococcus. These conflicting phylogenetic signals reveal a fuzzy species boundary that was postulated for prokaryotes (Lawrence 2002): under this model, only the neighborhood of genes conferring ecological distinctiveness is expected to conform to the biological species concept, whereas other genes recombine freely across the species boundary. However, fuzzy species boundaries are also found in eukaryotes without post-mating barriers. For example, in incipient species of Darwin's finches, frequent introgression can make some individuals characterized as belonging to the same species by morphology and mating behavior genetically more similar to a sister species (Grant et al. 2004).
Distribution of genes across functional categories shows that genes from all functional categories are transferred (see Figs. 6 and 7). We do not see a bias toward any biological function among the intraphylum HGT events (see Fig. 6), contradicting a recent report by Nakamura et al. (2004) and the complexity hypothesis (Jain et al. 1999).
The fact that we detect that ~50% of extended gene families putatively have a history of HGT (either between cyanobacteria and other phyla, or within cyanobacteria, or both) suggests that HGT plays an important role in the evolution of cyanobacteria, and the relationships among the taxa of this phylogenetic group cannot be represented by a strictly bifurcating tree. We find more conflicts within cyanobacteria than between cyanobacteria and other phyla, as did Beiko et al. (2005), using different methods of gene family selection, phylogenetic reconstruction, and HGT identification. Thus, our results are compatible with the hypothesis that HGT can reinforce coherence of a phylogenetic group.
Nevertheless, cyanobacteria are far from a fully coherent group (all genes supporting monophyly). Twenty-three percent of the 700 data sets for which monophyly was tested failed the test, showing non-cyanobacteria within the cyanobacterial clade, or cyanobacteria embedded within other phyla. Interestingly, even among those genes supporting cyanobacterial monophyly, a majority showed evidence of HGT within the cyanobacteria.
Methods
Quartet decomposition analyses
We analyzed 11 cyanobacterial genomes from NCBI and JGI databases: Anabaena sp. PCC7120, Trichodesmium erythraeum IMS101, Synechocystis sp. PCC6803, Prochlorococcus marinus CCMP1375 (also known as SS120), Prochlorococcus marinus MED4 (also known as CCMP1986), Prochlorococcus marinus MIT9313, marine Synechococcus WH8102, Thermosynechococcus elongatus BP-1, Gloeobacter violaceus PCC7421, Nostoc punctiforme ATCC29133, and Crocosphaera watsonii WH8501. We detected sets of orthologous protein-coding genes defined as mutual fully transitive reciprocal BLASTP (Altschul et al. 1997) hits (with E-value below 10−4) (see Zhaxybayeva and Gogarten 2002 for methodology). There are 3804 genes present in at least four of the genomes (see Table 1). In the analyses of the “relaxed core” presented, we used the 1128 genes present in at least nine of the genomes. Each data set was aligned using the CLUSTALW program version 1.83 (Thompson et al. 1994). For each data set, the shape parameter for a Γ distribution to approximate among-site rate variation (Yang 1994) was estimated using TREE-PUZZLE version 5.2 (Schmidt et al. 2002) with four discrete categories. One hundred bootstrap samples were generated using the SEQBOOT program from PHYLIP package version 3.6 (Felsenstein 1993), and for each bootstrap sample, a distance matrix was calculated using TREE-PUZZLE version 5.2 with the shape parameter set as estimated for the original data set. Phylogenetic trees from the distance matrices were calculated with the NEIGHBOR program from the PHYLIP package version 3.6 (Felsenstein 1993). For each data set of orthologous genes, we generated a list of embedded quartets (i.e., all possible combinations of four taxa contained in the data set). For each embedded quartet in each data set the bootstrap support vector was calculated, that is, the bootstrap support for each of the three alternative quartet topologies (see Zhaxybayeva and Gogarten 2003 for methodology and discussion of advantages of embedded quartet analyses). False inferences were screened as described in Results, and quartets with at least 80% bootstrap support for one of three possible unrooted tree topologies were summarized in a quartet spectrum diagram (see Fig. 2).
Plurality signal reconstruction
Quartet topologies that were supported by a plurality of data sets were used to reconstruct a supertree using the “matrix representation using parsimony” (MRP) method (Baum 1992; Ragan 1992) as implemented in Clann version 2.0.2 (Creevey and McInerney 2005). The resulting matrix was analyzed in PAUP* version 4.0beta10 (Swofford 1998) using an exhaustive tree space search to find the most parsimonious tree.
Functional category assignments
Functional categories were assigned to sets of orthologous genes by performing BLASTP searches of the COG database, March 2003 release (Tatusov et al. 2003), choosing the category of the top-scoring BLAST hit.
Simulations
We performed simulations of genome evolution using EvolSimulator (http://bioinformatics.org.au/evolsim/). Seven hundred genes were simulated for 10,000 generations in a dynamic population of genomes, with speciation balancing extinction (at a nominal rate of 0.015 events per generation) to maintain ~50 extant lineages at any given time following a brief initial phase of population growth. Disabling paralogous duplication as well as gene loss, each genome maintained exactly 700 genes whose orthology could thus be perfectly tracked. Parameters affecting sequence evolution were selected to reproduce a level of sequence divergence similar to that observed in the breadth of the cyanobacterial phylum. These parameters control mutation rates and biases at the gene and genome level, as well as per-residue substitution acceptabilities of the resulting proteins in a model that will be described elsewhere (R.G. Beiko and R.L. Charlebois, unpubl.).
In simulations to estimate false positives, no HGT was allowed. We selected 11 out of the 50 simulated genomes and performed the quartet decomposition analysis as described above. In addition, the same analysis was performed but using phylogenetic trees calculated instead with the PhyML program, version 2.4.4 (Guindon and Gascuel 2003).
To estimate false negatives (i.e., instances of HGT that are not detected), the second and third sets of simulations were conducted using the same parameters, but permitting relations-biased HGT (more transfer among more recently diverged genomes), at one of two rates (nominally 1.0 event per generation, and nominally 0.5 events, respectively, within the entire population). Each of the resulting 50 genomes had ~22%–25% of genes with a history of HGT in the one simulation, and 11%–13% of genes with a history of HGT in the other simulation. We selected the same 11 genomes and performed the quartet decomposition analysis as described above. Since not every family that had a history of HGT during a simulation would have an impact on the phylogeny of the subset of 11 genomes used in the analysis, we corrected the number of genes with a history of transfer only to include those whose history could cause phylogenetic incongruities in the 11-taxon subtree.
Extended data sets analyses
We added homologous sequences from 168 sequenced genomes (the list of genomes is available as Supplemental material) to each cyanobacterial data set by performing BLASTP searches and keeping the top-scoring hits for each cyanobacterial sequence in a data set with E-values below 10−20. Sequences within an extended data set (other than cyanobacterial) with 99% or higher identity at the amino acid level were excluded from further analyses (to reduce the size of an extended data set). The extended data sets were aligned using CLUSTALW version 1.83 (Thompson et al. 1994). Sites in which at least 50% of the taxa had a gap were removed. The shape parameter α for the Γ distribution was estimated using TREE-PUZZLE version 5.2 (Schmidt et al. 2002). One hundred bootstrap samples were generated for each extended data set using SEQBOOT from the PHYLIP package. For each bootstrap sample, a distance matrix was calculated in TREE-PUZZLE (Schmidt et al. 2002) using discrete approximation of the Γ distribution with four categories and with a pre-calculated value of the shape parameter α. Using these maximum likelihood distances, phylogenetic trees were calculated with the NEIGHBOR program of the PHYLIP package. Data sets were divided into three categories based on the topologies of corresponding phylogenetic trees: (1) cyanobacteria form a monophyletic group with at least 80% bootstrap support; (2) other taxa intersperse with cyanobacteria with at least 80% bootstrap support; (3) insufficiently resolved to support either of the above. The latter were considered phylogenetically uninformative and were excluded from further analyses.
Other software used
Most of the scripts for data analyses were written in Perl and Java (the scripts are available as Supplemental material). Java programs utilized the PAL library (Drummond and Strimmer 2001). The sets of orthologous genes were detected with the help of a MySQL database (http://www.mysql.com). Quartet spectra were plotted using GnuPlot (http://www.gnuplot.info). Combinations of M out of N taxa were generated using Chase's combinatorial algorithm (Chase 1970).
Acknowledgments
This work was supported through NASA Exobiology Program (NAG5-11470), NASA AISR (NNG04GP90G), and NSF Microbial Genetics Program (MCB-0237197) grants to J.P.G. and through CIHR (MOP-4467) and Genome Atlantic grants to W.F.D. O.Z. is supported through a CIHR Postdoctoral Fellowship and is an honorary Killam Postdoctoral Fellow at Dalhousie University.
Footnotes
Supplemental material is available online at http://www.genome.org. and http://carrot.mcb.uconn.edu/cyano/.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5322306.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.O., Sarchfield S.W., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Sarchfield S.W., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Sjogren A.M., Davis L.A., Embley T.M., Davis L.A., Embley T.M., Embley T.M. Gene transfers from Nanoarchaeota to an ancestor of diplomonads and parabasalids—Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Mol. Biol. Evol. 2005;22:85–90. doi: 10.1093/molbev/msh254. [DOI] [PubMed] [Google Scholar]
- Bapteste E., Boucher Y., Leigh J., Doolittle W.F., Boucher Y., Leigh J., Doolittle W.F., Leigh J., Doolittle W.F., Doolittle W.F. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 2004;12:406–411. doi: 10.1016/j.tim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Barker G.L., Handley B.A., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Handley B.A., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Stevens J.R., Hayes P.K., Hayes P.K. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (cyanobacteria) filaments in the Baltic Sea. Microbiol. 2000;146:2865–2875. doi: 10.1099/00221287-146-11-2865. [DOI] [PubMed] [Google Scholar]
- Baum B. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Beiko R.G., Harlow T.J., Ragan M.A., Harlow T.J., Ragan M.A., Ragan M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y., Douady C.J., Papke R.T., Walsh D.A., Boudreau M.E., Nesbo C.L., Case R.J., Doolittle W.F., Douady C.J., Papke R.T., Walsh D.A., Boudreau M.E., Nesbo C.L., Case R.J., Doolittle W.F., Papke R.T., Walsh D.A., Boudreau M.E., Nesbo C.L., Case R.J., Doolittle W.F., Walsh D.A., Boudreau M.E., Nesbo C.L., Case R.J., Doolittle W.F., Boudreau M.E., Nesbo C.L., Case R.J., Doolittle W.F., Nesbo C.L., Case R.J., Doolittle W.F., Case R.J., Doolittle W.F., Doolittle W.F. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- Boucher Y., Douady C.J., Sharma A.K., Kamekura M., Doolittle W.F., Douady C.J., Sharma A.K., Kamekura M., Doolittle W.F., Sharma A.K., Kamekura M., Doolittle W.F., Kamekura M., Doolittle W.F., Doolittle W.F. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 2004;186:3980–3990. doi: 10.1128/JB.186.12.3980-3990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois R.L., Doolittle W.F., Doolittle W.F. Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase P. Algorithm 382: Combinations of M out of N objects [G6] Commun. ACM. 1970;13 [Google Scholar]
- Chisholm S.W., Olson R.J., Zettler E.R., Goericke R., Waterbury J.B., Welschmeyer N.A., Olson R.J., Zettler E.R., Goericke R., Waterbury J.B., Welschmeyer N.A., Zettler E.R., Goericke R., Waterbury J.B., Welschmeyer N.A., Goericke R., Waterbury J.B., Welschmeyer N.A., Waterbury J.B., Welschmeyer N.A., Welschmeyer N.A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- Creevey C.J., McInerney J.O., McInerney J.O. Clann: Investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21:390–392. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- Drummond A., Strimmer K., Strimmer K. PAL: An object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics. 2001;17:662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- Dufresne A., Garczarek L., Partensky F., Garczarek L., Partensky F., Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony and compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
- Department of Genetics, University of Washington; Seattle: 1993. PHYLIP (Phylogeny Inference Package) Distributed by the author. ———. [Google Scholar]
- Inferring phylogenies. Sinauer; Sunderland, MA: 2004. ———. [Google Scholar]
- Ge F., Wang L.-S., Kim J., Wang L.-S., Kim J., Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S.J., Turner S., Olsen G.J., Barns S., Lane D.J., Pace N.R., Turner S., Olsen G.J., Barns S., Lane D.J., Pace N.R., Olsen G.J., Barns S., Lane D.J., Pace N.R., Barns S., Lane D.J., Pace N.R., Lane D.J., Pace N.R., Pace N.R. Evolutionary relationships among cyanobacteria and green chloroplasts. J. Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J.P., Townsend J.P., Townsend J.P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Gogarten J.P., Doolittle W.F., Lawrence J.G., Doolittle W.F., Lawrence J.G., Lawrence J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Grant P.R., Grant B.R., Markert J.A., Keller L.F., Petren K., Grant B.R., Markert J.A., Keller L.F., Petren K., Markert J.A., Keller L.F., Petren K., Keller L.F., Petren K., Petren K. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution Int. J. Org. Evolution. 2004;58:1588–1599. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gupta R.S., Pereira M., Chandrasekera C., Johari V., Pereira M., Chandrasekera C., Johari V., Chandrasekera C., Johari V., Johari V. Molecular signatures in protein sequences that are characteristic of cyanobacteria and plastid homologues. Int. J. Syst. Evol. Microbiol. 2003;53:1833–1842. doi: 10.1099/ijs.0.02720-0. [DOI] [PubMed] [Google Scholar]
- Hagen K.D., Meeks J.C., Meeks J.C. The unique cyanobacterial protein OpcA is an allosteric effector of glucose-6-phosphate dehydrogenase in Nostoc punctiforme ATCC 29133. J. Biol. Chem. 2001;276:11477–11486. doi: 10.1074/jbc.M010472200. [DOI] [PubMed] [Google Scholar]
- Harlow T.J., Gogarten J.P., Ragan M.A., Gogarten J.P., Ragan M.A., Ragan M.A. A hybrid clustering approach to recognition of protein families in 114 microbial genomes. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy M., Penny M., Penny M. Spectral analysis of phylogenetic data. J. Classif. 1993;10:5–24. [Google Scholar]
- Honda D., Yokota A., Sugiyama J., Yokota A., Sugiyama J., Sugiyama J. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 1999;48:723–739. doi: 10.1007/pl00006517. [DOI] [PubMed] [Google Scholar]
- Huang J., Xu Y., Gogarten J.P., Xu Y., Gogarten J.P., Gogarten J.P. The presence of a haloarchaeal type tyrosyl tRNA synthetase marks the opisthokonts as monophyletic. Mol. Biol. Evol. 2005;22:2142–2146. doi: 10.1093/molbev/msi221. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Tabata S., Tabata S. Synechocystis sp. PCC 6803—A useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 2001;70:73–83. doi: 10.1023/A:1013887908680. [DOI] [PubMed] [Google Scholar]
- Iwai M., Katoh H., Katayama M., Ikeuchi M., Katoh H., Katayama M., Ikeuchi M., Katayama M., Ikeuchi M., Ikeuchi M. Improved genetic transformation of the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 2004;45:171–175. doi: 10.1093/pcp/pch015. [DOI] [PubMed] [Google Scholar]
- Jain R., Rivera M.C., Lake J.A., Rivera M.C., Lake J.A., Lake J.A. Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Aravind L., Makarova K.S., Aravind L., Aravind L. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Goldovsky L., Darzentas N., Ouzounis C.A., Goldovsky L., Darzentas N., Ouzounis C.A., Darzentas N., Ouzounis C.A., Ouzounis C.A. The net of life: Reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G. Gene transfer in bacteria: Speciation without species? Theor. Popul. Biol. 2002;61:449–460. doi: 10.1006/tpbi.2002.1587. [DOI] [PubMed] [Google Scholar]
- Lawrence J.G., Hendrickson H., Hendrickson H. Genome evolution in bacteria: Order beneath chaos. Curr. Opin. Microbiol. 2005;8:572–578. doi: 10.1016/j.mib.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Lento G.M., Hickson R.E., Chambers G.K., Penny D., Hickson R.E., Chambers G.K., Penny D., Chambers G.K., Penny D., Penny D. Use of spectral analysis to test hypotheses on the origin of pinnipeds. Mol. Biol. Evol. 1995;12:28–52. doi: 10.1093/oxfordjournals.molbev.a040189. [DOI] [PubMed] [Google Scholar]
- Lerat E., Daubin V., Moran N.A., Daubin V., Moran N.A., Moran N.A. From gene trees to organismal phylogeny in prokaryotes: The case of the γ-Proteobacteria. PLoS Biol. 2003;1 doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D., Sullivan M.B., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W., Sullivan M.B., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W., Tolonen A.C., Rohwer F., Chisholm S.W., Rohwer F., Chisholm S.W., Chisholm S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodders N., Stackebrandt E., Nubel U., Stackebrandt E., Nubel U., Nubel U. Frequent genetic recombination in natural populations of the marine cyanobacterium Microcoleus chthonoplastes . Environ. Microbiol. 2005;7:434–442. doi: 10.1111/j.1462-2920.2005.00730.x. [DOI] [PubMed] [Google Scholar]
- Mann N.H., Cook A., Millard A., Bailey S., Clokie M., Cook A., Millard A., Bailey S., Clokie M., Millard A., Bailey S., Clokie M., Bailey S., Clokie M., Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424 doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- Millard A., Clokie M.R., Shub D.A., Mann N.H., Clokie M.R., Shub D.A., Mann N.H., Shub D.A., Mann N.H., Mann N.H. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.R., Augustine S., Olson T.L., Blankenship R.E., Selker J., Wood A.M., Augustine S., Olson T.L., Blankenship R.E., Selker J., Wood A.M., Olson T.L., Blankenship R.E., Selker J., Wood A.M., Blankenship R.E., Selker J., Wood A.M., Selker J., Wood A.M., Wood A.M. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc. Natl. Acad. Sci. 2005;102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin B.G., Fenner T.I., Galperin M.Y., Koonin E.V., Fenner T.I., Galperin M.Y., Koonin E.V., Galperin M.Y., Koonin E.V., Koonin E.V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3 doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M., Blaxter M.L., Bird D.M., McCarter J.P., Blaxter M.L., Bird D.M., McCarter J.P., Bird D.M., McCarter J.P., McCarter J.P. Comparative genomics of nematodes. Trends Genet. 2005;21:573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Morandi A., Zhaxybayeva O., Gogarten J.P., Graf J., Zhaxybayeva O., Gogarten J.P., Graf J., Gogarten J.P., Graf J., Graf J. Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J. Bacteriol. 2005;187:6561–6564. doi: 10.1128/JB.187.18.6561-6564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ru S., Katoh A., Dong S., Kuwabara T., Ru S., Katoh A., Dong S., Kuwabara T., Katoh A., Dong S., Kuwabara T., Dong S., Kuwabara T., Kuwabara T. Proceedings of 12th International Congress on Photosynthesis. 2001. An extracellular hemolysin homolog from cyanobacterium Synechocystis sp. PCC6803; pp. S36–S10. [Google Scholar]
- CSIRO Publishing, Melbourne, Australia. Nakamura Y., Itoh T., Matsuda H., Gojobori T., Itoh T., Matsuda H., Gojobori T., Matsuda H., Gojobori T., Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J.G., Groisman E.A., Lawrence J.G., Groisman E.A., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Olendzenski L., Zhaxybayeva O., Gogarten J.P., Zhaxybayeva O., Gogarten J.P., Gogarten J.P. Horizontal gene transfer: A new taxonomic principle? In: Syvanen M., Kado C., Kado C., editors. Horizontal gene transfer. Academic Press; New York: 2002. pp. 427–435. [Google Scholar]
- Otero A., Vincenzini M., Vincenzini M. Nostoc (Cyanophyceae) goes nude: Extracellular polysaccharides serve as a sink for reducing power under unbalanced C/N metabolism. J. Phycol. 2004;40:74–81. [Google Scholar]
- Partensky F., Hess W.R., Vaulot D., Hess W.R., Vaulot D., Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phylogenetic inference based on matrix representation of trees. Mol. Phylogenet. Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. ———. [DOI] [PubMed] [Google Scholar]
- Ragan M.A. Detection of lateral gene transfer among microbial genomes. Curr. Opin. Genet. Dev. 2001;11:620–626. doi: 10.1016/s0959-437x(00)00244-6. [DOI] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y., Waterbury J.B., Herdman M., Stanier R.Y., Herdman M., Stanier R.Y., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. [Google Scholar]
- Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Arellano A., Coleman M., Hauser L., Hess W.R., Coleman M., Hauser L., Hess W.R., Hauser L., Hess W.R., Hess W.R., et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rudi K., Skulberg O.M., Jakobsen K.S., Skulberg O.M., Jakobsen K.S., Jakobsen K.S. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bacteriol. 1998;180:3453–3461. doi: 10.1128/jb.180.13.3453-3461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Seo P.S., Yokota A., Yokota A. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, rpoC1 and rpoD1 gene sequences. J. Gen. Appl. Microbiol. 2003;49:191–203. doi: 10.2323/jgam.49.191. [DOI] [PubMed] [Google Scholar]
- Snel B., Bork P., Huynen M.A., Bork P., Huynen M.A., Huynen M.A. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Sullivan M.B., Coleman M.L., Weigele P., Rohwer F., Chisholm S.W., Coleman M.L., Weigele P., Rohwer F., Chisholm S.W., Weigele P., Rohwer F., Chisholm S.W., Rohwer F., Chisholm S.W., Chisholm S.W. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP* 4.0 beta version, phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Inc; Sunderland, MA: 1998. [Google Scholar]
- Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Mekhedov S.L., Nikolskaya A.N., Nikolskaya A.N., et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4 doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.S., Rocap G., King J., Chisholm S.W., Rocap G., King J., Chisholm S.W., King J., Chisholm S.W., Chisholm S.W. Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002;10:134–142. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- Turner S. Molecular systematics of oxygenic photosynthetic bacteria. Plant Syst. Evol. Suppl. 1997;11:13–52. [Google Scholar]
- Turner S., Pryer K.M., Miao V.P., Palmer J.D., Pryer K.M., Miao V.P., Palmer J.D., Miao V.P., Palmer J.D., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- Wilmotte A., Herdman M., Herdman M. Phylogenetic relationships among the cyanobacteria based on 16S rRNA sequences. In: Garrity G.M., editor. Bergey's manual of systematic bacteriology. Springer; New York: 2001. pp. 487–493. [Google Scholar]
- Wolf Y.I., Aravind L., Grishin N.V., Koonin E.V., Aravind L., Grishin N.V., Koonin E.V., Grishin N.V., Koonin E.V., Koonin E.V. Evolution of aminoacyl-tRNA synthetases—Analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Yap W.H., Zhang Z., Wang Y., Zhang Z., Wang Y., Wang Y. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 1999;181:5201–5209. doi: 10.1128/jb.181.17.5201-5209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidner G., Bielawski J.P., Shmoish M., Scanlan D.J., Sabehi G., Beja O., Bielawski J.P., Shmoish M., Scanlan D.J., Sabehi G., Beja O., Shmoish M., Scanlan D.J., Sabehi G., Beja O., Scanlan D.J., Sabehi G., Beja O., Sabehi G., Beja O., Beja O. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ. Microbiol. 2005;7:1505–1513. doi: 10.1111/j.1462-2920.2005.00833.x. [DOI] [PubMed] [Google Scholar]
- Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genomics. 2002;3 doi: 10.1186/1471-2164-3-4. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O., Gogarten J., Gogarten J. An improved probability mapping approach to assess genome mosaicism. BMC Genomics. 2003;4 doi: 10.1186/1471-2164-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O., Lapierre P., Gogarten J.P., Lapierre P., Gogarten J.P., Gogarten J.P. Genome mosaicism and organismal lineages. Trends Genet. 2004;20:254–260. doi: 10.1016/j.tig.2004.03.009. [DOI] [PubMed] [Google Scholar]