Abstract
The leading genetic cause of infant mortality is spinal muscular atrophy (SMA), a clinically and genetically heterogeneous group of disorders. Previously we described a domestic cat model of autosomal recessive, juvenile-onset SMA similar to human SMA type III. Here we report results of a whole-genome scan for linkage in the feline SMA pedigree using recently developed species-specific and comparative mapping resources. We identified a novel SMA gene candidate, LIX1, in an ~140-kb deletion on feline chromosome A1q in a region of conserved synteny to human chromosome 5q15. Though LIX1 function is unknown, the predicted secondary structure is compatible with a role in RNA metabolism. LIX1 expression is largely restricted to the central nervous system, primarily in spinal motor neurons, thus offering explanation of the tissue restriction of pathology in feline SMA. An exon sequence screen of 25 human SMA cases, not otherwise explicable by mutations at the SMN1 locus, failed to identify comparable LIX1 mutations. Nonetheless, a LIX1-associated etiology in feline SMA implicates a previously undetected mechanism of motor neuron maintenance and mandates consideration of LIX1 as a candidate gene in human SMA when SMN1 mutations are not found.
The spinal muscular atrophies (SMAs) are a genetically heterogeneous group of disorders that vary in clinical severity, from lethal in infancy to onset of mild weakness in adulthood, but all are characterized by neurogenic muscle atrophy due to degeneration of lower motor neurons of the spinal cord (Talbot and Davies 2001). The autosomal recessive SMAs with primary involvement of proximal musculature occur in one per 6000–10,000 live births and are the leading genetic cause of infant mortality (Pearn 1978; Ogino and Wilson 2002). The International SMA Consortium categorized proximal SMA by age of onset and clinical severity into types I through III (Munsat and Davies 1992). In studies restricted to well-characterized proximal SMA patients, ~97% show either deletions, gene conversions, or other subtle mutations in the telomeric copy of the survival of motor neuron gene 1 (SMN1), whereas the remaining ~3% fail to show any mutation in SMN1 (Lefebvre et al. 1995; Wirth et al. 1999; Wirth 2000). The latter category of patients with proximal SMA is usually characterized by two copies of SMN1 and most often exhibit the less severe phenotype categorized as type III (Wirth et al. 1999).
The high incidence of human SMA is mainly due to a human-specific (Rochette et al. 2001), inverted duplication of 500 kb on human chromosome (HSA) 5q13 that predisposes the SMN1 locus to de novo deletion and gene conversion events through interactions with the divergent and only partially functional centromeric copy of the gene (SMN2) (Wirth et al. 1997). The two copies of SMN differ in five nucleotide exchanges, only one of which is located in the coding region, and although it is translationally silent, it causes inefficient splicing of exon 7. Thus, 90% of transcripts generated by SMN2 lack exon 7 and only 10% are full length (Lefebvre et al. 1995; Lorson et al. 1999; Monani et al. 1999). While full-length SMN2 transcripts produce a protein identical to SMN1, the Δ7-SMN2 transcripts encode a truncated and dysfunctional protein. Increased SMN2 copy number (Wirth et al. 2006) and higher levels of full-length SMN expression from SMN2 correlate with milder clinical disease in human patients with SMN1 mutations (Coovert et al. 1997; Lefebvre et al. 1997) and in orthologous mouse models (Monani et al. 2000). The SMN1 gene product, SMN, is a ubiquitously expressed protein member of multiple ribonucleoprotein complexes with diverse roles in RNA metabolism, splicing, and transport in all cells (Meister et al. 2002; Bassell and Kelic 2004). Therefore, a central focus of SMA research remains to discern the disease mechanism(s) and thereby to understand why the primary disease pathology is localized to spinal lower motor neurons when SMN function is vital for all cells.
We recently described a domestic cat model of SMA exhibiting autosomal recessive, juvenile-onset skeletal muscle atrophy and weakness with onset at ~12 wk of age due to lower motor neuron loss (He et al. 2005). The affected cats exhibit progressive instability with gait and postural abnormalities attributable to symmetric weakness and atrophy of proximal muscles. Skeletal muscle atrophy is evident histologically by 3 mo and becomes palpable by 5 mo of age. In time, more distally placed muscles may become involved as well. Muscle changes correlate with loss of spinal lower motor neurons and axons of the ventral horn roots, but affected cats survive well into adulthood. He et al. (2005) also reported that SMN was excluded from the feline SMA disease locus. Thus, the affected cats are similar to type III SMA patients of the milder phenotype, with onset of signs after having gained the ability to walk and having two normal copies of SMN1. By implication, function of the disease gene underlying feline SMA is essential for lower motor neuron maintenance. Therefore, we undertook an unbiased whole-genome scan for linkage to the disorder and here report identification of LIX1, a gene largely expressed in motor neurons, but of as yet undescribed function, as the putative feline SMA disease gene.
Results
Whole-genome scan and linkage analysis
We genotyped 23 privately owned and 45 purpose-bred members of a domestic cat SMA pedigree (Fig. 1) with previously mapped, highly polymorphic short tandem repeat (STR) markers (Menotti-Raymond et al. 2003a) developed for the Feline Genome Project (http://www.ncbi.nlm.nih.gov/projects/genome/guide/cat/). As shown in Figure 1, the disorder segregates as an autosomal recessive trait, and all parents of affected cats can be traced to a single common ancestor. Therefore, we expected affected cats to be homozygous by descent for the same mutation and closely flanking marker alleles. We detected significant linkage with marker FCA689 on A1q, subsequently confirmed by other markers on A1q (Table 1), and multipoint analysis predicted that the cat SMA locus was between FCA071 and FCA770 (peak score 20.1, with all alternate placements scoring at least one LOD unit less).
Figure 1.
Domestic cat SMA pedigree. Cats above the dashed line were privately owned, purebred Maine coon cats, and those below were purposely bred for this investigation. Squares indicate males, circles indicate females, filled symbols indicated cats exhibiting the SMA phenotype, and half-filled symbols indicate obligate carriers. All cats indicated by numbers were genotyped, but cat 80 and those numbered >229 were genotyped only during fine-mapping of the established disease interval. The investigational breeding colony was derived by gamete collection (semen from cat 58 and oocytes from the unrelated mongrel cat 180), in vitro fertilization, and embryo transfer.
Table 1.
Integrated RH, comparative, and genetic linkage map of the feline SMA region
Comparison of the published order of feline markers (Murphy et al. 2000; Menotti-Raymond et al. 2003a, b) to the order of genes in the homologous chromosomal regions of human (HSA 5q), dog (CFA 3), and mouse (MMU 13) suggested the occurrence of an inversion within the feline disease interval relative to the other genomes or possible errors in gene order indicated by the cat radiation hybrid (RH) map. To address these alternative possibilities, we ordered sequence-tagged sites (STS) of eight protein-coding loci distributed across the region by RH mapping (Table 1, column 2). Ben-Dor and Chor (1997) had shown theoretically that larger panel sizes produce more robust RH maps, so the genotyping panel was expanded from 93 to 178 hybrid cell lines. As shown in Table 1, in the higher resolution RH map, the SMA disease interval displayed linear syntenic homology with the comparable region of the human genome (Table 1, column 3). The reordering of type I markers placing HEXB adjacent to SMN, rather than telomeric of FCA071, was further supported by comparison of the map to orthologous regions of the dog and mouse genomes and provided a high degree of confidence in the marker order across the disease interval.
Deletion abrogating LIX1 and LNPEP expression
For fine mapping the SMA locus, new STR markers within introns or2kb5′ or 3′ of six genes in the disease interval were developed and genotyped in the SMA pedigree (Table 1, columns 4 and 5). Four markers defining an ~5.2-Mb interval showed no recombination of alleles with disease locus alleles, but LIX1 was prominent among those because the marker appeared to be homozygous in all 28 SMA carriers and did not amplify in any of the 31 affected cats. These segregating, but nonamplifying allele(s) suggested the existence of a deletion, possibly disrupting LIX1 on the disease allele. In cats outside of this pedigree, the LIX1 marker probably behaves as a typical Mendelian microsatellite polymorphism in which alleles are defined by variable numbers of a short repeat unit. However, carriers in the SMA pedigree appeared to be homozygous, albeit for different alleles, but this was actually hemizygosity. In affected cats, no alleles of the LIX1 microsatellite were detected because both copies are deleted, whereas the other three intragenic microsatellites that had zero recombination (NR2F1, RHOBTB3, RGMB) did amplify in the affected cats.
Because the LIX1 microsatellite was deleted in affected cats, we avoided the usual challenge of choosing and sequencing positional candidate genes for mutations, but were instead faced with the challenge of determining the endpoints of the mutation and supplying further proof that the deletion disrupts LIX1 expression. Fifty-two additional sequence-tagged sites (STS) in LIX1 exons and at regular genomic intervals 3′ of LIX1 were analyzed in affected cat DNA, including the downstream genes LNPEP, LRAP, ARTS-1, and CAST. A deletion was confirmed, and the breakpoints were narrowed to intron 3 of LIX1 and intron 1 of LNPEP (Fig. 2A). LIX1 and LNPEP are transcribed from opposite strands, their most 3′ exons adjacent, so the data indicated loss of exons 4–6 of LIX1 and all but exon 1 of LNPEP. Based on comparison to the homologous region in the dog genome by BLAT analysis (http://genome.ucsc.edu/cgi-bin/hgBlat?), we estimated the deletion at the cat SMA disease locus to be ~140 kbp. We identified the precise deletion breakpoints by sequencing the PCR product amplified from affected cat genomic DNA with primers flanking the deletion. Two copies of the short sequence, AGTTTA, flanked the deletion site, a finding compatible with a deletion mechanism of nonhomologous recombination (Woods-Samuels et al. 1991).
Figure 2.
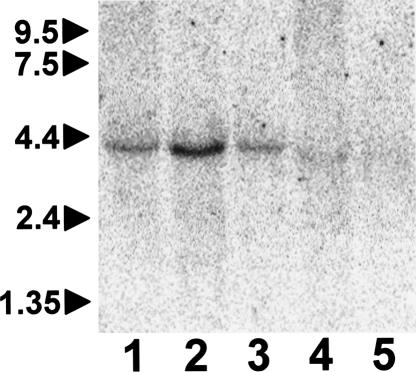
An ~140-kb deletion abrogates LIX1 and LNPEP expression in SMA affected cats. (A) A schematic of the genomic organization of LIX1 and LNPEP on FCA A1q. Arrows indicate the direction of transcription of each gene. The scale of chromosome coordinates is from the region of conserved synteny in the dog genome (CFA 3) (UCSC Genome Browser; July 2004 assembly). Sequence of the deleted and normal alleles are aligned showing the precise breakpoints below the schematic. The immediately flanking 6-bp sequence, AGTTTA, is in bold. (Note that the genome browser did not itself identify exon 1 of LNPEP by homology with RefSeq genes, but limited homology was identified by BLAT search of the dog genome with a cat sequence that was 82% identical over 300 bp of the human exon 1 and flanking 5′-UTR and intron 1 sequences). (B) PCR products produced from genomic DNA in a multiplex reaction designed to amplify across both deletion breakpoints when the normal allele (lane N) was present. A third product corresponding to the affected cat sequence shown in A amplified when the deleted allele was present, either in homozygous affected (lane A) or heterozygous carrier (lane C) cats. (C) RT–PCR products amplified from cervical spinal cord ventral horn RNA of a genetically normal cat (lane N), a clinically normal carrier (lane C), and an SMA affected cat (lane A). PCR primers for LIX1 were from exons 1 and 4, LNPEP primers were from exons 14 and 15, and GAPDH primers were from exons 6 and 8.
We confirmed that all 31 affected cats were homozygous for the deleted allele and all 28 obligate carriers were heterozygous by multiplex PCR assay amplifying products from each of the deletion breakpoints on the normal and deleted alleles of FCA A1q (Fig. 2B). One hundred fifty-two clinically normal purebred Maine coon cats, the breed from which the feline SMA family was derived, were heterozygous or homozygous for the normal allele. RT–PCR analysis demonstrated LIX1 and LNPEP mRNA expression in cervical spinal ventral horn gray matter of genetically normal and heterozygous carrier cats, but no expression of either gene was detected in the affected cats (Fig. 2C). Aberrant RNA transcripts were not observed in affected cat RNA by Northern blotting (data not shown). These data thus also confirmed that feline SMA in this kindred is an autosomal recessive trait caused by disruption of the LIX1/LNPEP locus.
Characterization of LIX1
LNPEP is a well-annotated gene, and based on reported functionality and expression profiles (discussed below), is an unlikely SMA disease gene. In contrast, LIX1 is poorly annotated; while it appears to be expressed largely in spinal cord (GNF Gene Expression Atlas 2, UCSC Genome Browser, Human 2004 assembly), its function is unknown in any species or tissue. Therefore, we undertook initial investigation of feline LIX1. Semi-quantitative RT–PCR (Fig. 3) and Northern blotting (Fig. 4) demonstrated that among tissues of juvenile cats, LIX1 mRNA is expressed primarily in the central nervous system (CNS) and most strongly in spinal cord. By comparison, LNPEP is more widely expressed and, except for pituitary gland and cerebellum, most strongly in the thymus, spleen, heart, and adrenal gland.
Figure 3.
Tissue expression of LIX1 and LNPEP. Shown are RT–PCR products of LIX1 and LNPEP amplified from multiple tissues of a genetically normal cat. (Lanes: 1, pancreas; 2, skeletal muscle; 3, fat; 4, liver; 5, spleen; 6, duodenum; 7, kidney cortex; 8, cardiac muscle; 9, adrenal; 10, thymus; 11, brainstem; 12, cerebellum; 13, brain cortex; 14, thalamus; 15, lumbosacral spinal cord; 16, pituitary; 17, cervical spinal cord.)
Figure 4.
LIX1 multiple tissue Northern blot. Twenty micrograms of total RNA isolated from each normal cat tissue was electrophoresed on a 0.8% formaldehyde-agarose gel, transferred to a nylon membrane, and hybridized to a random-primed labeled LIX1 cDNA probe that included 280 bp of 5′ UTR, the coding region, and 545 bp of 3′ UTR. Migration of RNA molecular weight markers (in kb) is indicated to the left. (Lanes: 1, thalamus; 2, spinal cord ventral horn; 3, kidney; 4, cardiac muscle; 5, skeletal muscle).
The nucleotide sequence of LIX1 cDNA amplified from cat spinal cord demonstrated an open reading frame (ORF) of 846 bp (GenBank accession no. DQ250154). The deduced amino acid sequence was 99%, 97%, 97%, 92%, 85%, 59%, and 51% identical to those of dog, cow, human, mouse, chicken, Danio rerio, and Drosophila melanogaster, respectively, and amino acid substitutions among these species are highly conservative. Protein sequence conservation across this broad range of species suggested that LIX1 function is essential. Of particular note was that computerized secondary structure analysis predicted amino acid residues 29–99 of LIX1 fold into a double-stranded RNA-binding domain, a domain common in proteins of RNA metabolism or transport and previously predicted in CG13139, the Drosophila LIX1 ortholog (Saunders and Barber 2003).
We observed additional conservation outside of the protein-coding sequence in LIX1 mRNA compatible with conservation of essential post-transcriptional cis regulatory elements. Sequence identity was highly concentrated in the ~400 bp before the start translation codon (5′ UTR), where the cat sequence was 97%, 95%, 95%, 88%, 69%, 52%, and 38% identical to the dog, cow, human, mouse, frog, chicken, and fruitfly sequences, respectively. In each of the mammals, the degree of nucleotide sequence conservation in the 5′ UTR was identical to or greater than that found in the ORF of LIX1. Included in this conserved segment are sequences predicted (Zuker 2003) to fold into relatively stable stem–loop structures (initial ΔĜ50 kcal/mol). In all but the chicken and fruitfly, these potential stem loops are part of or preceded by potential upstream ORFs. Examples of each of these kinds of features in mRNA have been implicated in modulating transcript stability, translation efficiency, or both (Mendell and Dietz 2001).
The LIX1 3′ UTR sequence is also highly conserved, further suggesting that post-transcriptional regulation may be an important element of LIX1 expression. The cat 3′ UTR sequence was 83%, 75%, 75%, 37%, and 39% identical to the dog, cow, human, mouse, and chicken, respectively, after excluding an Alu-J element from the human and two ruminant-specific SINEs from the cow sequences. The aligned sequences were 2.65–2.93 kb, except for the mouse sequence, which was 1.98 kb. In all but the mouse and chicken, there were four similarly placed poly(A) addition signals. We demonstrated the alternative use of two poly(A) addition signals in LIX1 mRNA isolated from cat lumbar ventral horn by RT–PCR and 3′-rapid amplification of cDNA ends (RACE) with polyadenylation beginning 1186 and 2642 bp 3′ of the stop translation codon, respectively. Among the five AAUAAA sequences in the cat LIX1 3′ UTR, the two we observed being used were those that were followed by a GU-rich region immediately after the polyadenylation site, conforming thereby to the bipartite consensus for efficiently used polyadenylation signals (Proudfoot 1991). However, multiple-tissue Northern blots revealed only a single mRNA species of 3.9–4.1 kb in each tissue of a juvenile cat (Fig. 4), compatible with message polyadenylated at the most 3′ site. This disparity of findings suggests that the shorter message determined by 3′ RACE is either not stable in these tissues or is not expressed at the developmental stage examined. The length of the stable LIX1 message and the linear extent of highly conserved sequence in the 5′ UTR also suggest that the 5′ UTR is longer than is annotated in sequences previously deposited in the GenBank and UniGene databases, none of which was determined experimentally.
Survey for human LIX1 mutations
The discovery of LIX1-associated etiology in the feline model raised the question as to its potential role in human SMA, particularly in cases that lack the common genetic cause of sequence variation in SMN1. Although they are not adjacent, SMN1 and LIX1 are genetically linked (θ = 0.25–0.30); so results of linkage analysis in small SMA families, even with multiple cases of SMA, might show no recombination between an informative marker of SMN1 when the true disease locus is LIX1.
To assess the possibility that mutations in LIX1 might also occur in cases of human SMA, we sequenced the LIX1-coding regions of 25 unrelated SMA patients (23 exhibited the type III phenotype) previously determined to be free of SMN1 deletions or subtle mutations (Wirth et al. 1999). We found that LIX1 mRNA expression is undetectable by RT–PCR in the human EBV-transformed lymphoblastoid cell lines, so the coding exons were amplified from whole-blood genomic DNA of these patients. While no mutational variant was found in exons or splice-site consensus sequences, we cannot conclusively exclude a LIX1 expression defect in these or other non-SMN1 SMA patients until LIX1-expressing tissue can be interrogated. Furthermore, populations other than that represented by our predominantly Caucasian patient sample may have higher prevalence of SMA caused by mutations at other loci or undetected SMN1 mutations (Stevens et al. 1999).
Discussion
We report here the molecular basis of type III SMA without SMN mutation in a cat model, the determination of which is the first discovery of a novel disease gene in the domestic cat by whole-genome scan and positional reasoning. Identification of LIX1 as the putative feline SMA gene and having function that is apparently required for motor neuron maintenance are findings of potential relevance to human SMA. This success demonstrates the current maturity of gene-mapping resources available for this species (Murphy et al. 2000; Menotti-Raymond et al. 2003a, b) as well as the utility of 2× whole-genome sequence coverage, even before assembly.
To date, there are reports of ~250 spontaneously occurring pathologies of likely genetic origin in cats (http://omia.angis.org.au/), and almost all have human counterparts. With the current sophistication of medical monitoring for pet cats, of which there are ~90 million in the United States alone (http://www.appma.org/press_industrytrends.asp), and the inbred nature of many cat breeds, new disease entities and new cases of previously described pathologies continue to be ascertained. With the development of genomic resources in the cat and the application of complementary comparative tools developed in other species, the domestic cat is emerging as a promising resource of pheno-typically defined genetic variation of biomedical significance (O'Brien et al. 2002). Exploration of similar resources in other species, particularly the dog and mouse, has provided important insight into otherwise unexplained biomedical disorders (e.g., Lin et al. 1999).
Strictly speaking, the question still remains as to which of the deleted genes is the SMA disease gene or whether loss of both, or perhaps of an unidentified nonprotein coding sequence is required to produce the disease. Based on reported functionality and/or expression profiles, we suggest that LIX1 alone is the disease gene. LNPEP is an aminopeptidase most highly expressed in placenta, heart, kidney, and small intestine that is variously secreted or a type II membrane protein, both forms having activity that degrade various peptides, including hormones such as oxytocin and vasopressin (Laustsen et al. 1997). The knockout mouse model revealed that Lnpep functions in some way to enhance mobilization of the glucose transporter, Glut4, to the cell surface in response to insulin stimulation, but the Lnpep − / − mice did not lose glucose homeostasis (Keller et al. 2002). Although CNS tissues of the knockout animals were not examined directly, overt neuromuscular disease was not reported, even though knockout mice over a year of age were investigated.
While LNPEP does not present a compelling case for significant expression or essential function in spinal cord, LIX1 expression appears to be highest in the spinal cord of humans (http://genome.ucsc.edu/cgi-bin/hgGene?hgsid=70251085&db=hg18&hgg_gene=NM_153234). LIX1 was first reported as transiently expressed in the chicken developing limb bud and facial primordia, the gene identifier standing for limb expression 1 (Swindell et al. 2001). The only other published report on the gene demonstrated transient Lix1 mRNA expression in the same structures as well as in various parts of the CNS in embryonic and fetal mice (Moeller et al. 2002). There was increasing expression from late gestation through adulthood in mouse spinal cord and dorsal root ganglia, with expression in spinal cord restricted to ventral horn motor neurons and some interneurons. Our data further indicate that in juvenile cats, expression of LIX1, but not LNPEP, is found mostly in the CNS and is strongest in spinal cord.
The high evolutionary conservation of LIX1 suggests that function of the gene product is essential, and the cat SMA phenotype caused by LIX1 disruption points to an important role in motor neuron development and/or maintenance. While LIX1 function remains to be determined, this putative disease gene identified by investigation of the cat SMA model exhibits an expression profile more restricted to the site of human and cat pathology than is the expression of SMN1, the most commonly altered gene in human SMA phenotypes. If the predicted double-stranded RNA-binding domain is a functional domain in this protein, it suggests that LIX1 may participate in RNA metabolism or transport, as does SMN (Meister et al. 2002; Bassell and Kelic 2004). One intriguing possibility is that the two gene products interact in a common pathway of motor neuron maintenance and that the tissue-specific nature of SMA pathology can be conferred by loss of LIX1 function by mutation at either locus. Determination of LIX1 function may well provide fresh insight into the mechanism of the disease, impetus for more targeted therapeutics, and answers to fundamental questions of motor neuron development, maintenance, and/or function.
Methods
Animals and patients
Privately owned, purebred Maine coon cats were investigated as previously described (He et al. 2005) to determine phenotype. We established the Michigan State University SMA breeding colony by semen collection of an affected male Maine coon (cat 58 in Fig. 1), in vitro fertilization of oocytes from a single unrelated domestic cat (cat 180 in Fig. 1), and embryo transfer using described methods (Swanson 2006). We performed designed matings, determination of phenotype, and euthanasia according to protocols approved by the All University Committee for Animal Use and Care of Michigan State University (protocol no. 01/04–014–00) and conforming to National Institutes of Health guidelines. Subsequently, colony cats mated and gave birth naturally without investigator assistance. To determine the SMA disease phenotype, we observed all colony offspring for clinical weakness beginning at ~10 wk and progressing through 5 mo of age. SMA patients or their parents gave informed consent for DNA analysis (University of Cologne protocol no. 04–138), according to the principles of the Declaration of Helsinki.
Genotyping
STR markers previously mapped in the cat genome and randomly distributed across the autosomes were chosen and genotyped as described (Menotti-Raymond et al. 2003a, b). We developed new gene-associated STRs for fine-mapping in the disease-interval (Supplemental Table 2) and STSs for RH and deletion breakpoint mapping (Supplemental Tables 1 and 3, respectively) by identifying the STRs or orthologous cat coding sequence in the growing database of unassembled and unannotated 2× Felis catus whole-genome shotgun (WGS) sequences (http://www.ncbi.nlm.nih.gov/blast/tracemb.shtml) by discontiguous MegaBLAST search (Zhang et al. 2000) with human or dog gene plus flanking sequence. To facilitate STS mapping and to construct a partial sequence assembly of LIX1, LNPEP, and the region between them, sequence similarity and overlap of sequences were determined with MegaBLAST (Zhang et al. 2000) and BLASTn (Altschul et al. 1997). Overlapping sequences were ordered and oriented to reflect the order and orientation found on HSA 5 and CFA 3. These were assembled into contigs using the TPF processor (http://www.ncbi.nlm.nih.gov/projects/zoo_seq/) with additional instructions to the processor to force overlaps considered valid despite low-quality end sequence.
Linkage analysis
We performed linkage computations with the Superlink program (Fishelson and Geiger 2004; available for download at http://bioinfo.cs.technion.ac.il/superlink) using assumptions of 0.001 disease allele frequency, simple recessive inheritance with full penetrance, and equal marker allele frequencies. Recombination fractions were estimated to within 0.01. Once we found an apparently linked marker, we used the existing cat maps (Menotti-Raymond et al. 2003a, b) to select nearby microsatellite markers for fine-mapping. For multipoint linkage analysis, intermarker genetic distances were estimated from the data on the SMA pedigree, and we used a 1-LOD unit support heuristic to determine the optimal marker order. Allele data for FCA microsatellites typed in the SMA pedigree and appearing in Table 1 are available at ftp://ftp.ncbi.nlm.nih.gov/pub/catSMA.
Radiation hybrid mapping
A radiation hybrid map of the portion of FCA A1q linked to the disease locus was computed using the software packages rh_tsp_ map (Agarwala et al. 2000), CONCORDE (Applegate et al. 1998), and QSopt (http://www.isye.gatech.edu/~wcook/qsopt) using the maximum likelihood criterion and the known reduction of the radiation hybrid mapping problem to the traveling salesman problem. We used methods similar to those used for the latest full cat map (Menotti-Raymond et al. 2003a) with the notable exceptions that we used vectors of size 178 (Murphy et al. 1999) and excluded markers with more than two uncertain entries. A flips test with window size 8 demonstrated that the determined marker order was at least three LOD units better than any alternative.
Mutation and gene expression analysis
We performed multiplex PCR analysis of SMA alleles using two primers flanking the deletion breakpoint in intron 3 of LIX1 and two flanking the deletion breakpoint in intron 1 of LNPEP. PCR reactions were 50 μL using primers 5′-TGGAGAGTGTGCAG GAAGCAGT-3′, 5′-TGGTGATCAAGGGCGTCTAAAA-3′, 5′-CGTCTGAGGTGTGGCTTGTCAG-3′, and 5′-TTTGGCCTTGA AGACCGCTAAA-3′, ~200 ng genomic DNA template, 2.5 mM each dNTP, 1.5 mM MgCl2, and 0.5 U Amplitaq DNA polymerase (Roche Diagnostics). Thermocycling was 35 cycles of 94°C and 62°C, each for 30 sec, and 72°C for 2 min.
Gene expression was analyzed by RT–PCR as previously described (He et al. 2005) using equal amounts of total RNA from multiple tissues isolated with Trizol (Invitrogen) according to the manufacturer's protocol. PCR primers for cat LIX1 exons 1 and 4 were 5′-ACTGCAAGAATTCAGGCATGAGG-3′ and 5′-TA GTGGTAGGCTCCAATGCTGGT-3′, respectively; for LNPEP exons 14 and 18 were 5′-TCTTTGATGATTGGGTGGCATCT-3′ and 5′-GCTACAGCCACCACGTCAGAGTT-3′, respectively; and for GAPDH exons 6 and 8 were 5′-TCATCTCTGCCCCTTCTGCTG AT-3′ and 5′-ATTCAGCTCTGGGATGACCTTGC-3′, respectively. Thermocycling was as above except that we performed these PCR reactions in 20-μL reactions for 30 cycles and the annealing temperature was 58°C. “No-RT” negative controls were included for all reactions.
The LIX1 mRNA sequence was determined by standard automated dideoxy chain termination cycle sequencing methods of RT–PCR products amplified from cat spinal cord RNA. We assembled overlapping sequences and carried out sequence analysis with the Lasergene suite of programs (DNAstar). Northern blots were prepared by standard protocols (Sambrook and Russell 2001), and 3′-RACE was performed according to the kit manufacturer's instructions (Invitrogen).
Primary and secondary protein structure prediction
We used the MegAlign program (DNAstar) to compare the deduced Cat LIX1 amino acid sequence with those of other species found in GenBank. Secondary-structure prediction was carried out using the four best-validated (Koh et al. 2003) prediction servers, PsiPRED, SABLE2, SAM-T2K, and PROFsec (http://cubic.bioc.columbia.edu/eva/sec/pairwise.html). Results were parsed, formatted, and visualized using in-house software (covered by the GNU General Public License and publicly available at http://proteins.msu.edu/Servers/Secondary_Structure/visualize_secondary_structure_predictions.html).
Human LIX1 sequencing
We amplified each LIX1 exon separately by PCR from SMA patient genomic DNA. Primer sequences are in Supplemental Table 4. Thermocycling was 31 cycles of 94°C for 15 sec, 55°C (63°C for exon 6) for 15 sec, and 72°C for 30 sec. PCR products were sequenced with dideoxy chain termination cycle sequencing methods.
RT–PCR analysis of RNA isolated from EBV-transformed lymphoblastoid cell lines of SMA patients was performed using the primers 5′-GATCCGGCTCTAGTCTTCAAA-3 and 5′-CCT CAGGGCCATTCGTAGC-3′ in exons 1 and 6, respectively. Positive controls included RT–PCR amplification of GAPDH and SFRS10 (Htra2-β) cDNA products from the sample lymphoblast RNA as well as LIX1 from RNA isolated from fetal human spinal cord.
Acknowledgments
We thank The Broad Institute and Agencourt Bioscience Corporation for public access to the 2× sequence assembly of the domestic cat genome. This work was supported in part by grants from the Association Française contre les Myopathies, National Institutes of Health (HD39888), Michigan State University Companion Animal Fund, Deutsche Forschungsgemeinschaft (Wi-945-12), Center for Molecular Medicine Cologne (TV98), and the Intramural Programs of the NIH, NCI, and NLM. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names imply endorsement by the U.S. Government.
Footnotes
Supplemental material is available online at www.genome.org. The sequence data from this study has been submitted to GenBank under accession no. DQ250154.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5268806.
References
- Agarwala R., Applegate D.L., Maglott D., Schuler G.D., Schäffer A.A., Applegate D.L., Maglott D., Schuler G.D., Schäffer A.A., Maglott D., Schuler G.D., Schäffer A.A., Schuler G.D., Schäffer A.A., Schäffer A.A. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate D., Bixby R., Chvátal V., Cook W., Bixby R., Chvátal V., Cook W., Chvátal V., Cook W., Cook W. Documenta Math. III. International Congress of Mathematics III; 1998. On the solution of traveling salesman problems; pp. 645–656. [Google Scholar]
- Bassell G.J., Kelic S., Kelic S. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr. Opin. Neurobiol. 2004;14:574–581. doi: 10.1016/j.conb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A., Chor B., Chor B. On constructing radiation hybrid maps. J. Comput. Biol. 1997;4:517–533. doi: 10.1089/cmb.1997.4.517. [DOI] [PubMed] [Google Scholar]
- Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M., Androphy E.J., Prior T.W., Burghes A.H.M., Prior T.W., Burghes A.H.M., Burghes A.H.M. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Fishelson M., Geiger D., Geiger D. Optimizing exact genetic linkage computations. J. Comput. Biol. 2004;11:263–275. doi: 10.1089/1066527041410409. [DOI] [PubMed] [Google Scholar]
- He Q., Lowrie C., Shelton G.D., Castellani R.J., Menotti-Raymond M., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., Lowrie C., Shelton G.D., Castellani R.J., Menotti-Raymond M., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., Shelton G.D., Castellani R.J., Menotti-Raymond M., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., Castellani R.J., Menotti-Raymond M., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., Menotti-Raymond M., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., Murphy W., O'Brien S.J., Swanson W.F., Fyfe J.C., O'Brien S.J., Swanson W.F., Fyfe J.C., Swanson W.F., Fyfe J.C., Fyfe J.C. Inherited motor neuron disease in domestic cats: A model of spinal muscular atrophy. Pediatr. Res. 2005;57:324–330. doi: 10.1203/01.PDR.0000153625.46892.6F. [DOI] [PubMed] [Google Scholar]
- Keller S.R., Davis A.C., Clairmont K.B., Davis A.C., Clairmont K.B., Clairmont K.B. Mice deficient in the insulin-regulated membrane aminopeptidase show substantial decreases in glucose transporter GLUT4 levels but maintain normal glucose homeostasis. J. Biol. Chem. 2002;277:17677–17686. doi: 10.1074/jbc.M202037200. [DOI] [PubMed] [Google Scholar]
- Koh I.Y.Y., Eyrich V.A., Marti-Renom M.A., Przybylski D., Madhusudhan M.S., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Eyrich V.A., Marti-Renom M.A., Przybylski D., Madhusudhan M.S., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Marti-Renom M.A., Przybylski D., Madhusudhan M.S., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Przybylski D., Madhusudhan M.S., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Madhusudhan M.S., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Eswar N., Graña O., Pazos F., Valencia A., Sali A., Graña O., Pazos F., Valencia A., Sali A., Pazos F., Valencia A., Sali A., Valencia A., Sali A., Sali A., et al. EVA: Evaluation of protein structure prediction servers. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laustsen P.G., Rasmussen T.E., Petersen K., Pedraza-Diaz S., Moestrup S.K., Gliemann J., Sottrup-Jensen L., Kristensen T., Rasmussen T.E., Petersen K., Pedraza-Diaz S., Moestrup S.K., Gliemann J., Sottrup-Jensen L., Kristensen T., Petersen K., Pedraza-Diaz S., Moestrup S.K., Gliemann J., Sottrup-Jensen L., Kristensen T., Pedraza-Diaz S., Moestrup S.K., Gliemann J., Sottrup-Jensen L., Kristensen T., Moestrup S.K., Gliemann J., Sottrup-Jensen L., Kristensen T., Gliemann J., Sottrup-Jensen L., Kristensen T., Sottrup-Jensen L., Kristensen T., Kristensen T. The complete amino acid sequence of human placental oxytocinase. Biochim. Biophys. Acta. 1997;1352:1–7. doi: 10.1016/s0167-4781(97)00036-5. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Benichou B., Cruaud C., Millasseau P., Zeviani M., Cruaud C., Millasseau P., Zeviani M., Millasseau P., Zeviani M., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J., Clermont O., Munnich A., Dreyfuss G., Melki J., Munnich A., Dreyfuss G., Melki J., Dreyfuss G., Melki J., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lin L., Faraco J., Li R., Kadotani H., Rogers W., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Faraco J., Li R., Kadotani H., Rogers W., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Li R., Kadotani H., Rogers W., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Kadotani H., Rogers W., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Rogers W., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Lin X., Qiu X., de Jong P.J., Nishino S., Mignot E., Qiu X., de Jong P.J., Nishino S., Mignot E., de Jong P.J., Nishino S., Mignot E., Nishino S., Mignot E., Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lorson C.L., Hahnen E., Androphy E.J., Wirth B., Hahnen E., Androphy E.J., Wirth B., Androphy E.J., Wirth B., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Eggert C., Fischer U., Eggert C., Fischer U., Fischer U. SMN-mediated assembly of RNPs: A complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Dietz H.C., Dietz H.C. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M., David V.A., Agarwala R., Schäffer A.A., Stephens R., O'Brien S.J., Murphy W.J., David V.A., Agarwala R., Schäffer A.A., Stephens R., O'Brien S.J., Murphy W.J., Agarwala R., Schäffer A.A., Stephens R., O'Brien S.J., Murphy W.J., Schäffer A.A., Stephens R., O'Brien S.J., Murphy W.J., Stephens R., O'Brien S.J., Murphy W.J., O'Brien S.J., Murphy W.J., Murphy W.J. Radiation hybrid mapping of 304 novel microsatellites in the domestic cat genome. Cytogenet. Genome Res. 2003a;102:272–276. doi: 10.1159/000075762. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M., David V.A., Roelke M.E., Chen Z.Q., Menotti K.A., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., David V.A., Roelke M.E., Chen Z.Q., Menotti K.A., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Roelke M.E., Chen Z.Q., Menotti K.A., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Chen Z.Q., Menotti K.A., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Menotti K.A., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Sun S., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Schäffer A.A., Tomlin J.F., Agarwala R., O'Brien S.J., Tomlin J.F., Agarwala R., O'Brien S.J., Agarwala R., O'Brien S.J., O'Brien S.J., et al. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus) J. Hered. 2003b;94:95–106. doi: 10.1093/jhered/esg008. [DOI] [PubMed] [Google Scholar]
- Moeller C., Yaylaoglu M.B., Alvarez-Bolado G., Thaller C., Eichele G., Yaylaoglu M.B., Alvarez-Bolado G., Thaller C., Eichele G., Alvarez-Bolado G., Thaller C., Eichele G., Thaller C., Eichele G., Eichele G. Murine Lix1, a novel marker for substantia nigra, cortical layer 5, and hindbrain structures. Brain Res. Gene Expr. Patterns. 2002;1:199–203. doi: 10.1016/s1567-133x(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D., Androphy E.J., Burghes A.H.M., McPherson J.D., Burghes A.H.M., McPherson J.D., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene. SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Monani U.R., Coovert D.D., Burghes A.H.M., Coovert D.D., Burghes A.H.M., Burghes A.H.M. Animal models of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- Munsat T.L., Davies K.E., Davies K.E. The International SMA consortium meeting. Neuromuscul. Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Mennotti-Raymond M., Lyons L.A., Thompson M.A., O'Brien S.J., Mennotti-Raymond M., Lyons L.A., Thompson M.A., O'Brien S.J., Lyons L.A., Thompson M.A., O'Brien S.J., Thompson M.A., O'Brien S.J., O'Brien S.J. Development of a feline whole-genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Sun S., Chen Z., Yuhki N., Hirschmann D., Menotti-Raymond M., O'Brien S.J., Sun S., Chen Z., Yuhki N., Hirschmann D., Menotti-Raymond M., O'Brien S.J., Chen Z., Yuhki N., Hirschmann D., Menotti-Raymond M., O'Brien S.J., Yuhki N., Hirschmann D., Menotti-Raymond M., O'Brien S.J., Hirschmann D., Menotti-Raymond M., O'Brien S.J., Menotti-Raymond M., O'Brien S.J., O'Brien S.J. A radiation hybrid map of the cat genome: Implications for comparative mapping. Genome Res. 2000;10:691–702. doi: 10.1101/gr.10.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien S.J., Menotti-Raymond M., Murphy W.J., Yuhki N., Menotti-Raymond M., Murphy W.J., Yuhki N., Murphy W.J., Yuhki N., Yuhki N. The Feline Genome Project. Annu. Rev. Genet. 2002;36:657–686. doi: 10.1146/annurev.genet.36.060602.145553. [DOI] [PubMed] [Google Scholar]
- Ogino S., Wilson R.B., Wilson R.B. Genetic testing and risk assessment for spinal muscular atrophy (SMA) Hum. Genet. 2002;111:477–500. doi: 10.1007/s00439-002-0828-x. [DOI] [PubMed] [Google Scholar]
- Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–428. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rochette C.F., Gilbert N., Simard L.R., Gilbert N., Simard L.R., Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens . Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W., Russell D.W. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- Saunders L.R., Barber G.N., Barber G.N. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- Stevens G., Yawitch T., Rodda J., Verhaart S., Krause A., Yawitch T., Rodda J., Verhaart S., Krause A., Rodda J., Verhaart S., Krause A., Verhaart S., Krause A., Krause A. Different molecular basis for spinal muscular atrophy in South African black patients. Am. J. Med. Genet. 1999;86:420–426. [PubMed] [Google Scholar]
- Swanson W.F. Application of assisted reproduction for population management in felids: The potential and reality for conservation of small cats. Theriogenology. 2006;66:49–58. doi: 10.1016/j.theriogenology.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Swindell E.C., Moeller C., Thaller C., Eichele G., Moeller C., Thaller C., Eichele G., Thaller C., Eichele G., Eichele G. Cloning and expression analysis of chicken Lix1, a founding member of a novel gene family. Mech. Dev. 2001;109:405–408. doi: 10.1016/s0925-4773(01)00535-4. [DOI] [PubMed] [Google Scholar]
- Talbot K., Davies K.E., Davies K.E. Spinal muscular atrophy. Semin. Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wirth B., Schmidt T., Hahnen E., Rudnik-Schoneborn S., Krawczak M., Muller-Myhsok B., Schonling J., Zerres K., Schmidt T., Hahnen E., Rudnik-Schoneborn S., Krawczak M., Muller-Myhsok B., Schonling J., Zerres K., Hahnen E., Rudnik-Schoneborn S., Krawczak M., Muller-Myhsok B., Schonling J., Zerres K., Rudnik-Schoneborn S., Krawczak M., Muller-Myhsok B., Schonling J., Zerres K., Krawczak M., Muller-Myhsok B., Schonling J., Zerres K., Muller-Myhsok B., Schonling J., Zerres K., Schonling J., Zerres K., Zerres K. De novo rearrangements found in 2% of index patients with spinal muscular atrophy: Mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am. J. Hum. Genet. 1997;61:1102–1111. doi: 10.1086/301608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B., Herz M., Wetter A., Moskau S., Hahnen E., Rudnik-Schoneborn S., Wienker T., Zerres K., Herz M., Wetter A., Moskau S., Hahnen E., Rudnik-Schoneborn S., Wienker T., Zerres K., Wetter A., Moskau S., Hahnen E., Rudnik-Schoneborn S., Wienker T., Zerres K., Moskau S., Hahnen E., Rudnik-Schoneborn S., Wienker T., Zerres K., Hahnen E., Rudnik-Schoneborn S., Wienker T., Zerres K., Rudnik-Schoneborn S., Wienker T., Zerres K., Wienker T., Zerres K., Zerres K. Quantitative analysis of survival motor neuron copies: Identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am. J. Hum. Genet. 1999;64:1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B., Brichta L., Schrank B., Lochmüller H., Blick S., Baasner A., Heller R., Brichta L., Schrank B., Lochmüller H., Blick S., Baasner A., Heller R., Schrank B., Lochmüller H., Blick S., Baasner A., Heller R., Lochmüller H., Blick S., Baasner A., Heller R., Blick S., Baasner A., Heller R., Baasner A., Heller R., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- Woods-Samuels P., Kazazian H.H., Jr., Antonarakis S.E., Kazazian H.H., Jr., Antonarakis S.E., Antonarakis S.E. Nonhomologous recombination on the human genome: Deletions in the human factor VIII gene. Genomics. 1991;10:94–101. doi: 10.1016/0888-7543(91)90489-2. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., Schwartz S., Wagner L., Miller W., Wagner L., Miller W., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]