Abstract
Array-CGH is a powerful tool for the detection of chromosomal aberrations. The introduction of high-density SNP genotyping technology to genomic profiling, termed SNP-CGH, represents a further advance, since simultaneous measurement of both signal intensity variations and changes in allelic composition makes it possible to detect both copy number changes and copy-neutral loss-of-heterozygosity (LOH) events. We demonstrate the utility of SNP-CGH with two Infinium whole-genome genotyping BeadChips, assaying 109,000 and 317,000 SNP loci, to detect chromosomal aberrations in samples bearing constitutional aberrations as well tumor samples at sub-100 kb effective resolution. Detected aberrations include homozygous deletions, hemizygous deletions, copy-neutral LOH, duplications, and amplifications. The statistical ability to detect common aberrations was modeled by analysis of an X chromosome titration model system, and sensitivity was modeled by titration of gDNA from a tumor cell with that of its paired normal cell line. Analysis was facilitated by using a genome browser that plots log ratios of normalized intensities and allelic ratios along the chromosomes. We developed two modes of SNP-CGH analysis, a single sample and a paired sample mode. The single sample mode computes log intensity ratios and allelic ratios by referencing to canonical genotype clusters generated from ∼120 reference samples, whereas the paired sample mode uses a paired normal reference sample from the same individual. Finally, the two analysis modes are compared and contrasted for their utility in analyzing different types of input gDNA: low input amounts, fragmented gDNA, and Phi29 whole-genome pre-amplified DNA.
A variety of chromosomal aberrations underlies developmental abnormalities (constitutional aberration) and cancer (acquired aberration) (Albertson and Pinkel 2003). Many of these aberrations are characterized by rearrangements in genomic DNA or changes in copy number such as deletions, duplications, and amplifications (Kallioniemi et al. 1992, 1994, 1996; Hayashizaki et al. 1993; Wang et al. 2002). Historically, two key techniques have been used to measure DNA copy number in DNA samples: comparative genomic hybridization (CGH) and loss of heterozygosity (LOH). CGH has been used extensively to detect amplifications and large homozygous deletions, and LOH has been used to detect regions of allelic homogeneity indicative of hemizygous deletions or copy-neutral LOH. LOH is typically assessed through the analysis of polymorphic genetic markers, traditionally either VNTRs or RFLPs (Singh et al. 1993; Dockhorn-Dworniczak et al. 1994), and more recently single nucleotide polymorphisms (SNPs) (Slater et al. 2005; Zheng et al. 2005). The importance of LOH is underscored by its extensive history in the discovery of many classical tumor-suppressor genes (TSGs) including RB1, WT1, and TP53, involved in the formation of retinoblastoma, Wilm's tumor, and Li-Fraumeni syndrome, respectively (Gray and Collins 2000; Hanahan and Weinberg 2000; Albertson and Pinkel 2003; Albertson et al. 2003).
CGH has been used widely to characterize DNA copy changes in tumors. Originally this technique was implemented using metaphase chromosomal spreads but has been adapted to array-CGH using BAC, cDNA, and oligonucleotide arrays (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Albertson and Pinkel 2003; Barrett et al. 2004; Ylstra et al. 2006). Development of high-density array-CGH technology has enabled 100-kb resolution using whole-genome BAC arrays containing >33,000 BAC clones (Ishkanian et al. 2004), or with oligonucleotide arrays containing >390,000 probes (Selzer et al. 2005). The effective resolution is not just a function of the number of probes on the array, but also depends on the signal-to-noise ratio (SNR) of the system. Typically, oligonucleotide probes have a much lower SNR than BAC arrays, and as a result oligonucleotide arrays require averaging over greater numbers of probes to achieve the same effective resolution (Ylstra et al. 2006). Nonetheless, oligonucleotide arrays are easily manufactured, and the technology is amenable to scaling improvements allowing ever-increasing feature density.
The ability to detect microdeletions and microduplications is essential in the study of constitutional disorders. Several disease states have been attributed to both microdeletion haploin-sufficiency and duplication-mediated overexpression in regions harboring known transcription factors and tumor-suppressor genes (Santarosa and Ashworth 2004). As such, microsatellite LOH and FISH have been instrumental in detecting microdeletions, such as the Williams-Beuren Syndrome (WBS) caused by a heterozygous deletion of a 1.5-Mb region on chromosome 7q11.23 (Francke 1999). Surprisingly, duplications in this region also lead to a phenotype that is nearly the inverse of the WBS phenotype (Somerville et al. 2005). In spite of the progress in this field, neither microsatellite LOH nor FISH analysis has the ability to quickly demarcate the extent and breakpoints of aberrations. There is a clear need for technology that can map breakpoints of these aberrations to delineate genes involved in a syndrome.
The ability of SNP-CGH to simultaneously measure both intensity differences and allelic ratios in a genomic sample allows both DNA copy number and copy-neutral LOH to be assessed. This is particularly important since copy-neutral LOH is receiving greater attention as a mechanism of possible tumor initiation (Langdon et al. 2006). In tumorigenesis, copy-neutral LOH is thought to arise from mitotic recombination, caused by the occurrence of a rare crossover event during mitotic cell division. The products of mitotic recombination are regions of the genome exhibiting uniparental disomy (UPD), in which both genomic regions originate from the same parent (Ledbetter and Engel 1995).
In this study, we demonstrate the utility of SNP-CGH with two Infinium whole-genome genotyping BeadChips, Human-1 and the HumanHap300, assaying 109K and 317K SNP loci respectively, to detect chromosomal aberrations in constitutional and tumor samples. The Whole Genome Genotyping (WGG) assay has previously been described and is composed of four basic components: a whole-genome amplification, hybridization capture on a 50mer probe array (BeadChip), array-based primer extension SNP scoring, and immunohistochemistry-based signal amplification (Gunderson et al. 2005; Steemers et al. 2006). This assay design allows almost any SNP to be assayed and virtually unlimited multiplexing dependent only on the array feature density. Using a model system of cell lines harboring from one to four X chromosomes, we show that this technology has very fine detection limits (sub-100 kb for the 317K BeadChip) and can detect single copy changes in chromosomal copy number with low variability levels. We demonstrate the detection of various chromosomal aberrations including both homozygous and hemizygous deletions as well as duplications with a set of constitutional aberrations and tumor cell lines previously characterized by FISH, BAC array-CGH, or SKY karyotyping. For the analysis of tumor samples, we address the effect of tumor heterogeneity and mosaicism on detection limits by showing that this assay can detect LOH in tumor samples mixed with ∼67% normal stroma. Finally, we compare single sample analysis to paired sample analysis and evaluate the pros and cons of both approaches.
Results
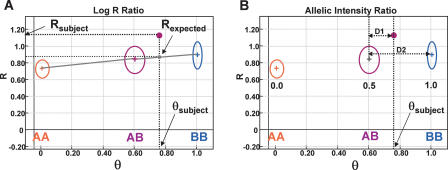
We present two genomic plots, one of the log-normalized intensity ratio and the other of an allelic intensity ratio for a single representative SNP from the genome (Fig. 1). These two parameters originate from a polar coordinate transformation of two-channel (two alleles) intensity data. This transformation generates a normalized intensity value, called R, and an allelic intensity ratio, called theta (θ). The intensity comparison between a subject sample and a reference sample is conducted in either a single sample mode using canonical genotype clusters derived from a large training set of normal samples, or in a paired sample mode in which the subject and reference sample are from the same individual and run side by side in one experiment.
Figure 1.
Analyzing SNP-CGH data. (A) The log2 R ratio compares the observed normalized intensity (Rsubject) of the subject sample to the expected intensity (Rexpected; gray dot) based on the observed allelic ratio, θsubject, through a linear interpolation (gray lines) of the canonical clusters AA, AB, and BB (shown as circles) in the GenoPlot. The normalized intensity value obtained from a single SNP is represented as a purple dot. The R and θ values for the subject are shown with thick black dotted lines. (B) The canonical clusters (shown as circles) are also used to convert θ values, that is, θsubject, to B allele frequency (allelic copy ratio). This is accomplished by a linear interpolation of the known allele frequencies assigned to each cluster (0.0, 0.5, and 1.0). The allele frequency for an observed θ value falling between two clusters is also calculated by linear interpolation with lines D1 and D2. In the example shown, a data point falling approximately a third of the distance from the AB to the BB cluster (e.g., θsubject ∼ 0.76) has an allele frequency of 0.5 + 0.33 * 0.5 = 0.67. These two transformed parameters, log2 R ratio and B allele frequency, are then plotted along the entire genome for all SNPs on the array.
In the “single sample” analysis mode, the observed normalized intensity of the subject sample (Rsubject) is compared to the expected intensity (Rexpected) computed from a linear interpolation of the observed allelic ratio (θsubject) with respect to the canonical genotype clusters (Fig. 1A). The three canonical genotype clusters were generated at one point in time by training on ∼120 normal samples, and serve as standards for all future experiments. In addition to computing Rexpected, the observed allelic intensity ratio (θsubject) is used to estimate a quantitative B allele frequency for the particular SNP in the given sample by using interpolation of the known B allele frequencies of the three canonical clusters (0, 0.5, and 1.0) (Fig. 1B). These two transformed parameters, log2 R ratio [log2(Rsubject/Rexpected)] and B allele frequency (AF), are then plotted along the entire genome for all SNPs on the array. It is these genomic profile plots that form the basis of detecting chromosomal aberrations with SNP-CGH.
X-copy cell lines as a model system for detection limits and sensitivity
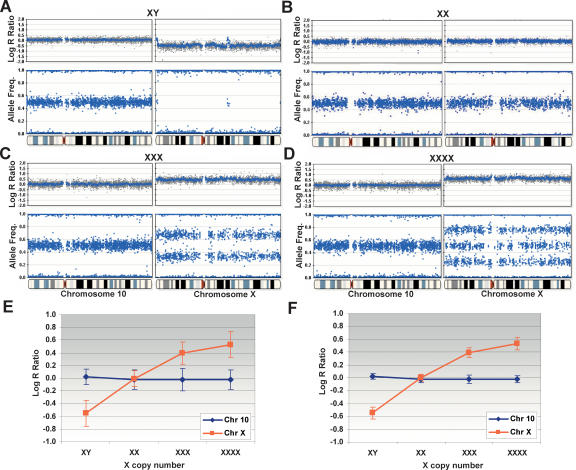
We evaluated the ability of the 109K SNP array to detect single-copy gains or losses using an X-copy model system. We analyzed amplified gDNA from several different cell lines containing one X (NA19160, male), two X (NA19172, female), three X (NA04626), and four X (NA01416) chromosomes (Mark et al. 1999; Bauters et al. 2005). The genome profiles of the log R ratios and AF (B allele frequencies) are shown for the X chromosome juxtaposed next to an autosome (chromosome 10) for comparison purposes (Fig. 2). These CGH genomic profiles are notable in that both copy number and genotyping allelic ratios are plotted as side-by-side profiles providing simple visualization of both physical and genetic abnormalities. Summary statistics for the X-copy cell line data are shown in Table 1 for reference. Single copy deletions (from two to one copy), monoallelic duplications (from two to three copies), and amplifications (two to four or more copies) are easily detected. In the case of XY, the presence of a single X chromosome is manifest as a decrease in the log R ratio from ∼0 to −0.55, and in the allele frequency plot, the heterozygous state completely collapses to the homozygous axis (Fig. 2A). Interestingly, the pseudoautosomal regions on the Y chromosome are also detected in the analysis of the male (XY) sample (Rouyer et al. 1986). In the XXX case, the log R ratio increases from ∼0 to ∼+0.40, and the heterozygous state splits into two populations, one located at 0.67 (2:1 ratio) and the other at 0.33 (1:2 ratio) (Fig. 2C). Finally, in the XXXX case, the log R ratio increases to +0.53, and the heterozygous state splits into three populations (0.25, 0.51, and 0.76) (Fig. 2D). The reason for the three clusters in XXXX derives from 3:1, 2:2, and 1:3 allelic ratios originating from the nondisjunction event in both the male and female gametes leading to four independently inherited chromosomes (Cammarata et al. 1999).
Figure 2.
X-copy cell lines as a model system. Single copy deletions, monoallelic duplications (trisomies), and amplifications are detected on the Human-1 (109K) BeadChip using cell lines with one to four X chromosomes. All plots are shown juxtaposed to normal genome profiles from chromosome 10. (A) In XY, the presence of a single X chromosome is shown as a decrease in the log2 R ratio from ∼0to −0.55, and in the AF plot where the heterozygous state completely collapses to the homozygous axis. Note the pseudoautosomal regions on the Y chromosome. (B) In XX, the presence of the expected two copies of the X chromosome show no deflection in the log R ratio (∼0), and the heterozygotes are clustered around +0.5. (C) In XXX, the log2 R ratio increases from ∼0 to +0.395, and the heterozygous state splits into two clusters, one located at 0.67 (2:1 ratio) and the other at 0.33 (1:2 ratio). (D) In XXXX, the log2 R ratio increases to +0.53, and the heterozygous state is divided into three populations (0.25, 0.51, and 0.76) with allelic ratio of 3:1, 2:2, and 1:3, respectively. (E) The response of the log R ratio for both the X chromosome and chromosome 10 for each X-copy number cell line. The log R ratio increases with increasing X-copy number for the X chromosome but not for chromosome 10. The corresponding standard deviation is shown for each data point. (F) The log R ratio response calculated for the X-copy cell lines with a 10-SNP moving average. Note that the standard deviation has been significantly reduced. For all genomic profiles, the blue line indicates a 500-kb moving median for the Human-1 (109K) BeadChip. The complete X-copy cell line data set, cluster information, and receiver operator plots are available in the Supplemental material.
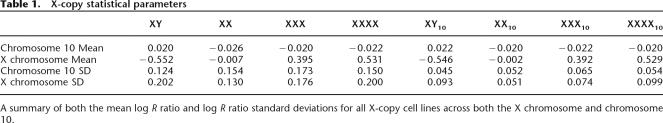
Table 1.
X-copy statistical parameters
We plotted the response of the log R ratio for both the X chromosome and an autosome of similar size, chromosome 10, for each X-copy number cell line. As expected, the log R ratio increases with increasing X-copy number for the X chromosome but not chromosome 10 (Fig. 2E). The corresponding standard deviation of the log R ratio is also shown. Statistical noise can be reduced by using a moving window average across neighboring SNPs. The corresponding log R ratio response plot is shown in Figure 2F for a 10-SNP moving average. In this case, the standard deviation of the moving average log R ratio is significantly reduced, and the effective SNR is correspondingly increased (Table 2). Interestingly, the response profile of the log R ratio is attenuated at higher copy levels potentially because of saturation effects in both the hybridization and immunohistochemistry staining steps in the assay. Receiver operator curves (ROC), which plot the true versus false positive rate for the detection of aberrations (deletions, duplications, and amplifications), were generated for both the raw X-copy data and data generated with a 5- and a 10-SNP moving average (see Supplemental material).
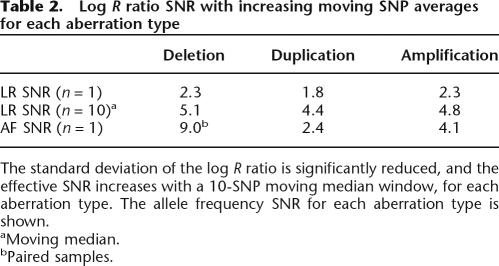
Table 2.
Log R ratio SNR with increasing moving SNP averages for each aberration type
The SNR for a single measurement point is much higher for the allele frequency (AF) measurement than the log R ratio parameter. The combination of both the log R ratio and AF is quite powerful since the precision of the allelic ratio measurement allows robust detection of the aberration and the log R ratio can be used to interpret the nature of the aberration. Furthermore, the statistical confidence of detecting an aberration can be greatly increased by using a moving window average of neighboring SNPs. We derived a 10-SNP “rule-of-thumb” based on analysis of the ROCs for various numbers of SNPs in a moving median window; a window size of 10 generated the most ideal ROC curves (see Supplemental material). This window size of 10 also encompasses, on average, three to four heterozygous SNPs per window based on an average heterozygosity of 30%–40%. Based on only the log R ratio parameter, we achieved a deletion detection rate of 90% (true positives) with a ∼1% false-positive rate for a single SNP. When we used a 10-SNP moving median, we achieved a remarkable detection rate of 99.9% with a 1.2% false-positive rate. This 10-SNP rule implies that the 109K BeadChip (26 kb mean spacing; 13 kb median spacing) has an effective resolution <260 kb, and the 317K BeadChip (9 kb mean spacing) has an effective resolution of <90 kb. As such, we typically plot a smoothing function for the log R ratio profile using a moving average of 300 kb for the 109K array, and 100 kb for the 317K array.
Examples of common aberrations using HL-60
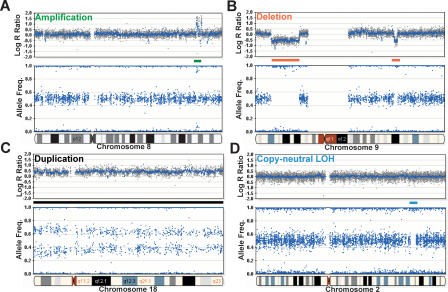
After assessing the ability to detect aberrations using the X-copy model system, we analyzed a common tumor cell line, HL-60 (human promyelocytic leukemia cell line; ATCC No. HL60). HL-60 contains a modicum of several well-characterized chromosomal deletions and amplifications (Volpi et al. 1996; Cottier et al. 2004). In addition, HL-60 was recently analyzed on the Affymetrix 10K array-CGH platform by the Sanger Institute as part of the Cancer Genome Project (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/10kCGHviewer.cgi?dna=HL-60). Karyotype analysis on HL-60 has also been performed using the SKY karyotype 6 (∼2 Mb) with LOH is noted by the lack of heterozygotes in the AF, but no change is seen in the log R ratio (blue bar). For all log R ratio plots, the blue line indicates a 500-kb moving median for the Human-1 (109K) BeadChip. For all aberrations contained in HL-60, refer to the Supplemental material. method and is available at NCI and NCBI's SKY/M-FISH and CGH Database (http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi). Such karyotype analyses have proven successful in analyzing various types of chromosomal aberrations (Liyanage et al. 1996; Macville et al. 1997).
Using the 109K BeadChip, we successfully detected all of the aberrations previously detected by both platforms (SKY karyo-type and array-CGH) in the HL-60 cell line. Several types of such aberrations are shown in Figure 3. A 1.9-Mb amplification, denoted by a green arrow in Figure 3A, was found on chromosome 8. This amplification is detected by an increase in the log R ratio and a split in the heterozygous B allele frequency from a normal value of 0.5 (1:1 allelic ratio) to ∼0.1 and ∼0.9 (roughly 1:9, 9:1 allelic ratios), indicating allelic amplification in this region. Examples of two hemizygous deletions were found on chromosome 9, and they are shown in Figure 3B (denoted with red arrows). The two deletions, 21 Mb and 2.4 Mb in size, were visually detected by observing a negative deflection in the log R ratio plot and the lack of heterozygotes in the allele frequency plot corroborating the reduction in copy number. The smallest aberration observed in the HL-60 cell line was an ∼25-kb homozygous deletion on chromosome 17 in the ATP1B2 gene (Supplemental material). The HL-60 cell line also presents an example of a monoallelic duplication as illustrated by the trisomy present on the majority of chromosome 18 (∼76 Mb). This duplication is characterized by an upward deflection in the log R ratio plot and a split in the heterozygous allele frequencies corresponding to a 2:1 and 1:2 allelic ratio ( Fig. 3C) similar to the results with the XXX cell line. Finally, an illustration of detecting a copy-neutral LOH event is depicted in the genomic profile from the Coriell cell line NA09208 (Fig. 3D, blue arrow). We find a small region on chromosome 2 (∼2 Mb) containing LOH as noted by the lack of heterozygotes in the AF without an observed change in the log R ratio. This analysis demonstrates the power of SNP-CGH to differentiate between LOH events with a reduction in copy number from copy-neutral events; this should prove particularly valuable in evaluating genome-wide aberrations in tumor samples.
Figure 3.
Examples of aberrations using HL-60 on the 109K BeadChip. The human promyelocytic leukemia cell line (HL-60) contains several well-characterized chromosomal aberrations. (A) An example of several discrete monoallelic amplifications across an ∼4.5-Mb region on chromosome 8 (green bar). The monoallelic amplification is evidenced by an increase in the log R ratio and the large split in the allele frequency. Based on the allelic ratios of ∼0.1 and ∼0.9, the level of amplification is on the order of five-to 10-fold. (B) Two deletions found on chromosome 9 (red bars). The first, which is ∼21 Mb, is detected by a deflection in the log R ratio and the collapse of heterozygotes in the allele frequency. The second deletion (∼2.4 Mb) was also detected using the same parameters. (C) The entire length of chromosome 18 (∼76 Mb) is duplicated (black bar), inferring a total copy number of 3. Notice the increase of the log R ratio to ∼0.5 and the cluster split in AF. (D) SNP-CGH arrays can detect a copy-neutral LOH event such as recombination or gene conversion. A small region on chromosome
Detection of constitutional chromosomal aberrations
As illustrated by the previous examples, high-density SNP-CGH BeadChips should have the power to easily detect most common constitutional chromosomal aberrations. This has particular importance in the field of neo and postnatal analysis of chromosomal aberrations. Currently, most clinical tests of aberrations use a combination of karyotyping and fluorescence in situ hybridization (FISH). These karyotypes are useful for outlining the overall chromosomal structures and detecting aberrations including translocations and inversions. However, karyotyping resolution is limited to 10–20 Mb. As such, follow-up studies on patients usually employ FISH probes for detection of small deletions and duplications. Clinics currently use panels of FISH probes for ∼25 microdeletion syndromes including DiGeorge, Prader-Willi (Angleman), Williams-Beuren, and Wolf-Hirschhorn syndromes (Scambler et al. 1991; Driscoll et al. 1992; Clayton-Smith et al. 1993; Wright et al. 1997; Francke 1999). In contrast, genomic profiling with SNP-CGH arrays enables discovery of aberrations throughout the genome and allows for a precise localization of chromosomal breakpoints of those aberrations.
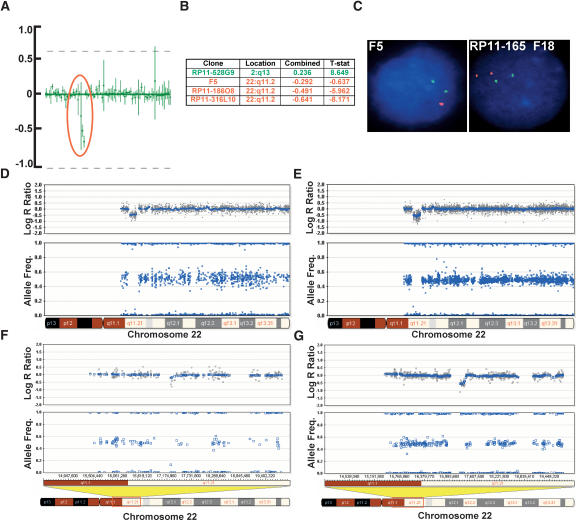
To demonstrate the feasibility of using high-density Bead-Chips for detection of congenital chromosomal aberrations in patient samples, we conducted a blinded study on six DNA samples collected from patients with diagnosed developmental clinical phenotypes. The DNA was previously characterized by karyotype, FISH, and BAC array-CGH analysis. All documented chromosomal aberrations (n = 10) ranging in size from 72 kb (duplication) to entire chromosomes were detected on the 317K BeadChip. The 72-kb duplication was detected on the 109K primarily because of the split of the heterozygous cluster. The 109K array detected nine out of 10 aberrations (Fig. 4F), and the 317K detected the aberration missed by the 109K array (a 115-kb deletion) (Fig. 4G). An example of one such aberration, a small 1.5-Mb deletion on chromosome 22q11.2, detected by BAC array-CGH (Fig. 4A,B), FISH analysis (Fig. 4C), was also detected by SNP-CGH on the 109K and 317K (Fig. 4D,E, respectively). In the blinded study, numerous (n = 12) other small deletions and duplications ranging in size from 46 kb to 470 kb were discovered using either the 109K or 317K arrays (data not shown). These results demonstrate the utility of both the 109K and 317K Bead-Chips for the detection of various chromosomal aberrations in constitutional samples. In particular, the 317K array, with its greater density, allowed discovery of more small aberrations than the 109K.
Figure 4.
Verification of a chromosomal deletion with BAC array-CGH and FISH. We performed a blinded study on samples collected from patients with developmental clinical phenotypes previously characterized by karyotype, FISH, and BAC array-CGH analysis. (A) Data from a chromosomal BAC microarray showing the mean values of signal to noise (T/R) ratio and error bars of data from two separate hybridizations. The profile shown here represents an enlarged section of a chromosomal microarray showing a loss of three clones in the DiGeorge syndrome I critical region (encircled in red). (B) List of BAC clones, their location, and the log2 R ratio, indicating a loss of copy number in this region (three clones denoted in red). One additional clone shows a potential amplification present in another position in the genome (denoted in green; plots not shown). (C) FISH analysis using the F5 clone (for the DiGeorge region) showing one signal in red while the control probe in green shows two signals, confirming a deletion in the DiGeorge critical region. FISH analysis using the RP11-165F18 clone (distal to F5) shows no deletion. (D) The same aberration, an ∼1.5-Mb deletion on chromosome 22q11.2, detected by SNP-CGH on the Human-1 (109K) array as seen by the deflection in the log R ratio and the loss of heterozygote data points in the AF. (E) The same deletion on chromosome 22q11.2 detected by SNP-CGH on the HumanHap300 (317K) array. Notice the higher density of SNPs in this region on the HumanHap300 BeadChip. This finding confirms the deletion known to be present in the critical region of the DiGeorge syndrome. (F) Another deletion detected on chromosome 22q11.21 that is difficult to discern with the Human-1, which was not detected with any other method. (G) The same deletion can clearly be visualized with the HumanHap300 BeadChip, especially by the deflection in the log R ratio. For all plots, the blue line indicates a 500-kb and a 100-kb moving median, for the Human-1 and HumanHap300 BeadChips, respectively.
Analyzing heterogeneous tumor samples
Analysis of tumor samples obtained from patients typically involves some form of microdissection to extract a specific population of cells since contamination by surrounding tissues containing normal genomic DNA can result in decreased accuracy for LOH calls (Hata et al. 2006). Current estimates suggest that contamination with normal stroma from 10% to 50% of the total genomic DNA content can severely affect measurements of LOH. On the other hand, even in samples with high levels of normal tissues, it may still be possible to detect changes in copy number and homozygous deletions, but LOH resolution is still lost. With the development of laser capture microdissection (LCM), the ability to gather samples of high homogeneity increases, which should improve LOH measurements in the genome (Rook et al. 2004). Other techniques such as flow cytometry can also increase the accuracy of LOH calls in tumor samples (Abeln et al. 1994).
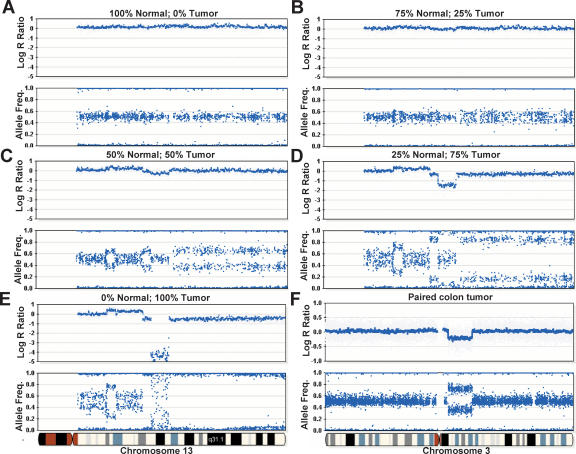
We evaluated the effect of tumor heterogeneity on the detectability of various types of aberrations. This was accomplished by mixing gDNA harvested from cell line pairs (blood and tumor; ATCC: CRL-2325D, CRL-2324D) derived from the same patient in various ratios of 0%, 25%, 50%, 75%, and 100% tumor (Fig. 5A–E, respectively). The samples were combined and gDNA was amplified as described in the Methods section and then analyzed on 109K BeadChips. With relatively low levels of normal contamination (0%–25%) (see Fig. 5D), the allele frequency difference between tumor and normal is clearly distinguishable. Even in the case of 50% normal contamination, various aberrations can easily be detected (Fig. 5C); however, their interpretation remains ambiguous as to whether they are deletions or duplications (Fig. 5B). Finally, at 75% normal contamination, aberrations are not visible. At 100% tumor, homozygous deletions are easily detected in both the log R ratio and the allele frequency profile (Fig. 5E). Here, the AF profile exhibits “scatter” since the target loci are not present with homozygous deletions. Detection of homozygous deletions is made challenging in the presence of contaminating normal gDNA since the allele frequency is indistinguishable from normal; the only detectable signal is a slight dip in the log R ratio profile. In addition, since the AF profile is characteristically more responsive than the log R ratio, an LOH event or duplication may be occurring on the proximal region of the p-arm of chromosome 13 although no change in the log R ratio is observed (Fig. 5E). Overall, these results suggest that the fraction of normal tissue in heterogeneous tumor samples should be kept well below 50% for sensitive detection of aberrations with a goal of 25% or less.
Figure 5.
Analyzing heterogeneous tumor samples. DNA from a tumor cell line was mixed with matched normal DNA at ratios of 0%, 25%, 50%, 75%, and 100% and analyzed on the Human-1 (109K) BeadChip. The genome profile of chromosome 13 is shown as an example. (A) No aberrations are seen in the sample containing 100% normal gDNA. (B) No discernable differences in the log R ratio are seen in a sample composed of 75% normal and 25% tumor. It is also difficult to determine if there are any changes in the allelic frequency. (C) At 50% normal and 50% tumor DNA, deflections in the log R ratio appear. Changes in the AF are also seen, although it is difficult to establish the nature of each aberration. At these levels, an allelic duplication event and allelic LOH bear resemblance to each other. (D) At 25% normal and 75% tumor gDNA, the nature of each type of aberration becomes more apparent. For example, the large decrease in the log R ratio in the center of the plot denotes a potential region with a deletion, which was not easily discernable in C. (E) The genoplot from a pure (100%) tumor sample. A homozygous deletion can be seen in the center of the plot, visualized by a decrease in the log R ratio. (F) A region exhibiting LOH is observed on chromosome 3 in a paired colon tumor patient sample analyzed on the HumanHap300 BeadChip. A decrease in the log R ratio indicates a loss of copy number. The AF is divided into two populations(∼0.33 and ∼0.67), suggesting that this sample contains ∼67% normal cells. The blue line shown for all log R ratio profiles indicates a 250-kb and a 100-kb moving median for the Human-1 and HumanHap300 BeadChips, respectively.
Owing to the extensive aneuploidy often present in tumor cell lines that confounds normalization and interpretation of data, we also evaluated patient tumor samples containing endogenous levels of normal contamination. In cases with a modest number of aberrations, it can be straightforward to clearly detect LOH in samples containing ∼67% normal cells. Shown in Figure 5F is a profile from a paired colon tumor patient sample analyzed on the 317K BeadChip. The deflection downward in the log2 R ratio indicates a loss of copy number. In the AF profile, the heterozygous state splits into two populations (∼0.33 and ∼0.67), suggesting that this sample contains ∼67% normal cells since the heterozygotes have not completely collapsed to the homozygote axes. Regardless, in this case, the LOH event is clearly detectable even in the presence of a high percentage of normal cells. In spite of the challenges analyzing such samples, the ever-increasing density of array technology, such as the HumanHap550 BeadChip, should further improve the sensitivity with heterogeneous samples, especially since one can average over greater numbers of probes.
Effect of varying DNA input amount and fragmentation on SNP-CGH data
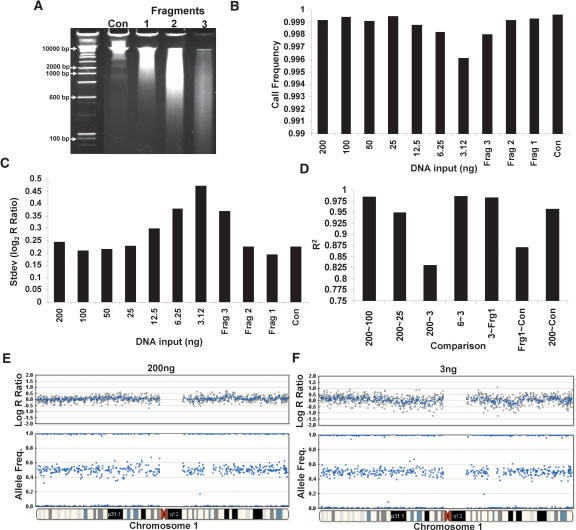
The requirement to obtain relatively pure tumor samples may place limits on the amount and quality of starting gDNA available for the assay. As such, we evaluated the effect of decreased gDNA input amounts and the effect of fragmented DNA on the quality of the data from the SNP-CGH assay. For these experiments, we used a multisample BeadChip format allowing analysis of ∼10,000 SNP assays across 12 samples (subset of 109K product). We titrated gDNA from the standard 1× input (750 ng equivalent) down to 1/64th input (∼12 ng), and a “zero DNA” control was included (data not shown). The effect of degraded DNA was evaluated by artificially digesting DNA with a titration of DNase I (Fig. 6A, Fragments 1, 2, and 3) for a fixed input level 1× (750 ng equivalent). Interestingly, we find that the overall call rate is relatively insensitive to quantity and fragmentation length across the entire range of DNA inputs (Fig. 6B). The call rates for all levels of input DNA were above 0.996 even at the lowest input amount. Additionally, the total variation (standard deviation, SD) in the allelic ratios was minimally affected by input amount; however, the variation in the log R ratio increased dramatically with a decrease in input amount from 200 ng (0.24) to 12 ng (0.47) (Fig. 6C,E,F). The log R ratio and AF genomic profiles are shown for chromosome 1 at input levels of 1× and (1/64)×, respectively (Fig. 6E,F). Similarly to reduced input amounts, fragmented DNA also exhibited an increase in the variation of the log R ratio from 0.19 (Fragment 1) to 0.37 (Fragment 3) (Fig. 6C). It is not known, however, how these fragmentation results correlate with data produced with FFPE (formalin-fixed paraffin-embedded) samples, which often contain high levels of cross-linking and depurination.
Figure 6.
Effect of gDNA quantity and fragmentation on SNP-CGH data. SNP-CGH data quality as a function of the quantity and fragmentation length of gDNA in the amplification reactions was tested using a multisample 10K BeadChip format. (A) Various lengths of fragmented DNA were used as starting input for the whole-genome amplification reaction (Fragments 1, 2, and 3). (B) The call rate is relatively insensitive to input amount across the entire range of 200 ng to 3 ng or fragment length. The call rates for input DNA ranging from 25 to 200 ng were all above 0.999, and the call rates for the 3–12.5 ng were all above 0.996. (C) Genomic DNA was titrated from the standard 1× input (200 ng in a one-quarter scale reaction) down to 1/64th input (3 ng in a one-quarter scale reaction) as well as Fragments 1, 2, and 3. As the levels of input DNA decreased, the variability in the log R ratio noticeably increased, whereas the allelic ratio was relatively insensitive to input amounts. (D) The R 2 correlation between samples remains high when similar amounts of gDNA are used for input regardless if it falls into either high or low levels; however, the R 2 decreases dramatically between inputs that differ substantially in amount. (E) An example genome profile from chromosome 1 showing both the log R ratio and AF from the sample using 200 ng of input DNA. (F) The same plot as in E but with 3 ng of input DNA. Notice the slight increase in log R ratio variability with the lower amount of DNA input.
We suspected that the increase in log R ratio variation was due to systematic amplification bias. As such, we computed the regression r 2 for normalized intensity values between various input levels of gDNA (Fig. 6D). The r 2 correlation was high between similar input amounts of gDNA, but decreased dramatically between both high and low input amounts or between intact and degraded DNA (Fig. 6D). Here, we conclude that the variation levels can be greatly reduced in the log R ratio parameter when paired samples of similar quantity (defined as starting input amount) or fragment length are used, or when the canonical reference is generated from samples of similar quantity/quality to the subject sample. Regardless, these results suggest that only limited amounts of sample are required for LOH or copy number analyses if performed in the “paired analysis” mode. Currently, we use a canonical reference derived from “clustering” on 120 samples with a 1× input (750 ng in a full reaction; see Methods), and as such, single sample genomic profiling exhibits optimal performance when high-quality DNA is used at this input level. When using a lower input amount of gDNA, it is recommended that the user “re-cluster” on normal samples run at the corresponding reduced input levels of gDNA.
Single versus paired sample analysis
The analysis of paired gDNA samples (tumor tissue and adjacent normal tissue or blood) is a common experimental design in cancer research that allows detection of LOH using a limited number of polymorphic markers (Zauber et al. 1999). The power to detect LOH is greatly increased with paired sample analysis since LOH can be confidently asserted for even a single SNP differing in state between the pairs. Without normal or parental genotype information, however, one can only rely on statistical probabilities of obtaining runs of homozygosity. In general, a run of 10–20 contiguous homozygous SNPs is required to statistically assert LOH (assuming an average heterozygosity of 30%–40% and that SNPs are independent of one another) (Huang et al. 2004), and even then it might be explained by chance haplotype sharing (Altshuler et al. 2005).
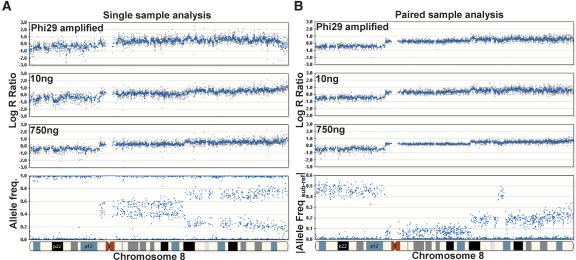
To evaluate the differences between single and paired sample modes, we used paired breast tumor cell lines obtained from ATCC (CRL-2325D, CRL-2324D). From the analysis in the previous section on input amounts, we speculated that paired analysis would reduce the variability (standard deviation) in the log R ratio. We tested this effect by comparing the log R ratio variation generated when using the standard 750 ng of input gDNA versus 10 ng of input. In addition, we tested pre-amplification of gDNA with Phi29 (Repli-G; Qiagen) starting with 10 ng of input gDNA in the Phi29 WGA pre-reaction and using 750 ng of this product in the Infinium amplification reaction. We analyzed these samples on a 12-sample BeadChip using a 30,000 subset of loci from the 317K BeadChip.
As expected, we find that paired analysis greatly decreases the variation in the log R ratio profile when using nonstandard DNA input (reduced amounts, phi29 pre-amplified, etc.) (Fig. 7B). In the single sample mode, the variation in the log R ratio across a region with normal genotypes on chromosome 6 is 0.422 for Phi29-amplified gDNA, 0.430 for 10-ng input DNA, and 0.236 for 750-ng input gDNA. In the paired sample mode, the variation in the log R ratio across the same region is significantly lower; 0.135 for Phi29-amplified gDNA, 0.162 for 10-ng input gDNA, and 0.106 for 750-ng input gDNA. In general, the data obtained with paired sample analysis are of higher quality than single sample analysis (Fig. 7B, see 750 ng). However, there are a few exceptions to this observation. If the reference sample is dramatically different in quality or quantity than the tumor sample, paired analysis may generate inferior results (data not shown). Thus it is of paramount importance when conducting paired sample analysis that the DNA samples be of similar quality as well as accurately quantified, preferably with a PicoGreen assay.
Figure 7.
Paired versus single sample analysis. Varying starting inputs of DNA (10 ng and 750 ng) from a paired breast tumor cell line were hybridized to a multisample BeadChip containing a subset of loci (∼33,000) from the HumanHap300 product. In addition, Phi29 was used to amplify 10 ng of gDNA, and 750 ng of this amplified product was used in the initial whole-genome amplification step. Overall, we find that the effect of different inputs of gDNA on the resulting genomic profiles is ameliorated by paired-sample analysis. (A) In the single sample mode, the variability (standard deviation) in the log R ratio is shown as gray dots for Phi29-amplified gDNA, 10 ng of input DNA, and 750 ng of input gDNA (from top to bottom). (B) In the paired-sample mode, the variation in the log R ratio across chromosome 8 is reduced for the same samples. For reference, the AF for the tumor sample is shown on the bottom left and the |Allele Freq subject-reference| for the same tumor sample with paired analysis is shown on the bottom right. Using paired-sample analysis, the allele frequency difference between normal and tumor genotypes is very distinct. Where applicable, a 500-kb moving median was used (blue line).
Discussion
Our results demonstrate the utility of using Infinium SNP-CGH bead arrays for detection of chromosomal aberrations including both homozygous and hemizygous deletions, copy-neutral LOH, monoallelic duplications, and amplifications. A major strength of SNP-CGH arrays compared to conventional array-CGH is the ability to simultaneously profile the genome for both physical and genetic anomalies (Bignell et al. 2004; Zhao et al. 2004; Koed et al. 2005). Detection of copy-neutral events such as uniparental disomy (UPD), in which both alleles of a locus are derived from the same parent, is essential to understanding both developmental disorders (DeBaun et al. 2002; Fukuzawa et al. 2003; Bruce et al. 2005) as well as tumorigenesis.
A frequent cause of UPD in somatic cells (i.e., tumors) is rare but significant mitotic recombination events (Raghavan et al. 2005; Teh et al. 2005; Langdon et al. 2006). In fact, it is hypothesized that mitotic recombination (Tischfield 1997) may be responsible for the second hit in Knudson's two-hit model describing the inactivation of tumor-suppressor genes (TSGs) (Knudson Jr. 1975, 2001). An example of this type of TSG inactivation has been documented in leukemia, in which a first-hit mutation is made homozygous through mitotic recombination (Fitzgibbon et al. 2005). As such, the widespread application of SNP-CGH arrays should facilitate the unearthing of novel tumor-suppressor genes. In addition to UPD, genetic information collected by SNPCGH is valuable for assessing the allelic origin of deletions, duplications, and amplifications (LaFramboise et al. 2005).
We used an X chromosome titration model system to assess the signal to variability ratio of our Infinium SNP-CGH platform for various types of chromosomal aberrations. Averaging data from neighboring SNPs along the genome greatly increases the effective SNR. The average variation in the log2 intensity ratio for the Infinium SNP-CGH platform is relatively low (∼0.13 to ∼0.20) as compared to other BAC or oligonucleotide array-CGH platforms, which exhibit variation levels of ∼0.16 (BAC arrays) and ∼0.26 and ∼0.66 for two different oligonucleotide array formats (Ylstra et al. 2006). We find, overall, that the allele frequency parameter is much more precise (standard deviation of ∼0.04) and is less affected by DNA quantity and quality than the log2 intensity ratio. However, we found that the variability in the log R ratio parameter can be greatly reduced by analyzing either paired samples or analyzing a single sample with appropriate GenTrain clusters derived from DNA samples of similar quantity (starting input amount) and fragment length. As such, for optimal performance with low input amounts such as 10 ng (vs. the standard 750 ng), the matching sample or the samples used to create GenTrain clusters should also be at this input level. Given the limited input requirement, this approach should be of use for analyzing samples generated from laser capture microdissection (LCM), permitting analysis of relatively homogeneous tumor samples (Curran et al. 2000).
Practical validation of the SNP-CGH platform was accomplished by a blinded study of six congenital samples containing various constitutional aberrations. We identified all previously documented chromosomal aberrations, as assessed by karyotype, FISH, and BAC array-CGH analysis. The detected aberrations ranged in size from a duplication of 72 kb to an entire duplicated chromosome. These results reveal the utility of using high-density SNP-CGH to screen for various chromosomal aberrations in constitutional samples. For future clinical applications, one can envision a custom SNP-CGH array containing perhaps from 20,000 to 50,000 elements designed to known regions of constitutional aberrations.
We find that the use of SNP-CGH for constitutional analysis of congenital samples is relatively straightforward. However, the heterogeneity typically found in tumor samples can make analysis and interpretation of results more challenging. Nonetheless, we have demonstrated sufficient sensitivity in mixed tumor–normal samples to detect single copy changes in tumor samples contaminated by as much as 50% normal background and LOH events in paired samples with ∼67% normal background. Robust identification of both various aberrations and copy number changes, in addition to the precise localization of breakpoints in tumor samples, will likely require refined statistical analysis algorithms (Lai et al. 2005; Nannya et al. 2005). Furthermore, identification of the exact aberration type may need to be followed up with supplemental approaches such as FISH or real-time PCR (Brinkschmidt et al. 1997; Shearer et al. 1999).
In conclusion, we have shown that SNP-CGH arrays offer several distinct advantages over conventional array-CGH. SNPCGH arrays profile the genome for both physical and genetic anomalies allowing detection of copy-neutral genetic events. Additionally, these arrays provide allelic information on deletions, duplications, and amplifications, which is important since many high-level amplification events are monoallelic. Finally, a significant advantage of SNP-CGH bead arrays is their ease of manufacture and intrinsic scalability. Just recently, the density of SNPCGH beads has been extended to >550,000 SNPs (Illumina HumanHap550), and future innovations may drive this density even higher. Given these advantages, SNP-CGH may replace array-CGH as a standard for measuring genome-wide chromosomal aberrations.
Methods
High-density SNP genotyping arrays for SNP-CGH
Illumina currently manufactures multiple formats of high-density SNP genotyping arrays (BeadChips); from 109K to 650K SNPs. A brief description of their content is provided in this section. All experimental processes for SNP-CGH described were completed with Infinium I and Infinium II WGG reagents from Illumina.
Sentrix Genotyping BeadChips
The genotyping data contained in this paper were collected either on the Sentrix Human-1 (109,000 exon-centric SNPs) or HumanHap300 (317,000 tag SNPs) Genotyping BeadChips from Illumina. The principles described within apply to the 550K array (HumanHap550). The Human-1 BeadChip employs the Infinium WGG assay to interrogate >109,000 single nucleotide polymorphism (SNP) loci on a single BeadChip. The Infinium assay generates high genotyping quality as assessed by call rates and accuracy on 120 DNA samples. The overall call rate was 99.94% and the concordance with HapMap data was 99.6%. The SNP assays were chosen from a combination of exon-centric, highly conserved, and putative functional SNPs. These include SNPs located directly within coding, promoter, and highly conserved, non-coding regions. It is probable that a large fraction of disease-related variants, including nonsynonymous, splice site, and regulatory variants, will lie in conserved and exon-rich regions. More than 85% of all SNPs on this BeadChip are within 10 kb of an exon or within evolutionarily conserved sequences. In addition, “gene deserts” were filled in with at least 5 SNPs per 200 kb density. The genome-wide average spacing between SNPs is 26 kb, and the median spacing is 13 kb (higher density in gene-rich regions).
The 10K linkage multisample BeadChip contains uniformly spaced SNPs that are a subset of the SNPs from the 109K Bead-Chip, and the average spacing between each SNP probe is ∼300 kb, which should allow for the detection of typical amplifications and deletions of several megabases. The 317K BeadChip (Human-Hap300) consists of >300,000 tagSNP assays derived from the HapMap project with a 9-kb mean spacing between SNPs, enabling an effective resolution of 90 kb.
DNA samples
The three canonical genotype clusters for both the Human-1 and the HumanHap 300 BeadChips were created from ∼120 individuals representing a subset of the HapMap samples (which may contain structural polymorphisms) collected from the Yoruba population (YRI), the CEPH project in Utah (CEU; Northern and Western European ancestry), the Han Chinese population of Beijing (CHB), and the Japanese population (JPT) (http://www.illumina.com/general/support/downloads/SuplMat_GenRes_Peiffer.zip).
DNA from cell lines X (NA19160, male), 2X (NA12236, female), NA04626 (3X), and NA01416 (4X) were obtained from Coriell. The paired breast tumor cell lines (CRL-2325D and CRL-2324D) were obtained from American Type Culture Collection (ATCC). The acute promyelocytic leukemia cell line (HL-60) was also obtained from ATCC (ATCC No. HL60). The paired colon tumor genomic DNA was obtained from BioChain (A704198). For the DNA fragmentation experiments, cell line NA60136 was obtained from Coriell as was the cell line exhibiting the copy-neutral LOH event (NA09208). Genomic DNA from patient samples was isolated from peripheral blood lymphocytes, cultured amniocytes, or fibroblasts by using a PureGene DNA-purification kit according to the manufacturer's protocol (Gentra Systems).
Sample amplification and hybridization for BeadChips
The whole-genome amplification process requires 250–750 ng of input gDNA and creates a sufficient quantity of DNA (1000× amplification) to be used on a single BeadChip in the Infinium assay (Illumina MP1 and AMM). After amplification, the product is fragmented using a proprietary reagent (FRG), precipitated with 2-propanol (plus precipitating reagent; PA1), and resus-pended in formamide-containing hybridization buffer (RA1). The DNA samples are denatured at 95°C for 20 min, loaded into the Tecan Flow Through Chambers (Tecan Part No. 760,810), and placed in a humidified container for a minimum of 16 h at 48°C allowing SNP loci to hybridize to the 50mer capture probes (Steemers and Gunderson 2005).
For the DNA fragmentation experiments, Coriell NA60136 was digested with 0.0125 U, 0.025 U, and 0.05 U of DNase I (Invitrogen), respectively. Briefly, 2 μg of gDNA was digested with DNase I in a 20-μL reaction, using 1-Phor-All Plus buffer and 0.5 mM CaCl2. The reactions were incubated at 37°C for 15 min, inactivated by 1 μL of 0.5 M EDTA, purified with MultiScreen PCR Plate (Millipore), resuspended in 40 μL of TE, and visualized on a 6% TBE/urea gel with SYBR Gold Staining (Invitrogen). For Phi29 amplifications, 10 μg of genomic DNA was amplified with the Repli-G kit from Qiagen (#59045) according to the manufacturer's protocol.
Infinium I (ASPE) and Infinium II (SBE) on BeadChips
Following hybridization, the BeadChip/Te-Flow chamber assembly was placed on the temperature-controlled Tecan Flowthrough Rack, and all subsequent washing, extension, and staining were performed by addition of reagents to the Te-Flow chamber.
For the allele-specific primer extension (ASPE; Infinium I) assay, the BeadChip was first washed (Illumina RA1 and PB1) to remove unhybridized and nonspecifically hybridized DNA. Next the BeadChip was blocked for 10 min prior to addition of the extension mix (XB1). The extension step extends correctly matched probes hybridized to DNA on the BeadChip and incorporates biotin-labeled nucleotides (EMM). After extension, a formamide wash removed the hybridized DNA to reduce extraneous signal. The array then underwent a multilayer staining process to signal amplify and detect the incorporated label (LMM, ASM, and XB3). Finally, the BeadChips were washed (PB1) and dried before subsequent imaging.
For the single-base extension (SBE) (Infinium II) assay, primers were extended with a polymerase and labeled nucleotide mix (TEM), and stained with repeated application of LTM (staining reagent) and ASM (anti-staining reagent). After staining was complete, the slides were washed with low salt wash buffer (PB1), immediately coated with XC4, and then imaged on the Illumina BeadArray Reader.
Imaging the BeadChip and data analysis
The Illumina BeadArray Reader is a two-color (543 nm/643 nm) confocal fluorescent scanner with 0.84-μm pixel resolution. The scanner excites the fluorophors generated during signal amplification/staining of the allele-specific (one color) or single-base (two colors) extension products on the BeadChips. The image intensities are extracted using Illumina's BeadScan software.
The data for each BeadChip is self-normalized using information contained within the array. This normalization algorithm removes outliers, adjusts for channel-dependent background and global intensity differences, and also scales the data. The X and Y color channels undergo an affine coordinate transformation to make the data appear as canonical as possible with the homozygotes lying along the transformed x-and y-axes. The following five steps are applied: (1) outlier removal; (2) a translation correction in which the asymptotes are fitted to candidate AA and BB homozygotes; the intersection of these fit lines defines the translated origin; (3) rotational correction: the angle of the AA homozygote asymptote with respect to the translated X-axis is used to define the rotational correction; (4) shear correction: the angle of the BB homozygote asymptote with respect to the translated and rotated y-axis is used to define the shear correction; (5) scaling correction: statistical centroids are computed for the candidate AA homozygotes to define an x-axis scaling parameter, and for candidate BB homozygotes to define a y-axis scaling parameter. The translated, rotated, shear-corrected data are normalized to a scale of ∼1 using the scaling parameters.
After normalization, the genotyping data are transformed to a polar coordinate plot of normalized intensity R = X norm + Y norm and allelic intensity ratio θ = (2/π) * arctan(Y norm/X norm), where X norm and Y norm represent transformed normalized signals from alleles A and B for a particular locus. The log R ratio of signal intensities is shown as log2(Rsubject/Rexpected) and is the base 2 logarithm of the ratio of observed intensity versus expected intensity (see Results section). Genomic profiles were created using the Illumina Genome Viewer (IGV) and Chromosome Browser (ICV) of Illumina's BeadStudio2.0 software, which plots SNP genotyping data to view, identify, and annotate chromosomal aberrations.
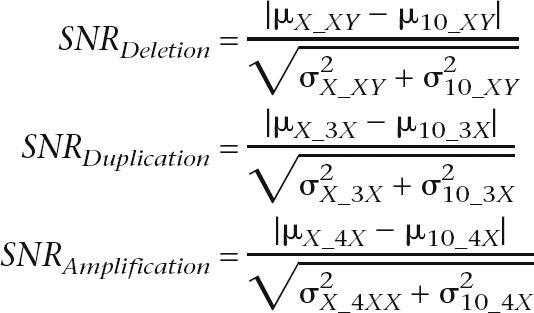
The SNR for both the log R ratio and allele frequency (AF) were calculated with the following equations (where μX_XY represents the mean log R ratio intensity or mean heterozygous (nonhomozygous) AF for the X chromosome of the XY sample. The other parameters are similarly defined.
 |
BAC array-CGH
The targeted chromosomal BAC microarrays contain 860 FISH-verified BAC clones that covered >70 known genetic disorders, 41 subtelomeric regions, and 43 pericentromeric regions.
Patient genomic DNA was isolated as described above. Control DNA was from a healthy individual male and female using the same DNA extraction method. For CGH BAC-arrays, genomic DNA was fragmented to a smear between 600 and 2000 bp by sonication with a microcup horn (Sonics & Materials) and then purified (Zymo Research).
The fragmented 500 ng of genomic DNA from patient and control was differentially labeled with cy3 and cy5 (Perkin Elmer) using Bioprime labeling (Invitrogen). To avoid dye bias, we performed dye swap experiments on each sample. Sixty micrograms of human cot-1 were added to each sample to block repetitive sequences (Invitrogen). The labeled DNA probe was dissolved in 30 μL of ULTRAhyb buffer (Ambion) and hybridized to the arrays in a sealed moisture hybridization chamber at 37°C in a rocking incubator for 22 h. After hybridization, washes were performed (2× SSC brief rinse, 50% formamide/2× SSC twice for 15 min, 0.5× SSC for 5 min, 0.07× SSC rinse twice, and nitrogen gas dry).
Arrays were scanned into 16-bit tiff image files with an Axon 4000B microarray scanner and quantified using GenePix Pro 6.0 (Axon Instruments). Data analysis was performed as previously described (Cheung et al. 2005). In addition to the uniform cutoff value of 0.2 for the test/reference ratios, a single clone T-statistic and permutation-based P-value were computed, providing additional criteria to determine whether a clone significantly deviated from the Mean.
FISH analysis
Metaphase spreads for detection of deletions were prepared from amniocytes or CVS using standard procedures. Interphase nuclei were analyzed for duplications/copy number gains. Miniprep BAC DNA (100 ng) was labeled with Spectrum Orange-dUTP or Spectrum Green-dUTP (Vysis) according to the manufacturer's protocol and used as probes for FISH analysis using established protocols (Trask 1991).
Acknowledgments
We thank our many Illumina colleagues involved in the development of the Infinium genotyping products including those in assay development, high-throughput oligonucleotide synthesis, bead production, bioinformatics and software, marketing, and manufacturing. This work was supported, in part, by a grant from the NIH/NCI (2 R44 CA103406-02). Illumina, Sentrix, BeadArray, and Infinium are registered trademarks or trademarks of Illumina, Inc.
Footnotes
Supplemental material is available online at http://www.genome.org.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5402306. Freely available online through the Genome Research Open Access option.
References
- Abeln E.C., Corver W.E., Kuipers-Dijkshoorn N.J., Fleuren G.J., Cornelisse C.J., Corver W.E., Kuipers-Dijkshoorn N.J., Fleuren G.J., Cornelisse C.J., Kuipers-Dijkshoorn N.J., Fleuren G.J., Cornelisse C.J., Fleuren G.J., Cornelisse C.J., Cornelisse C.J. Molecular genetic analysis of flow-sorted ovarian tumour cells: Improved detection of loss of heterozygosity. Br. J. Cancer. 1994;70:255–262. doi: 10.1038/bjc.1994.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D.G., Pinkel D., Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12:R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Collins C., McCormick F., Gray J.W., Collins C., McCormick F., Gray J.W., McCormick F., Gray J.W., Gray J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Collins F.S., Daly M.J., Donnelly P., Daly M.J., Donnelly P., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M.T., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Tsang P., Curry B., Baird K., Meltzer P.S., Curry B., Baird K., Meltzer P.S., Baird K., Meltzer P.S., Meltzer P.S., et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl. Acad. Sci. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters M., Van Esch H., Marynen P., Froyen G., Van Esch H., Marynen P., Froyen G., Marynen P., Froyen G., Froyen G. X chromosome array-CGH for the identification of novel X-linked mental retardation genes. Eur. J. Med. Genet. 2005;48:263–275. doi: 10.1016/j.ejmg.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Bignell G.R., Huang J., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Huang J., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Grigorova M., Jones K.W., Wei W., Stratton M.R., Jones K.W., Wei W., Stratton M.R., Wei W., Stratton M.R., Stratton M.R., et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkschmidt C., Christiansen H., Terpe H.J., Simon R., Boecker W., Lampert F., Stoerkel S., Christiansen H., Terpe H.J., Simon R., Boecker W., Lampert F., Stoerkel S., Terpe H.J., Simon R., Boecker W., Lampert F., Stoerkel S., Simon R., Boecker W., Lampert F., Stoerkel S., Boecker W., Lampert F., Stoerkel S., Lampert F., Stoerkel S., Stoerkel S. Comparative genomic hybridization (CGH) analysis of neuroblastomas—An important methodological approach in paediatric tumour pathology. J. Pathol. 1997;181:394–400. doi: 10.1002/(SICI)1096-9896(199704)181:4<394::AID-PATH800>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Bruce S., Leinonen R., Lindgren C.M., Kivinen K., Dahlman-Wright K., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Leinonen R., Lindgren C.M., Kivinen K., Dahlman-Wright K., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Lindgren C.M., Kivinen K., Dahlman-Wright K., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Kivinen K., Dahlman-Wright K., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Dahlman-Wright K., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Lipsanen-Nyman M., Hannula-Jouppi K., Kere J., Hannula-Jouppi K., Kere J., Kere J. Global analysis of uniparental disomy using high density genotyping arrays. J. Med. Genet. 2005;42:847–851. doi: 10.1136/jmg.2005.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata M., Di Simone P., Graziano L., Giuffre M., Corsello G., Garofalo G., Di Simone P., Graziano L., Giuffre M., Corsello G., Garofalo G., Graziano L., Giuffre M., Corsello G., Garofalo G., Giuffre M., Corsello G., Garofalo G., Corsello G., Garofalo G., Garofalo G. Rare sex chromosome aneuploidies in humans: Report of six patients with 48,XXYY, 49,XXXXY, and 48,XXXX karyotypes. Am. J. Med. Genet. 1999;85:86–87. doi: 10.1002/(sici)1096-8628(19990702)85:1<86::aid-ajmg16>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Cheung S.W., Shaw C.A., Yu W., Li J., Ou Z., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Shaw C.A., Yu W., Li J., Ou Z., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Yu W., Li J., Ou Z., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Li J., Ou Z., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Ou Z., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Patel A., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Yatsenko S.A., Cooper M.L., Furman P., Stankiewicz P., Cooper M.L., Furman P., Stankiewicz P., Furman P., Stankiewicz P., Stankiewicz P., et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet. Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J., Webb T., Cheng X.J., Pembrey M.E., Malcolm S., Webb T., Cheng X.J., Pembrey M.E., Malcolm S., Cheng X.J., Pembrey M.E., Malcolm S., Pembrey M.E., Malcolm S., Malcolm S.1993Duplication of chromosome 15 in the region 15q11–13 in a patient with developmental delay and ataxia with similarities to Angelman syndrome J. Med. Genet. 30: 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier M., Tchirkov A., Perissel B., Giollant M., Campos L., Vago P., Tchirkov A., Perissel B., Giollant M., Campos L., Vago P., Perissel B., Giollant M., Campos L., Vago P., Giollant M., Campos L., Vago P., Campos L., Vago P., Vago P. Cytogenetic characterization of seven human cancer cell lines by combining G- and R-banding, M-FISH, CGH and chromosome- and locus-specific FISH. Int. J. Mol. Med. 2004;14:483–495. [PubMed] [Google Scholar]
- Curran S., McKay J.A., McLeod H.L., Murray G.I., McKay J.A., McLeod H.L., Murray G.I., McLeod H.L., Murray G.I., Murray G.I. Laser capture microscopy. Mol. Pathol. 2000;53:64–68. doi: 10.1136/mp.53.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBaun M.R., Niemitz E.L., McNeil D.E., Brandenburg S.A., Lee M.P., Feinberg A.P., Niemitz E.L., McNeil D.E., Brandenburg S.A., Lee M.P., Feinberg A.P., McNeil D.E., Brandenburg S.A., Lee M.P., Feinberg A.P., Brandenburg S.A., Lee M.P., Feinberg A.P., Lee M.P., Feinberg A.P., Feinberg A.P. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am. J. Hum. Genet. 2002;70:604–611. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockhorn-Dworniczak B., Poremba C., Dantcheva R., Stucker A., Brommelkamp E., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Poremba C., Dantcheva R., Stucker A., Brommelkamp E., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Dantcheva R., Stucker A., Brommelkamp E., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Stucker A., Brommelkamp E., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Brommelkamp E., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Blasius S., Mellin W., Roessner A., Yandell D.W., Bocker W., Mellin W., Roessner A., Yandell D.W., Bocker W., Roessner A., Yandell D.W., Bocker W., Yandell D.W., Bocker W., Bocker W. Rapid detection of loss of heterozygosity of chromosome 17p by polymerase chain reaction-based variable number of tandem repeat analysis and detection of single-strand conformation polymorphism of intragenic p53 polymorphisms. Virchows Arch. 1994;424:337–342. doi: 10.1007/BF00190553. [DOI] [PubMed] [Google Scholar]
- Driscoll D.A., Budarf M.L., Emanuel B.S., Budarf M.L., Emanuel B.S., Emanuel B.S. A genetic etiology for DiGeorge syndrome: Consistent deletions and microdeletions of 22q11. Am. J. Hum. Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J., Smith L.L., Raghavan M., Smith M.L., Debernardi S., Skoulakis S., Lillington D., Lister T.A., Young B.D., Smith L.L., Raghavan M., Smith M.L., Debernardi S., Skoulakis S., Lillington D., Lister T.A., Young B.D., Raghavan M., Smith M.L., Debernardi S., Skoulakis S., Lillington D., Lister T.A., Young B.D., Smith M.L., Debernardi S., Skoulakis S., Lillington D., Lister T.A., Young B.D., Debernardi S., Skoulakis S., Lillington D., Lister T.A., Young B.D., Skoulakis S., Lillington D., Lister T.A., Young B.D., Lillington D., Lister T.A., Young B.D., Lister T.A., Young B.D., Young B.D. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–9154. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- Francke U.1999Williams-Beuren syndrome: Genes and mechanisms. Hum. Mol Genet. 8: 1947–1954. [DOI] [PubMed] [Google Scholar]
- Fukuzawa R., Hata J., Hayashi Y., Ikeda H., Reeve A.E., Hata J., Hayashi Y., Ikeda H., Reeve A.E., Hayashi Y., Ikeda H., Reeve A.E., Ikeda H., Reeve A.E., Reeve A.E. Beckwith-Wiedemann syndrome-associated hepatoblastoma: wnt signal activation occurs later in tumorigenesis in patients with 11p15.5 uniparental disomy. Pediatr. Dev. Pathol. 2003;6:299–306. doi: 10.1007/s10024-003-1009-1. [DOI] [PubMed] [Google Scholar]
- Gray J.W., Collins C., Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis. 2000;21:443–452. doi: 10.1093/carcin/21.3.443. [DOI] [PubMed] [Google Scholar]
- Gunderson K.L., Steemers F.J., Kuhn K., Ren H., Zhou L., Ng P., King C., Lee G., Tsan C., Chang W., Steemers F.J., Kuhn K., Ren H., Zhou L., Ng P., King C., Lee G., Tsan C., Chang W., Kuhn K., Ren H., Zhou L., Ng P., King C., Lee G., Tsan C., Chang W., Ren H., Zhou L., Ng P., King C., Lee G., Tsan C., Chang W., Zhou L., Ng P., King C., Lee G., Tsan C., Chang W., Ng P., King C., Lee G., Tsan C., Chang W., King C., Lee G., Tsan C., Chang W., Lee G., Tsan C., Chang W., Tsan C., Chang W., Chang W., et al. Genetic variance detection: Technologies for pharmacogenomics. DNA Press; Eagleville, PA: 2005. pp. 221–235. [Google Scholar]
- Hanahan D., Weinberg R.A., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hata N., Yoshimoto K., Yokoyama N., Mizoguchi M., Shono T., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Yoshimoto K., Yokoyama N., Mizoguchi M., Shono T., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Yokoyama N., Mizoguchi M., Shono T., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Mizoguchi M., Shono T., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Shono T., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Guan Y., Tahira T., Kukita Y., Higasa K., Nagata S., Tahira T., Kukita Y., Higasa K., Nagata S., Kukita Y., Higasa K., Nagata S., Higasa K., Nagata S., Nagata S., et al. Allelic losses of chromosome 10 in glioma tissues detected by quantitative single-strand conformation polymorphism analysis. Clin. Chem. 2006;52:370–378. doi: 10.1373/clinchem.2005.060954. [DOI] [PubMed] [Google Scholar]
- Hayashizaki Y., Hirotsune S., Okazaki Y., Hatada I., Shibata H., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Hirotsune S., Okazaki Y., Hatada I., Shibata H., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Okazaki Y., Hatada I., Shibata H., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Hatada I., Shibata H., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Shibata H., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Kawai J., Hirose K., Watanabe S., Fushiki S., Wada S., Hirose K., Watanabe S., Fushiki S., Wada S., Watanabe S., Fushiki S., Wada S., Fushiki S., Wada S., Wada S., et al. Restriction landmark genomic scanning method and its various applications. Electrophoresis. 1993;14:251–258. doi: 10.1002/elps.1150140145. [DOI] [PubMed] [Google Scholar]
- Huang J., Wei W., Zhang J., Liu G., Bignell G.R., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Wei W., Zhang J., Liu G., Bignell G.R., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Zhang J., Liu G., Bignell G.R., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Liu G., Bignell G.R., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Bignell G.R., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Stratton M.R., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Futreal P.A., Wooster R., Jones K.W., Shapero M.H., Wooster R., Jones K.W., Shapero M.H., Jones K.W., Shapero M.H., Shapero M.H. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum. Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Albertson D.G., Pinkel D., Marra M.A., Pinkel D., Marra M.A., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Kallioniemi O.P., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Smith H.S., Pinkel D., Gray J.W., Pinkel D., Gray J.W., Gray J.W. Detection of retinoblastoma gene copy number in metaphase chromosomes and interphase nuclei by fluorescence in situ hybridization. Cytogenet. Cell Genet. 1992;60:190–193. doi: 10.1159/000133333. [DOI] [PubMed] [Google Scholar]
- Kallioniemi O.P., Kallioniemi A., Piper J., Isola J., Waldman F.M., Gray J.W., Pinkel D., Kallioniemi A., Piper J., Isola J., Waldman F.M., Gray J.W., Pinkel D., Piper J., Isola J., Waldman F.M., Gray J.W., Pinkel D., Isola J., Waldman F.M., Gray J.W., Pinkel D., Waldman F.M., Gray J.W., Pinkel D., Gray J.W., Pinkel D., Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Visakorpi T., Karhu R., Pinkel D., Kallioniemi O.P., Visakorpi T., Karhu R., Pinkel D., Kallioniemi O.P., Karhu R., Pinkel D., Kallioniemi O.P., Pinkel D., Kallioniemi O.P., Kallioniemi O.P. Gene copy number analysis by fluorescence in situ hybridization and comparative genomic hybridization. Methods. 1996;9:113–121. doi: 10.1006/meth.1996.0015. [DOI] [PubMed] [Google Scholar]
- Knudson A.G., Jr. The genetics of childhood cancer. Cancer. 1975;35:1022–1026. doi: 10.1002/1097-0142(197503)35:3+<1022::aid-cncr2820350726>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Knudson A.G., Jr. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Koed K., Wiuf C., Christensen L.L., Wikman F.P., Zieger K., Moller K., von der Maase H., Orntoft T.F., Wiuf C., Christensen L.L., Wikman F.P., Zieger K., Moller K., von der Maase H., Orntoft T.F., Christensen L.L., Wikman F.P., Zieger K., Moller K., von der Maase H., Orntoft T.F., Wikman F.P., Zieger K., Moller K., von der Maase H., Orntoft T.F., Zieger K., Moller K., von der Maase H., Orntoft T.F., Moller K., von der Maase H., Orntoft T.F., von der Maase H., Orntoft T.F., Orntoft T.F. High-density single nucleotide polymorphism array defines novel stage and location-dependent allelic imbalances in human bladder tumors. Cancer Res. 2005;65:34–45. [PubMed] [Google Scholar]
- LaFramboise T., Weir B.A., Zhao X., Beroukhim R., Li C., Harrington D., Sellers W.R., Meyerson M., Weir B.A., Zhao X., Beroukhim R., Li C., Harrington D., Sellers W.R., Meyerson M., Zhao X., Beroukhim R., Li C., Harrington D., Sellers W.R., Meyerson M., Beroukhim R., Li C., Harrington D., Sellers W.R., Meyerson M., Li C., Harrington D., Sellers W.R., Meyerson M., Harrington D., Sellers W.R., Meyerson M., Sellers W.R., Meyerson M., Meyerson M. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput. Biol. 2005;1 doi: 10.1371/journal.pcbi.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.R., Johnson M.D., Kucherlapati R., Park P.J., Johnson M.D., Kucherlapati R., Park P.J., Kucherlapati R., Park P.J., Park P.J. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon J.A., Lamont J.M., Scott D.K., Dyer S., Prebble E., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Lamont J.M., Scott D.K., Dyer S., Prebble E., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Scott D.K., Dyer S., Prebble E., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Dyer S., Prebble E., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Prebble E., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Bown N., Grundy R.G., Ellison D.W., Clifford S.C., Grundy R.G., Ellison D.W., Clifford S.C., Ellison D.W., Clifford S.C., Clifford S.C. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes Chromosomes Cancer. 2006;45:47–60. doi: 10.1002/gcc.20262. [DOI] [PubMed] [Google Scholar]
- Ledbetter D.H., Engel E., Engel E. Uniparental disomy in humans: Development of an imprinting map and its implications for prenatal diagnosis. Hum. Mol. Genet. 1995;4:1757–1764. doi: 10.1093/hmg/4.suppl_1.1757. [DOI] [PubMed] [Google Scholar]
- Liyanage M., Coleman A., du Manoir S., Veldman T., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., Coleman A., du Manoir S., Veldman T., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., du Manoir S., Veldman T., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., Veldman T., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., Wynshaw-Boris A., Janz S., Wienberg J., Janz S., Wienberg J., Wienberg J., et al. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- Macville M., Veldman T., Padilla-Nash H., Wangsa D., O'Brien P., Schrock E., Ried T., Veldman T., Padilla-Nash H., Wangsa D., O'Brien P., Schrock E., Ried T., Padilla-Nash H., Wangsa D., O'Brien P., Schrock E., Ried T., Wangsa D., O'Brien P., Schrock E., Ried T., O'Brien P., Schrock E., Ried T., Schrock E., Ried T., Ried T. Spectral karyotyping, a 24-colour FISH technique for the identification of chromosomal rearrangements. Histochem. Cell Biol. 1997;108:299–305. doi: 10.1007/s004180050169. [DOI] [PubMed] [Google Scholar]
- Mark H.F., Bai H., Sotomayor E., Mark S., Zolnierz K., Airall E., Sigman M., Bai H., Sotomayor E., Mark S., Zolnierz K., Airall E., Sigman M., Sotomayor E., Mark S., Zolnierz K., Airall E., Sigman M., Mark S., Zolnierz K., Airall E., Sigman M., Zolnierz K., Airall E., Sigman M., Airall E., Sigman M., Sigman M. A variant Klinefelter syndrome patient with an XXY/XX/XY karyotype studied by GTG-banding and fluorescence in situ hybridization. Exp. Mol. Pathol. 1999;67:50–56. doi: 10.1006/exmp.1999.2244. [DOI] [PubMed] [Google Scholar]
- Nannya Y., Sanada M., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Sanada M., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Chiba S., Bailey D.K., Kennedy G.C., Bailey D.K., Kennedy G.C., Kennedy G.C., et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Collins C., Kuo W.L., Chen C., Zhai Y., Kuo W.L., Chen C., Zhai Y., Chen C., Zhai Y., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Raghavan M., Lillington D.M., Skoulakis S., Debernardi S., Chaplin T., Foot N.J., Lister T.A., Young B.D., Lillington D.M., Skoulakis S., Debernardi S., Chaplin T., Foot N.J., Lister T.A., Young B.D., Skoulakis S., Debernardi S., Chaplin T., Foot N.J., Lister T.A., Young B.D., Debernardi S., Chaplin T., Foot N.J., Lister T.A., Young B.D., Chaplin T., Foot N.J., Lister T.A., Young B.D., Foot N.J., Lister T.A., Young B.D., Lister T.A., Young B.D., Young B.D. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–378. [PubMed] [Google Scholar]
- Rook M.S., Delach S.M., Deyneko G., Worlock A., Wolfe J.L., Delach S.M., Deyneko G., Worlock A., Wolfe J.L., Deyneko G., Worlock A., Wolfe J.L., Worlock A., Wolfe J.L., Wolfe J.L. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am. J. Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F., Simmler M.C., Vergnaud G., Johnsson C., Levilliers J., Petit C., Weissenbach J., Simmler M.C., Vergnaud G., Johnsson C., Levilliers J., Petit C., Weissenbach J., Vergnaud G., Johnsson C., Levilliers J., Petit C., Weissenbach J., Johnsson C., Levilliers J., Petit C., Weissenbach J., Levilliers J., Petit C., Weissenbach J., Petit C., Weissenbach J., Weissenbach J. The pseudoautosomal region of the human sex chromosomes. Cold Spring Harb. Symp. Quant. Biol. 1986;51:221–228. doi: 10.1101/sqb.1986.051.01.027. [DOI] [PubMed] [Google Scholar]
- Santarosa M., Ashworth A., Ashworth A. Haploinsufficiency for tumour suppressor genes: When you don't need to go all the way. Biochim. Biophys. Acta. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Scambler P.J., Carey A.H., Wyse R.K., Roach S., Dumanski J.P., Nordenskjold M., Williamson R., Carey A.H., Wyse R.K., Roach S., Dumanski J.P., Nordenskjold M., Williamson R., Wyse R.K., Roach S., Dumanski J.P., Nordenskjold M., Williamson R., Roach S., Dumanski J.P., Nordenskjold M., Williamson R., Dumanski J.P., Nordenskjold M., Williamson R., Nordenskjold M., Williamson R., Williamson R. Microdeletions within 22q11 associated with sporadic and familial DiGeorge syndrome. Genomics. 1991;10:201–206. doi: 10.1016/0888-7543(91)90501-5. [DOI] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Nair P., Brothman A.R., Stallings R.L., Brothman A.R., Stallings R.L., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Shearer P.D., Valentine M.B., Grundy P., DeCou J.M., Banavali S.D., Komuro H., Green D.M., Beckwith J.B., Look A.T., Valentine M.B., Grundy P., DeCou J.M., Banavali S.D., Komuro H., Green D.M., Beckwith J.B., Look A.T., Grundy P., DeCou J.M., Banavali S.D., Komuro H., Green D.M., Beckwith J.B., Look A.T., DeCou J.M., Banavali S.D., Komuro H., Green D.M., Beckwith J.B., Look A.T., Banavali S.D., Komuro H., Green D.M., Beckwith J.B., Look A.T., Komuro H., Green D.M., Beckwith J.B., Look A.T., Green D.M., Beckwith J.B., Look A.T., Beckwith J.B., Look A.T., Look A.T. Hemizygous deletions of chromosome band 16q24 in Wilms tumor: Detection by fluorescence in situ hybridization. Cancer Genet. Cytogenet. 1999;115:100–105. doi: 10.1016/s0165-4608(99)00093-x. [DOI] [PubMed] [Google Scholar]
- Singh S., Simon M., Meybohm I., Jantke I., Jonat W., Maass H., Goedde H.W., Simon M., Meybohm I., Jantke I., Jonat W., Maass H., Goedde H.W., Meybohm I., Jantke I., Jonat W., Maass H., Goedde H.W., Jantke I., Jonat W., Maass H., Goedde H.W., Jonat W., Maass H., Goedde H.W., Maass H., Goedde H.W., Goedde H.W. Human breast cancer: Frequent p53 allele loss and protein overexpression. Hum. Genet. 1993;90:635–640. doi: 10.1007/BF00202481. [DOI] [PubMed] [Google Scholar]
- Slater H.R., Bailey D.K., Ren H., Cao M., Bell K., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Bailey D.K., Ren H., Cao M., Bell K., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Ren H., Cao M., Bell K., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Cao M., Bell K., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Bell K., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Nasioulas S., Henke R., Choo K.H., Kennedy G.C., Henke R., Choo K.H., Kennedy G.C., Choo K.H., Kennedy G.C., Kennedy G.C. High-resolution identification of chromosomal abnormalities using oligonucleotide arrays containing 116,204 SNPs. Am. J. Hum. Genet. 2005;77:709–726. doi: 10.1086/497343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas-Toldo S., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Benner A., Dohner H., Cremer T., Lichter P., Dohner H., Cremer T., Lichter P., Cremer T., Lichter P., Lichter P. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- Somerville M.J., Mervis C.B., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Mervis C.B., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Loo W., Lilley M., Perez-Jurado L.A., Lilley M., Perez-Jurado L.A., Perez-Jurado L.A., et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemers F.J., Gunderson K.L., Gunderson K.L. Illumina, Inc. Pharmacogenomics. 2005;6:777–782. doi: 10.2217/14622416.6.7.777. [DOI] [PubMed] [Google Scholar]
- Steemers F.J., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L., Lee G., Barker D.L., Shen R., Gunderson K.L., Barker D.L., Shen R., Gunderson K.L., Shen R., Gunderson K.L., Gunderson K.L. Whole-genome genotyping with the single-base extension assay. Nat. Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- Teh M.T., Blaydon D., Chaplin T., Foot N.J., Skoulakis S., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Blaydon D., Chaplin T., Foot N.J., Skoulakis S., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Chaplin T., Foot N.J., Skoulakis S., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Foot N.J., Skoulakis S., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Skoulakis S., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Raghavan M., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Harwood C.A., Proby C.M., Philpott M.P., Young B.D., Proby C.M., Philpott M.P., Young B.D., Philpott M.P., Young B.D., Young B.D., et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 2005;65:8597–8603. doi: 10.1158/0008-5472.CAN-05-0842. [DOI] [PubMed] [Google Scholar]
- Tischfield J.A. Loss of heterozygosity or: How I learned to stop worrying and love mitotic recombination. Am. J. Hum. Genet. 1997;61:995–999. doi: 10.1086/301617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask B.J. Fluorescence in situ hybridization: Applications in cytogenetics and gene mapping. Trends Genet. 1991;7:149–154. doi: 10.1016/0168-9525(91)90378-4. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Vatcheva R., Labella T., Gan S.U., Vatcheva R., Labella T., Gan S.U., Labella T., Gan S.U., Gan S.U. More detailed characterization of some of the HL60 karyotypic features by fluorescence in situ hybridization. Cancer Genet. Cytogenet. 1996;87:103–106. doi: 10.1016/0165-4608(95)00214-6. [DOI] [PubMed] [Google Scholar]
- Wang T.L., Maierhofer C., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Maierhofer C., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Kinzler K.W., Velculescu V.E., Velculescu V.E. Digital karyotyping. Proc. Natl. Acad. Sci. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright T.J., Ricke D.O., Denison K., Abmayr S., Cotter P.D., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Ricke D.O., Denison K., Abmayr S., Cotter P.D., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Denison K., Abmayr S., Cotter P.D., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Abmayr S., Cotter P.D., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Cotter P.D., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Hirschhorn K., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., Keinanen M., McDonald-McGinn D., Somer M., Spinner N., McDonald-McGinn D., Somer M., Spinner N., Somer M., Spinner N., Spinner N., et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum. Mol. Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- Ylstra B., van den Ijssel P., Carvalho B., Brakenhoff R.H., Meijer G.A., van den Ijssel P., Carvalho B., Brakenhoff R.H., Meijer G.A., Carvalho B., Brakenhoff R.H., Meijer G.A., Brakenhoff R.H., Meijer G.A., Meijer G.A. BAC to the future! or Oligonucleotides: A perspective for micro array comparative genomic hybridization (array CGH) Nucleic Acids Res. 2006;34:445–450. doi: 10.1093/nar/gkj456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber N.P., Sabbath-Solitare M., Marotta S.P., McMahon L., Bishop D.T., Sabbath-Solitare M., Marotta S.P., McMahon L., Bishop D.T., Marotta S.P., McMahon L., Bishop D.T., McMahon L., Bishop D.T., Bishop D.T. Comparison of allelic ratios from paired blood and paraffin-embedded normal tissue for use in a polymerase chain reaction to assess loss of heterozygosity. Mol. Diagn. 1999;4:29–35. doi: 10.1016/s1084-8592(99)80047-2. [DOI] [PubMed] [Google Scholar]
- Zhao X., Li C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Li C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Girard L., Minna J., Christiani D., Leo C., Minna J., Christiani D., Leo C., Christiani D., Leo C., Leo C., et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- Zheng H.T., Peng Z.H., Li S., He L., Peng Z.H., Li S., He L., Li S., He L., He L. Loss of heterozygosity analyzed by single nucleotide polymorphism array in cancer. World J. Gastroenterol. 2005;11:6740–6744. doi: 10.3748/wjg.v11.i43.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]