Abstract
Mouse odorant receptors (ORs) are encoded by >1000 genes dispersed throughout the genome. Each olfactory neuron expresses one single OR gene, while the rest of the genes remain silent. The mechanisms underlying OR gene expression are poorly understood. Here, we investigated if OR genes share common cis-regulatory sequences in their promoter regions. We carried out a comprehensive analysis in which the upstream regions of a large number of OR genes were compared. First, using RLM-RACE, we generated cDNAs containing the complete 5′-untranslated regions (5′-UTRs) for a total number of 198 mouse OR genes. Then, we aligned these cDNA sequences to the mouse genome so that the 5′ structure and transcription start sites (TSSs) of the OR genes could be precisely determined. Sequences upstream of the TSSs were retrieved and browsed for common elements. We found DNA sequence motifs that are overrepresented in the promoter regions of the OR genes. Most motifs resemble O/E-like sites and are preferentially localized within 200 bp upstream of the TSSs. Finally, we show that these motifs specifically interact with proteins extracted from nuclei prepared from the olfactory epithelium, but not from brain or liver. Our results show that the OR genes share common promoter elements. The present strategy should provide information on the role played by cis-regulatory sequences in OR gene regulation.
Mammalian olfactory sensory neurons select, from >1000 possible choices, one single olfactory receptor (OR) allele to express (Ressler et al. 1993; Vassar et al. 1993; Chess et al. 1994; Malnic et al. 1999; but see Mombaerts 2004). The receptor type that is chosen will not only determine the range of odorants to which this neuron will respond, but also axonal targeting to specific glomeruli in the olfactory bulb (Mombaerts et al. 1996; Wang et al. 1998). OR gene choice is therefore fundamental for the functional organization of the olfactory system. How this choice is accomplished is, however, still unclear.
Little is known about the role of cis-regulatory sequences in the regulation of OR gene expression. In studies using transgenic mice, different sizes of genomic DNA segments containing OR genes were tested for their ability to drive an OR expression similar to that of the endogenous gene. It was demonstrated that short pieces of DNA located upstream of the coding region, ranging from 460 bp to 6.7 kb, are sufficient for expression of the ORs M4, M71, and MOR23 (Qasba and Reed 1998; Vassalli et al. 2002). However, large segments of ~200 kb are required to obtain expression of MOR28 (Serizawa et al. 2000). Sequence comparison of the mouse and human genome revealed a 2-kb conserved sequence located ~75 kb upstream of the MOR28 gene cluster. This region, denominated the H region, was proposed to work as a cis-acting locus control region (LCR) that would activate the expression of one single OR gene member from within the MOR28 gene cluster (Serizawa et al. 2003). Altogether, these results indicate that cis-regulatory sequences may play important roles in OR gene choice.
To date, different combinations of transcription factor binding sites (TFBSs) have been identified in promoters of OR genes (Hoppe et al. 2000, 2003; Sosinsky et al. 2000; Lane et al. 2001; Vassalli et al. 2002); however, there is no evidence yet that these sites are directly involved in OR gene choice. A strong consensus sequence, the Olf-1 site (O/E-like site), was identified in the promoters of several olfactory specific genes, such as GnaI (formerly known as Golf), adenylyl cyclase III (AcIII), olfactory cyclic nucleotide gated channel (Cnga2), and olfactory marker protein (Omp) (Kudrycki et al. 1993; Wang et al. 1993), and was also found in the promoter regions from some OR genes (Glusman et al. 2000b; Sosinsky et al. 2000; Vassalli et al. 2002; Hoppe et al. 2003) but not from other OR genes (Hoppe et al. 2000; Lane et al. 2001).
There are >1000 OR genes dispersed throughout the genome (Young et al. 2002; Zhang and Firestein 2002; Godfrey et al. 2004). A genomic approach to identify potential regulatory cis-acting sequences is to search for DNA sequence elements that are conserved in a large number of OR gene promoters. Promoter sequences, which are usually located proximal to and upstream of the transcription start site (TSS), can be retrieved from the available mouse genome sequence. This can be done by aligning full-length OR mRNA sequences with their counterpart genomic sequences. It is important that full-length mRNA sequences are used, because the transcriptional start sites can be located far away from the translational start sites. It has been demonstrated, for example, that ORs in the mouse P2 OR cluster have 5′-untranslated regions (5′-UTRs) that range from 1.7 to 9 kb (Lane et al. 2001). Besides, it is known that many OR genes have 5′ non-coding exons, and some undergo 5′ alternative splicing (Sosinsky et al. 2000; Lane et al. 2001).
To date, full-length 5′ cDNA sequences are available only for ~30 different mouse OR genes, distributed over five different chromosomes (Bulger et al. 2000; Hoppe et al. 2000, 2003; Sosinsky et al. 2000; Lane et al. 2001; Vassalli et al. 2002). Another study produced cDNA sequences representing >400 OR genes, many of them containing 5′-UTR sequences. However, they may not be full-length sequences, since they originated from a regular cDNA library, and not from a full-length cDNA library (Young et al. 2003).
Here we have used RLM-RACE with degenerate primers to produce cDNAs containing the complete 5′-UTRs for a total number of 198 mouse OR genes. Then, we aligned these cDNA sequences to the mouse genome so that sequences corresponding to one same OR gene are organized in one same cluster, making a total of 198 clusters. For each OR gene cluster, we determined the 5′-gene structure (exon and intron distribution) and TSS. The 198 promoter sequences were retrieved and browsed for common elements. Motifs that are common to a large percentage of the OR genes were found. Most motifs resemble O/E- and homeodomain-like sites. The O/E-like sites are localized within 200 bp upstream of the TSS and specifically interact with proteins extracted from nuclei prepared from the olfactory epithelium, but not with proteins extracted from brain or liver.
Results
Amplification of OR cDNA 5′-ends
To obtain 5′-end sequences from OR cDNAs, we performed RNA ligase-mediated rapid amplification of 5′ cDNA ends (RLM-RACE) using total RNA purified from mouse olfactory epithelium. This method has the advantage that only full-length transcripts (authentic 5′-end capped mRNAs) are amplified (Fig. 1A). In order to obtain 5′ cDNA ends for a large number of OR genes, we used degenerate primers matching to conserved regions in ORs (Fig. 1B) because these primers can amplify the majority of the members of the OR family (Buck and Axel 1991; Malnic et al. 1999). The initial PCR reaction was performed using the 5′ GeneRacer primer together with the reverse P27 degenerate primer. In order to eliminate PCR artifacts, a secondary (nested) PCR was performed using the first PCR product as a template and the 5′ GeneRacer nested primer together with the P26R degenerate primer (Fig. 1A). The primary PCR product contained a heterogeneous mixture of cDNA fragments ranging from 0.8 to 2 kb in length (Fig. 1C, lane 2). This result was expected, since different OR cDNAs must have different 5′-UTR sizes. The secondary PCR reaction produced a similar range of cDNA fragments, except that their sizes were ~350 bp smaller (Fig. 1C, lane 3). This was also expected, since the P26R primer matches to a region in the OR coding sequence that is located ~350 bp upstream to the region matched by P27 (Fig. 1B). Since the region between the first AUG codon and TM-III in OR genes is ~380 bp long, only the nested PCR products ≥380 bp long were gel-purified, cloned, and sequenced.
Figure 1.
The strategy used for the generation of OR complete 5′-end cDNA sequences. (A) Truncated mRNAs were dephosphorylated using calf intestine phosphatase (CIP) so that they cannot participate in subsequent ligation reactions. The RNA was then treated with tobacco acid pyrophosphatase (TAP) to remove the 5′-cap structure (represented by black balls) from intact full-length mRNA, and the GeneRacer RNA oligo-nucleotide was ligated to the decapped mRNA. Reverse transcription was performed using oligo(dT) primers. To obtain 5′-ends, PCR was done using the GeneRacer 5′-primer and a degenerate primer directed toward conserved OR regions (P8 or P27). Nested PCR was then done using the GeneRacer 5′ nested primer and another OR degenerate reverse primer (P27 or P26R). (B) Schematic representation of an OR coding region showing the seven transmembrane regions (I–VII) and the regions matched by the degenerate primers used in this study. (C) 1.5% agarose gel showing the PCR products obtained using different combinations of primers. (Lane 1) Negative control for reaction in lane 2 (no DNA added); (lane 2) GeneRacer 5′-primer and P27; (lane 3) GeneRacer 5′ nested primer and P26R; (lane 4) negative control for reaction in lane 5 (no DNA added); (lane 5) GeneRacer 5′-primer and P8; (lane 6) GeneRacer 5′ nested primer and P27. (M) Molecular weights are given in kilobases.
The same procedure was also performed using primers 5′ GeneRacer and P8 degenerate primer for the primary PCR reaction (Fig. 1C, lane 5), and primers 5′ GeneRacer nested and P27 for the secondary PCR reaction (Fig. 1C, lane 6). In this case, since the region between the first AUG codon and TM-VI in ORs is ~750 bp long, only the nested PCR products ≥750 bp were analyzed.
OR gene clusters
We sequenced 1012 clones from their 5′-ends, and 80% of them correspond to ORs, indicating that our strategy preferentially amplifies OR sequences. In addition, 96% of the OR cDNAs contain 5′ sequences upstream of the predicted initial AUG codon, indicating that full-length mRNAs were amplified. Sequence analysis showed that 5′-RACE products were obtained for a total number of 198 different OR genes, corresponding to ~17% of the complete mouse OR gene repertoire (Table 1). Only nine of the OR genes are pseudogenes.
Table 1.
Chromosomal distribution of OR genes
OR sequences are classified into two phylogenetic classes, referred to as Class I (fish-like) and Class II (terrestrial-specific) ORs (Ngai et al. 1993; Freitag et al. 1995; Glusman et al. 2000a). The Class I ORs constitute ~12% of the mouse OR repertoire (Zhang and Firestein 2002). Nine of the 198 ORs (4.5%) are Class I ORs, indicating that the method we used amplifies members of the two OR Classes, although it may favor amplification of Class II ORs.
The OR genes for which cDNA sequences were obtained are distributed among all of the mouse chromosomes previously shown to contain OR genes (Godfrey et al. 2004), except for chromosome 3 (Table 1). The majority of the sequences correspond to genes in chromosomes 2 and 7, which had been previously shown to contain the higher numbers of OR genes (Young et al. 2002; Zhang and Firestein 2002; Godfrey et al. 2004). In addition, the 198 OR genes can be subdivided into 102 (41%) out of the total 248 mouse OR subfamilies (where all members of a subfamily are ≥60% identical to all other members in amino acid sequence, as described by Godfrey et al. 2004; see Supplemental Table 1). We therefore believe that our sequences are representative of a random sample of the mouse OR genes. However, it is important to note that the present method may favor the amplification of OR cDNAs that have short 3′-UTRs or that are highly expressed in the olfactory epithelium.
We next used BLAST (Altschul et al. 1990) and Sim4 (Florea et al. 1998) to align all of the cDNA sequences with the mouse genome sequence. The previously annotated mouse OR genes (Young et al. 2002; Zhang and Firestein 2002) were also included in the alignment, to help with the localization of the cDNAs 5′-UTR regions. Each cDNA sequence aligned to one single genomic region, and the cDNAs that aligned to the same genomic region as one of the annotated ORs were considered to correspond to that particular OR gene. In this way, we obtained a total of 198 OR clusters, where each cluster corresponds to one different OR gene. Each one of the clusters contains at least one cDNA sequence, the largest cluster contains 71 sequences, and 54% of the clusters contain more than two sequences (Supplemental Table 1).
The structural organization of each one of the 198 clusters can be visualized using the Olfactory Receptor cDNA Clusters Viewer (http://gbrowser.compbio.ludwig.org.br/or/) by entering the corresponding cluster numbers shown in Supplemental Table 1. In summary, of the 198 OR gene clusters, only two do not have introns in their 5′-ends (Supplemental Tables 1 and 2), and 39 showed alternative splicing (in this case, only clusters containing more than two cDNA sequences were analyzed).
5′-structure of the OR genes
On average, the 5′-UTR is 189 bp long, ranging from 32 to 659 bp. The 5′-UTR exons range from 9 to 403 bp (Supplemental Table 2). Most of the OR genes (72%) have only one 5′-UTR exon, 23% have two, and 3% have three (Supplemental Tables 1 and 2). The 5′-UTR introns show a wide variation in size, ranging from 91 bp to 22.5 kb. A large fraction of the OR genes (46%) have introns with sizes between 2 kb and 4 kb, and 34% of the genes have introns >4 kb.
It is predicted that in most eukaryotic mRNAs translation initiates at the first AUG starting from the 5′-cap (Kozak 1999). Therefore, one would expect not to find AUG codons or upstream ORFs (uORFs) in the 5′-UTRs. However, we found that 108 of the 198 OR genes have uORFs at least 10 codons long. It has been suggested that uORFs could be involved in down-regulation of protein translational efficiency (Pesole et al. 2001). Further analysis should clarify whether these uORFs play a role in OR gene regulation.
We also found that a significant percentage of the OR genes (18%) have an in-frame upstream AUG, indicating that these genes code for OR proteins with a longer N-terminal region than the one originally predicted from their genomic sequences.
OR promoter regions
The generation of 5′ full-length cDNAs allowed us to precisely determine the TSSs for the 198 OR genes. On average, the TSSs are located 4.3 kb upstream of the initial AUG codon, the furthest TSSs being located 22.5 kb away and the closest only 18 bp away. Sequences (600 bp) upstream of each TSS were excised from the mouse genomic sequence and analyzed. We first screened the sequences with RepeatMasker (http://www.repeatmasker.org/) and found that only 7% of the total sequences contain repeats or low-complexity regions. Typical TATA-boxes were found in only a small number of the OR gene promoters (35%), consistent with previous reports (Hoppe et al. 2000; Sosinsky et al. 2000; Lane et al. 2001).
Because most TFBS are usually short, they can occur very frequently in the sequences, making it difficult to identify significant sites. In order to reduce the false-positive predictions, we decided to search for motifs that are common to a large fraction of the promoter sequences and thereby identify elements that are more likely to be functionally important. To do this, we used the Gibbs recursive sampler (Thompson et al. 2003), Consensus (Hertz and Stormo 1999), and Weeder (Pavesi et al. 2004) tools, which were designed to locate common elements in collections of unaligned DNA sequences. We found several motifs that are shared by the OR gene promoter sequences (Fig. 2). A closer inspection of the motifs revealed that although they are diverse, the majority of them resemble Olf-1 (O/E) like sites (Figs. 2and3; Wang et al. 1993). The O/E-like motifs can be divided into four groups, denominated M1–M4. Motifs in each one of these four groups show different conserved nucleotide sequences (Fig. 3). We also found motifs that resemble the homeodomain sites, previously shown to be located in proximity to O/E-like binding sites in OR promoter genes (Fig. 2; Vassalli et al. 2002; Rothman et al. 2005).
Figure 2.
Motif patterns found in OR promoter regions. Logo representations were created using the software from http://weblogo.berkeley.edu/logo.cgi. Motifs were identified by using Gibbs Recursive Sampler, Consensus (*) or Weeder (†). Motifs that resemble O/E-like sites show a conserved CN6G sequence. Motifs 1–4 were further analyzed as described in Figures 3–5.
Figure 3.
Nucleotide sequence alignment of the conserved sequences in O/E-like motifs M1–M4 and comparison with Olf-1 binding sites. Ten motif sequences were randomly selected for each one of the motifs (M1– M4) and manually aligned to the Olf-1 sites (from Omp, AcIII, Cnga2, and GnaI) (Wang et al. 1993). Identical nucleotide positions in each group of sequences are indicated by asterisks. The names of the OR genes from which the motifs were retrieved are indicated (e.g., M1_720: motif 1 from olfr720).
The spatial distribution of O/E-like motifs M1–M4 in the promoter sequences is shown in Figure 4A. All of the motifs are concentrated near the TSSs (between +1 and −200 bp). Differently, the homeodomain-like sites show a broader distribution over the entire extent of the 600-bp sequence (Fig. 4B).
Figure 4.
Localization of the motifs with respect to the transcription start sites. (A) The number of sites for motifs M1–M4 found across the OR gene promoter regions. The positions of the TSS (+1) and 600 bp upstream of the TSS (−100 to −600) are indicated. (B) The number of total O/E-like sites (sum of M1–M4 motifs) and homeodomain-like sites (based on the HAATTA consensus) found across the OR promoter regions. The distribution of TAATTG homeodomain sites, previously shown to be present in many OR promoter regions (Vassalli et al. 2002) and to be involved in the regulation of M71 expression (Rothman et al. 2005), is also shown.
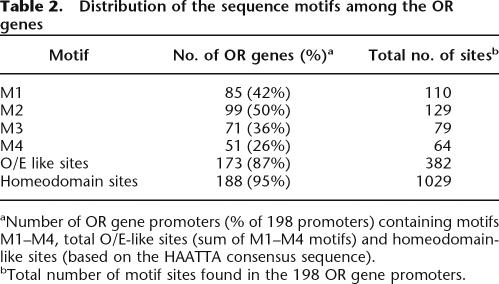
The O/E-like motifs and the homeodomain-like motifs were found, respectively in 87% and 95% of the OR promoter sequences (Table 2).
Table 2.
Distribution of the sequence motifs among the OR genes
DNA motifs form complexes with olfactory nuclear proteins
To evaluate the biological significance of the motifs, we performed gel shift assays. As shown in Figure 5, motifs M1–M4 formed DNA–protein complexes in the presence of the olfactory nuclear extracts, but not in the presence of brain and liver nuclear extracts. The different complexes show similar electro-phoretic mobilities, but the intensities of the shifted bands vary. Formation of these complexes was inhibited by pre-incubation of the binding reaction with a 100-fold excess of the corresponding specific unlabeled oligonucleotides (Fig. 5A). We next evaluated the specificity of the DNA–protein complexes. Mutated motifs M1–M4, where the conserved nucleotides were changed into different ones, were unable to form stable DNA–protein complexes with olfactory nuclei proteins (Supplemental material 3). Altogether, these results show that proteins present in nuclei of olfactory epithelium cells, but not in liver and brain extracts, specifically bind to motifs M1–M4.
Figure 5.
Binding of nuclear proteins to the DNA motifs. Labeled double-stranded oligonucleotides corresponding to motifs M1–M4 were incubated with nuclear extracts from (A) olfactory epithelium, (B) brain, or (C) liver, as indicated. The Olf-1 binding site (Olf-1s) was used as a positive control. DNA–olfactory epithelium nuclear protein complexes are observed for motifs M1–M4. The DNA–protein interactions are completed by the addition of a 100-fold molar excess of the corresponding unlabeled specific oligonucleotide (A). The positions of the free probes are indicated by filled circles.
Discussion
The role of cis-regulatory sequences in the regulation of OR gene expression is not understood. In the present study, we carried out a comprehensive analysis in which the promoter regions of 198 OR genes were compared.
Common OR gene promoter motifs
By comparing a variable set of OR genes, we aimed to identify common promoter elements that may be involved in the general mechanisms of OR gene regulation. Strikingly, 87% of the OR gene promoter regions contain O/E-like sites, and 95% contain homeodomain-like sites. No other types of motifs were found to be overrepresented in these sequences, although we cannot exclude the possibility that the algorithms we used may have precluded us from finding additional sites.
The roles of the O/E-like and homeodomain-like proteins in OR gene expression are still unclear. It has been demonstrated that expression of the M71 OR gene can be driven by a short region upstream of the TSS (a minimal promoter region) containing an O/E-like site and a homeodomain-like site (Vassalli et al. 2002). Mutational studies using transgenic mice indicate that both sites are required for normal OR gene expression (Rothman et al. 2005). Yet, disruption of olf-1-like genes does not alter OR gene expression (Lin and Grosschedl 1995; Wang et al. 2003), possibly because of the functional redundancy of the multiple O/E family members expressed in the olfactory epithelium (O/E1, O/E2, O/E3, and O/E4) (Wang et al. 1997, 2002). Nevertheless, it was demonstrated that O/E2 and O/E3 mutant mice show defects in the projection of olfactory neurons to the olfactory bulb, indicating that the O/E genes’ function may not be completely redundant (Wang et al. 2003).
The LIM-homeodomain protein Lhx2 was shown to bind to the homeodomain site in the M71 OR promoter region (Hirota and Mombaerts 2004). Lhx2 knockout mice do not express ORs, but since they also lack mature olfactory neurons, there is no evidence so far that this homeodomain protein is directly involved in OR gene expression (Hirota and Mombaerts 2004; Kolterud et al. 2004). It is also possible that other homeodomain proteins than Lhx2 bind to the homeodomain sites in the OR gene promoters.
Although the vast majority of OR gene promoters have O/E-like sites, the structure of these sites is variable (see Figs. 2and3). It has been previously shown that the different O/E proteins possess similar DNA-binding properties (Wang et al. 1997). Our results indicate that the M1–M4 motifs interact differently with proteins from olfactory epithelium nuclei (Fig. 5). Several possibilities could explain the different DNA–protein complex affinities. The motifs could bind to different O/E proteins, or to alternatively spliced versions of these proteins (Wang et al. 1997, 2002). It is also possible that the same O/E protein types could bind to the M1–M4 motifs, but with different affinities.
Our findings suggest that different OR gene promoters are bound by different combinations or amounts of O/E-like proteins. The consequences of these differential interactions for OR gene regulation are unknown. It is known that different ORs are expressed in different levels (Young et al. 2003). One interesting possibility is that the types of O/E-like sites in an OR gene promoter region may determine its probability of being transcribed. The identification of the proteins that interact with each one of the motifs and the analysis of the expression patterns of OR genes that have different motifs should clarify the role of these O/E-like sites in OR gene regulation.
Promoter DNA elements and OR gene regulation
Different models for OR gene regulation have been considered to date (for review, see Sosinsky et al. 2000; Mombaerts 2004; Serizawa et al. 2005; Shykind 2005). It has been recently demonstrated that the monoallelic expression of an OR gene is regulated by a negative feedback mechanism that requires a functional OR protein (Serizawa et al. 2003; Lewcock and Reed 2004). In addition, it was shown that immature olfactory neurons that express a given odorant receptor can switch receptor expression at a low frequency, while neurons expressing a mutant (nonfunctional) OR can switch expression with a greater probability (Shykind et al. 2004). Based on these results, a new model has been proposed (Serizawa et al. 2004; Shykind 2005). In this model, after an OR gene is stochastically selected for expression by a limiting factor, its corresponding OR protein product mediates a feedback signal that results in the maintenance of the receptor choice.
Here we show that a collection of random OR genes will have the same types of cis-regulatory elements, suggesting that these common promoter elements are likely to play an important role in OR gene expression. It is possible that enhancers or LCRs interact with elements in one OR gene promoter to select that specific OR for expression. Interestingly, it was shown that the H region, which works as an LCR and is located 75 kb upstream of the MOR28 gene cluster (Serizawa et al. 2003), also contains at least one set of homeodomain- and O/E-like sites (Hirota and Mombaerts 2004). Alternatively, cis-elements and protein factors that bind to these elements could bring one given OR gene promoter to a single expression site body in the nucleus (Borst 2002; Voss et al. 2006).
However, it is important to note that other olfactory genes that are expressed in all mature olfactory neurons, such as Omp, GnaI, and AcIII, also have O/E-like sites (Fig. 3). Therefore, the mere presence of O/E-like sites in the promoter regions does not explain the mosaic pattern of OR expression in the olfactory epithelium.
In conclusion, our results indicate that intraspecies comparisons of promoter sequences are likely to be a useful strategy for identifying common regulatory motifs that may be involved in regulation of OR gene expression. A similar strategy can also be applied to other multigene families whose members are coordinately regulated, such as the pheromone receptor families (Dulac and Torello 2003).
Methods
5′ RLM-RACE
Total RNA was purified from C57BL/6J mice (6–8 wk old) olfactory epithelium using TRIzol reagent (Invitrogen), following the manufacturer's instructions. RLM-RACE was performed using the GeneRacer kit (Invitrogen) and 4 μg of total RNA. Twenty-fivemicroliter PCR reactions containing 1 μL of RLM-RACE cDNA, 0.2 mM dNTP, 1.5 mM MgCl2, 0.5 μM each forward and reverse primer (or 2 μM degenerate primers), 1.25 U of Platinum Taq DNA polymerase (Invitrogen) were heated to 95°C for 2 min, followed by 40 thermal cycles of 95°C for 1 min, 50°C for 3 min, 72°C for 2 min, and a final incubation at 72°C for 10 min.
Twenty-five-microliter nested PCR reactions were done using 1 μL of a 200-fold dilution of the primary PCR product and 25 cycles as above.
Degenerate oligonucleotide primers
The following degenerate OR primers were used in this study:
Reverse primers
P8 (TM-VII): (GA)TTIC(TG)IA(AG)I(GC)(TA)(GA)TA IAT(AG)AAIGG(GA)TT
P27 (TM-VI): ACIACIGAIAG(GA)TGIGAI(GC)C(GA)CAIGT
-
P26R (TM-III): CAIATIGCIAC(AG)TAICG(GA)TCAIGTAIGC
Forward primer
P26 (TM-III): GCITA(CT)GA(CT)CGITA(CT)GTIGCIATITG
Cloning and sequencing
The RACE nested PCR products were gel-purified and cloned into the pCRII vector (Invitrogen). Colonies containing OR cDNAs were selected by colony PCR using the OR degenerate primers P26 and P27, and their orientation was determined also by colony PCR using the pair of primers T7/P27 or SP6/P27. Plasmid DNA was prepared from positive colonies using Filter Plate for high-throughput separations (Multiscreen Millipore). DNA was sequenced with the ABI PRISM Big Dye Terminator V3.1 Cycle sequencing kit using T7 or SP6 primers on an ABI PRISM 3100 Genetic Analyzer (Hitachi). Four percent of the OR sequences were truncated OR RNAs, probably because the CIP reaction during RLM-RACE was not 100% efficient.
Genomic alignment of cDNA sequences
We aligned the cDNA sequences against the mouse genome using BLAST. Only the alignments with percent identities >93% were considered. The position of each alignment was calculated, and the flanking 50-kb genomic sequences were extracted from the corresponding genomic contigs. Each sequence was realigned with its corresponding extracted genomic sequence using the Sim4 program (Florea et al. 1998). Only the Sim4 alignments showing average percent identity >93%, entire sequence alignment >50%, and with the best score (based on the nucleotide identity over the entire alignment) were selected. A MySQL database was loaded with the alignment information.
Clustering of cDNA sequences
The cDNA sequences were clustered based on their genomic coordinates. Sequences that share at least one same exon/intron boundary were included in the same cluster. When no exon/intron boundaries were defined, sequences with at least 30-bp overlap in one same genomic location were included in the same cluster. The Olfactory Receptor cDNA Clusters Viewer site was generated using the Generic Genome Browser (Stein et al. 2002; http://www.gmod.org/ggb/).
Promoter sequence analysis
Promoter sequences were analyzed using the Gibbs Recursive Sampler (Thompson et al. 2003). A FASTA sequence file containing the 198 promoter sequences (600 bp upstream of the TSS) (Supplemental material 1) was analyzed using the eukaryotic default values for all parameters and motif lengths 8, 8, 6, 8, 8; 12, 12, 10, 12, 12; or 14, 14, 12, 14, 14. The parameters used to identify some of the motifs are shown in the Gibbs output files (Supplemental material 2). The promoter sequences were also analyzed using Consensus (Hertz and Stormo 1999) and Weeder (Pavesi et al. 2004) (in both cases, motif widths were set to 6, 8, 10, or 12). Potential TATA-box sequences were predicted using HCtata (http://l25.itba.mi.cnr.it/~webgene/wwwHC_tata.html). The location of the motifs within the promoter regions was determined using SiteSeer (http://rocky.bms.umist.ac.uk/SiteSeer/). Motifs were searched in both strands of the input sequences.
Preparation of nuclear extracts
Nuclear extracts were prepared using the method described by Kudrycki et al. (1993) from olfactory epithelium dissected from 30 4–7-wk-old C57BL/6J mice. The extract was first concentrated using a Microcon centrifugal filter device (Millipore), and then the buffer was exchanged with binding buffer (10 mM Tris-HCl at pH 7.9, 1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 10% glycerol, 3 mM DTT, 0.3 mM PMSF) using a Micro Bio-Spin P-6 chromatography column (Bio-Rad). Aliquots were stored at −80°C. Protein concentration was determined using the Bradford assay (Bio-Rad).
Gel shift assay
The digoxigenin (DIG) gel shift kit (Roche Applied Science) was used for gel shift assays. Binding reactions contained 2 μg of poly[d(I-C)], 0.1 μg of poly-L-lysine, 1.2 ng of labeled oligo-nucleotide, and 15 μg of nuclear protein extract. After a 10-min incubation on ice and a 15-min incubation at room temperature, the mixture was added with 5 μL of loading buffer (60% 0.25× TBE buffer, 40% glycerol, 0.2% bromophenol blue) and electro-phoresed in 0.5× TBE on a nondenaturing 4% polyacrylamide gel in 0.5× TBE containing a 2-cm 15% acrylamide layer at the bottom to retain the unbound probe in the gel, as described by Bell et al. (1999). The gel was pre-electrophoresed for 1 h at 80 V before the samples were applied. Competition experiments were performed by incubating the binding reaction mixtures with a 100× excess of unlabeled competitor oligonucleotide for 5 min before the addition of the labeled oligonucleotide. Blotting was performed using a Bio-Rad electroblotting system, and chemiluminescence detection of the DIG-labeled DNA–protein complexes was performed using anti-digoxigenin antibody conjugated to alkaline phosphatase and the CSPD substrate (Roche Applied Science).
The following pairs of complementary oligonucleotides were used as double-stranded DNA probes for the gel shift reactions (motif sequences are underlined, and the OR genes from which sequences were extracted are indicated):
M1 motif (from olfr720): 5′-TCTCAGACTTTTCTCCTGGGAGACATCTCAG-3′ and 5′ CCTGAGATGTCTCCCAGGAGAAAAGTCTGAG-3′;
M2 motif (olfr165): 5′-TAAGATGCTAAATTCCCTGGAGAAATTGTAA-3′ and 5′-TTTACAATTTCTCCAGGGAATTTAGCATCTT-3′;
M3 motif (olfr211): 5′-CCTGGCATCTCCCACTGGGGCTTATATTCTG-3′ and 5′-ACAGAATATAAGCCCCAGTGGGAGATGCCAG-3′;
M4 motif (olfr1339): 5′-CTTCAGCTTCATCCTCCCTGAGGACAGGGAG-3′ and 5′-GCTCCCTGTCCTCAGGGAGGATGAAGCTGAA-3′.
Acknowledgments
We thank Hiroaki Matsunami for critically reading the manuscript. We are grateful to Sandro José de Souza and Noboro Jo Sakabe for providing bioinformatics assistance. We also thank Erica Bandeira and Luci Navarro for technical assistance. This work was supported by research grants from FAPESP and CNPq (to B.M.) and fellowships from FAPESP (to J.S.M. and P.A.F.G.).
Footnotes
Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. DR065530–DR065963.
Article is published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5185406.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bell A., Feng X., Reder A., Feng X., Reder A., Reder A. Improved band resolution, loading reliability and reduced 32P contamination in mobility shift assays by retention of unbound probe. Biotechniques. 1999;27:1122–1126. doi: 10.2144/99276bm07. [DOI] [PubMed] [Google Scholar]
- Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bulger M., Bender M.A., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Bender M.A., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Felsenfeld G., Groudine M., Hardison R., Groudine M., Hardison R., Hardison R. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse β-globin gene clusters. Proc. Natl. Acad. Sci. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., Axel R., Simon I., Cedar H., Axel R., Cedar H., Axel R., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Dulac C., Torello A.T., Torello A.T. Molecular detection of pheromone signals in mammals: From genes to behaviour. Nat. Rev. Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W., Hartzell G., Zhang Z., Rubin G.M., Miller W., Zhang Z., Rubin G.M., Miller W., Rubin G.M., Miller W., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag J., Krieger J., Strotmann J., Breer H., Krieger J., Strotmann J., Breer H., Strotmann J., Breer H., Breer H. Two classes of olfactory receptors in Xenopus laevis . Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Glusman G., Bahar A., Sharon D., Pilpel Y., White J., Lancet D., Bahar A., Sharon D., Pilpel Y., White J., Lancet D., Sharon D., Pilpel Y., White J., Lancet D., Pilpel Y., White J., Lancet D., White J., Lancet D., Lancet D. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm. Genome. 2000a;11:1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]
- Glusman G., Sosinsky A., Ben-Asher E., Avidan N., Sonkin D., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Sosinsky A., Ben-Asher E., Avidan N., Sonkin D., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Ben-Asher E., Avidan N., Sonkin D., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Avidan N., Sonkin D., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Sonkin D., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Bahar A., Rosenthal A., Clifton S., Roe B., Ferraz C., Rosenthal A., Clifton S., Roe B., Ferraz C., Clifton S., Roe B., Ferraz C., Roe B., Ferraz C., Ferraz C., et al. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics. 2000b;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- Godfrey P.A., Malnic B., Buck L.B., Malnic B., Buck L.B., Buck L.B. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G., Stormo G., Stormo G. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hirota J., Mombaerts P., Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory neurons. Proc. Natl. Acad. Sci. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe R., Weimer M., Beck A., Breer H., Strotmann J., Weimer M., Beck A., Breer H., Strotmann J., Beck A., Breer H., Strotmann J., Breer H., Strotmann J., Strotmann J. Sequence analyses of the olfactory receptor gene cluster mOR37 on mouse chromosome 4. Genomics. 2000;66:284–295. doi: 10.1006/geno.2000.6205. [DOI] [PubMed] [Google Scholar]
- Hoppe R., Frank M.E., Breer H., Strotmann J., Frank M.E., Breer H., Strotmann J., Breer H., Strotmann J., Strotmann J. The clustered olfactory receptor gene family 262: Genomic organization, promoter elements, and interacting transcription factors. Genome Res. 2003;13:2674–2685. doi: 10.1101/gr.1372203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud A., Alenius M., Carlsson L., Bohm S., Alenius M., Carlsson L., Bohm S., Carlsson L., Bohm S., Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131:5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kudrycki K., Stein-Izsak C., Behn C., Grillo M., Akeson R., Margolis F., Stein-Izsak C., Behn C., Grillo M., Akeson R., Margolis F., Behn C., Grillo M., Akeson R., Margolis F., Grillo M., Akeson R., Margolis F., Akeson R., Margolis F., Margolis F. Olf-1 binding site: Characterization of an olfactory neuron-specific promoter motif. Mol. Cell. Biol. 1993;13:3002–3014. doi: 10.1128/mcb.13.5.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane R., Cutforth T., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Cutforth T., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Evans G., Axel R., Hood L., Trask B.J., Axel R., Hood L., Trask B.J., Hood L., Trask B.J., Trask B.J. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl. Acad. Sci. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock J.W., Reed R.R., Reed R.R. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl. Acad. Sci. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Grosschedl R., Grosschedl R. Failure of B cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Malnic B., Hirono J., Sato T., Buck L.B., Hirono J., Sato T., Buck L.B., Sato T., Buck L.B., Buck L.B. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: The one receptor–one neuron hypothesis revisited. Curr. Opin. Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Wang F., Dulac C., Chao S., Nemes A., Mendelsohn M., Edmondson J., Axel R., Wang F., Dulac C., Chao S., Nemes A., Mendelsohn M., Edmondson J., Axel R., Dulac C., Chao S., Nemes A., Mendelsohn M., Edmondson J., Axel R., Chao S., Nemes A., Mendelsohn M., Edmondson J., Axel R., Nemes A., Mendelsohn M., Edmondson J., Axel R., Mendelsohn M., Edmondson J., Axel R., Edmondson J., Axel R., Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Ngai J., Dowling M.M., Buck L., Axel R., Chess A., Dowling M.M., Buck L., Axel R., Chess A., Buck L., Axel R., Chess A., Axel R., Chess A., Chess A. The family of genes encoding odorant receptors in the channel catfish. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesoli G., Mereghetti P., Mauri G., Pesoli G., Mauri G., Pesoli G., Pesoli G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S., Gissi C., Grillo G., Licciulli F., Liuni S., Grillo G., Licciulli F., Liuni S., Licciulli F., Liuni S., Liuni S. Structural and functional features of eukaryotic mRNA untranslated region. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Qasba P., Reed R.R., Reed R.R. Tissue and zonal-specific expression of an olfactory receptor transgene. J. Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler K.J., Sullivan S.L., Buck L.B., Sullivan S.L., Buck L.B., Buck L.B. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Rothman A., Feinstein P., Hirota J., Mombaerts P., Feinstein P., Hirota J., Mombaerts P., Hirota J., Mombaerts P., Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol. Cell. Neurosci. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Serizawa S., Ishii T., Nakatani H., Tsuboi A., Nagawa F., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Ishii T., Nakatani H., Tsuboi A., Nagawa F., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Nakatani H., Tsuboi A., Nagawa F., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Tsuboi A., Nagawa F., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Nagawa F., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Asano M., Sudo K., Sakagami J., Sakano H., Ijiri T., Sudo K., Sakagami J., Sakano H., Ijiri T., Sakagami J., Sakano H., Ijiri T., Sakano H., Ijiri T., Ijiri T., et al. Mutually exclusive expression of odorant receptor transgenes. Nat. Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- ———2003Negative feedback regulation ensures the one receptor–one olfactory neuron rule in the mouse Science 3022088–2094. [DOI] [PubMed] [Google Scholar]
- Serizawa S., Miyamichi K., Sakano H., Miyamichi K., Sakano H., Sakano H. One neuron–one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- ———2005Negative feedback regulation ensures the one neuron–one receptor rule in the mouse olfactory system Chem. Senses 30i99–i100. [DOI] [PubMed] [Google Scholar]
- Shykind B.M. Regulation of odorant receptors: One allele at a time. Hum. Mol. Genet. 2005;14:R33–R39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- Shykind B.M., Rohani S.C., O'Donnel S., Nemes A., Mendelsohn M., Sun Y., Axel R., Barnea G., Rohani S.C., O'Donnel S., Nemes A., Mendelsohn M., Sun Y., Axel R., Barnea G., O'Donnel S., Nemes A., Mendelsohn M., Sun Y., Axel R., Barnea G., Nemes A., Mendelsohn M., Sun Y., Axel R., Barnea G., Mendelsohn M., Sun Y., Axel R., Barnea G., Sun Y., Axel R., Barnea G., Axel R., Barnea G., Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sosinsky A., Glusman G., Lancet D., Glusman G., Lancet D., Lancet D. The genomic structure of human olfactory receptor genes. Genomics. 2000;70:49–61. doi: 10.1006/geno.2000.6363. [DOI] [PubMed] [Google Scholar]
- Stein L., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., Caudy M., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., Day A., Nickerson E., Stajich J., Harris T., Arva A., Nickerson E., Stajich J., Harris T., Arva A., Stajich J., Harris T., Arva A., Harris T., Arva A., Arva A., et al. The Generic Genome Browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W., Rouchka E., Lawrence C., Rouchka E., Lawrence C., Lawrence C. Gibbs recursive sampler: Finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A., Rothman A., Feinstein P., Zapotocky M., Mombaerts P., Rothman A., Feinstein P., Zapotocky M., Mombaerts P., Feinstein P., Zapotocky M., Mombaerts P., Zapotocky M., Mombaerts P., Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R., Ngai J., Axel R., Ngai J., Axel R., Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Voss T.S., Healer J., Marty A.J., Duffy M.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Healer J., Marty A.J., Duffy M.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Marty A.J., Duffy M.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Duffy M.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F., Reeder J.C., Crabb B.S., Cowman A.F., Crabb B.S., Cowman A.F., Cowman A.F. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Wang M.M., Tsai R.Y.L., Schrader K.A., Reed R.R., Tsai R.Y.L., Schrader K.A., Reed R.R., Schrader K.A., Reed R.R., Reed R.R. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol. Cell. Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tsai R., Reed R., Tsai R., Reed R., Reed R. The characterization of the Olf-1/EBF-like HLH transcription factor family: Implications in olfactory gene regulation and neuronal development. J. Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nemes A., Mendelsohn M., Axel R., Nemes A., Mendelsohn M., Axel R., Mendelsohn M., Axel R., Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang S., Betz A., Reed R., Betz A., Reed R., Reed R. Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol. Cell. Neurosci. 2002;20:404–414. doi: 10.1006/mcne.2002.1138. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R., Feinstein P., Mombaerts P., Reed R.R., Mombaerts P., Reed R.R., Reed R.R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2003;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Young J.M., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Ross J.A., Tonnes-Priddy L., Trask B.J., Tonnes-Priddy L., Trask B.J., Trask B.J. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Young J.M., Shykind B.M., Lane R.P., Tonnes-Priddy L., Ross E.M., Walker M., Williams E.M., Trask B.J., Shykind B.M., Lane R.P., Tonnes-Priddy L., Ross E.M., Walker M., Williams E.M., Trask B.J., Lane R.P., Tonnes-Priddy L., Ross E.M., Walker M., Williams E.M., Trask B.J., Tonnes-Priddy L., Ross E.M., Walker M., Williams E.M., Trask B.J., Ross E.M., Walker M., Williams E.M., Trask B.J., Walker M., Williams E.M., Trask B.J., Williams E.M., Trask B.J., Trask B.J. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Firestein S., Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]