Abstract
The potential for genetic change arising from specific single types of DNA lesion has been thoroughly explored, but much less is known about the mutagenic effects of DNA lesions present in clustered damage sites. Localized clustering of damage is a hallmark of certain DNA-damaging agents, particularly ionizing radiation. We have investigated the potential of a non-mutagenic DNA base lesion, 5,6-dihydrothymine (DHT), to influence the mutagenicity of 8-oxo-7,8-dihydroguanine (8-oxoG) when the two lesions are closely opposed. Using a bacterial plasmid-based assay we present the first report of a significantly higher mutation frequency for the clustered DHT and 8-oxoG lesions than for single 8-oxoG in wild-type and in glycosylase-deficient strains. We propose that endonuclease III has an important role in the initial stages of processing DHT/8-oxoG clusters, removing DHT to give an intermediate with an abasic site or single-strand break opposing 8-oxoG. We suggest that this mutagenic intermediate is common to several different combinations of base lesions forming clustered DNA damage sites. The MutY glycosylase, acting post-replication, is most important for reducing mutation formation. Recovered plasmids commonly gave rise to both wild-type and mutant progeny, suggesting that there is differential replication of the two DNA strands carrying specific forms of base damage.
INTRODUCTION
The damaging potential of ionizing radiations on biological materials has been suggested to arise largely from the formation of clusters of lesions within localized regions of DNA (1,2). A cluster is defined as two or more lesions formed within a few tens of base pairs by a single radiation track. Recent computer simulations of radiation track–DNA interactions have predicted that a large fraction of damage sites consist of closely opposed base lesions, and that the complexity of these clusters increases as the ionization density of the radiation increases (3). Consistent with this proposal, biological effects such as lethality, mutation induction and irrepairability of DNA double strand breaks (DSBs) have been shown to increase as the ionization density of the radiation increases (4,5). It has been hypothesized, therefore, that radiation-induced clustered damage sites are less repairable than the isolated base lesions caused by aerobic metabolism and are particularly harmful to cells.
Sites of clustered DNA damage induced by radiation in cells commonly consist of closely associated base damages, such as 8-oxo-7,8-dihydroguanine (8-oxoG) with other types of base lesion or apurinic/apyrimidinic (AP) sites, as verified by both in vitro and in vivo analyses (6–8). In this study we have chosen to examine the biological consequences of 8-oxoG associated with 5,6-dihydrothymine (DHT), known to be produced in irradiated DNA and cells under anaerobic conditions (9,10). It has been found from experiments using cell extracts or purified proteins that there is a complex interplay between different repair activities in the processing of specific forms of base lesion within a clustered damage site, and this interplay determines the outcome of attempted repair. Thus, a lesion such as DHT placed 3′ or 5′ on the opposite strand at 1, 3 or 5 bp away from 8-oxoG has little or no effect on the efficiency of excision of 8-oxoG by formamido pyrimidine DNA glycosylase (Fpg) (11). Similarly excision of DHT by endonuclease III (Nth) is not significantly affected by a neighbouring 8-oxoG on the complementary strand (12). In contrast, an AP site or a single-strand break (SSB) placed close on the opposite strand to either 8-oxoG or DHT can reduce the efficiency of excision of 8-oxoG by Fpg or DHT by Nth >10-fold (11–13). Similarly, studies on DNA carrying clustered base lesions exposed to mammalian cell extracts have shown that the cluster may present difficulties for repair enzymes to resolve (13–16).
To determine the biological consequences of clustered damage, we and others have investigated the mutagenic potential of clustered AP and/or 8-oxoG lesions in Escherichia coli. When compared to a single base damage, it was found that clustered damage led to a 2- to 3-fold increase in mutations (17–19). However, with some types of clustered damage sites, it was found that a lethal DSB is formed during attempted repair of the site (20–22). In the present study, we report the mutagenic consequences of 8-oxoG when closely opposed to a DHT lesion. 8-OxoG is a mutagenic lesion, and E.coli is known to employ at least two DNA glycosylases to prevent mutagenesis by this lesion: one is Fpg, which excises 8-oxoG residues from DNA, and the other is MutY, which excises adenine residues incorporated opposite 8-oxoG after replication (23). DHT, on the other hand, is neither mutagenic nor lethal (24,25). Nth is known to excise DHT (26,27), although data on the reaction efficiency vary (11,28). Despite the non-mutagenic nature of DHT we hypothesized that the competition between, or the inhibition of, these glycosylases when excising lesions in clustered damage would have a significant influence on mutagenic consequences. Using wild-type and glycosylase-deficient (fpg, mutY and nth) strains of E.coli, as well as a combination of these deficiencies, we have examined the effect of lesion clustering on the induction of mutation and the importance of individual glycosylases for repair of clustered damage sites in E.coli.
MATERIALS AND METHODS
E.coli strains
Isogenic strains CC104 (29), BH540 (fpg::KanR), BH980 (mutY::KanR) and BH990 (fpg::KanR mutY::KanR) double mutant strain were a gift from Dr S. Boiteux (CEA, France). Strains CC104 nth::KanR and CC104 fpg::TetR mutY::KanR nth::CmR were kindly provided by Dr S. Yonei, Kyoto University, Japan.
Preparation of oligonucleotides
Oligonucleotides (40mer) carrying a DHT lesion (or an 8-oxoG lesion in one set of experiments) at different positions or an 8-oxoG lesion at a fixed position were synthesized by MWG Biotech and purified by high-performance liquid chromatography. To generate double-stranded oligonucleotides (Table 1), 20 pmol of each of the complementary strands were annealed in Tris–EDTA buffer at pH 8 through heating at 80°C, before cooling the DNA to room temperature for over 2–3 h. The annealed oligonucleotides were phosphorylated at the 5′ termini using 10 U of T4 polynucleotide kinase in 2.5 mM ATP with 5× Forward exchange buffer (Invitrogen) at 37°C for 30 min. The oligonucleotides were purified by passage through a Qiagen nucleotide extraction column.
Table 1.
Double-stranded oligonucleotides used in this study
| Name | Position of DHT | Sequence of the oligonucleotide |
|---|---|---|
| 8G/D−5 | −5 | 5′-CTCTTAGTCAGGAAYAT GTCTC TATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| 8G/D−3 | −3 | 5′-CTCTTAGTCAGGAATAY GTCTC TATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| 8G/D−1 | −1 | 5′-CTCTTAGTCAGGAATAT GYCTC TATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| 8G/D+1 | +1 | 5′-CTCTTAGTCAGGAATAT GTCYC TATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| 8G/D+3 | +3 | 5′-CTCTTAGTCAGGAATAT GTCTC YATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| 8G/D+5 | +5 | 5′-CTCTTAGTCAGGAATAT GTCTC TAYGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ | ||
| Position of second 8-oxoG | ||
| 8G/8G−2 | −2 | 5′-CTCTTAGTCAGGAATAT XTCTC TATGCTGGGAGCAAAGGC-3′ |
| 3′-GAGAATCAGTCCTTATA CAXAG ATACGACCCTCGTTTCCG-5′ |
X, 8-oxoG; Y, DHT. −5 to −1, position on the complementary strand of the X base when 3′ from the Y or the second X base. +1 to +5, position on the complementary strand of the X base when 5′ from the Y base. The recognition sequence of BsmAI is underlined. Oligonucleotide with DHT alone was named as DHT(−5), DHT(−3), DHT(−1), DHT(+1), DHT(+3) or DHT(+5), according to the position of DHT.
Plasmid preparation and ligation
pUC18 plasmid DNA (∼10 µg) in Tris–EDTA was linearized with 100 U of SmaI (NEB) at room temperature for 3 h. Following electrophoresis, the DNA was purified using a Qiagen gel purification system and dephosphorylated with 20 U of calf intestine phosphatase (NEB) for 30 min at 37°C. The plasmid DNA was again gel purified and 200 fmol of linearized pUC18 DNA was ligated to 5 pmol of the annealed oligonucleotide in a reaction volume of 20 µl for 16 h at 16°C using T4 Ligase and ligation reaction buffer (NEB), followed by dialysis using 0.025 µm millipore nitrocellulose filters.
Bacterial transformation
Ligation product (5 µl) was mixed with 60 µl of electrocompetent bacteria [10% (v/v) glycerol, distilled water], and electroporated using a Bio-Rad E.coli pulser, set at 1.8 kV, 200 Ω, 25 µF. Following electroporation, the transformed bacteria were incubated for 1 h at 37°C in 1 ml of SOC (20 g/l bacto-tryptone, 5 g/l bacto-yeast extract, 0.5 g/l NaCl, 0.186 g/l KCl, 0.95 g/l MgCl2 and 20 mM glucose). Transformants were then grown in 2 ml Luria broth (LB) with ampicillin (100 µg/ml) at 37°C for 16 h. Transformation frequencies were very similar with all plasmid constructs (undamaged or damaged) for all bacterial strains, indicating that there was little or no loss of plasmid due to the damage present in the oligonucleotide.
Quantification of mutation
The mutation assay is based on inability to cut a BsmAI restriction site within the cloned oligonucleotide (18). Plasmid DNA was retrieved from 2 ml overnight mini-culture using a QIAprep spin miniprep kit. To achieve complete cutting of unmutated plasmid DNA, 5 µl plasmid DNA was incubated with 5 U BsmAI in a total volume of 20 µl for 3 h at 55°C, followed by incubation with a further 5 U BsmAI at 55°C for 16 h. The samples were electrophoresed for 6.5 h at room temperature on a 1% agarose gel (1 µg/ml ethidium bromide) at 4 V/cm. Following electrophoresis, the gel image was captured under UV light using a CCD camera and analysed using Alphainnotech AlphaEase™ Software.
When mutated, the expected length of the plasmid fragment carrying the BsmAI site contained within the ligated oligomer is 1755 bp, whereas digested (=unmutated) fragments are 1352 and 403 bp (Figure 1). Thus, the mutation frequency of the clustered damage site was determined using the following equation (18):
where I is total pixel intensity of the band minus local background.
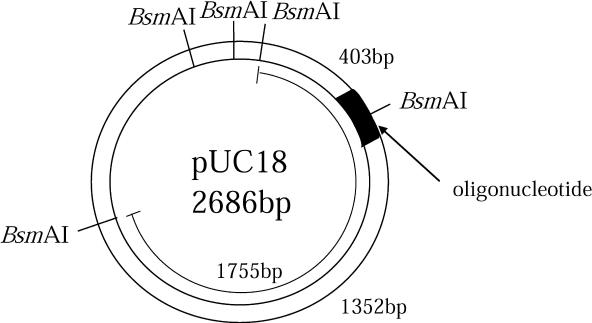
Figure 1.
Plasmid with BsmAI restriction site and fragment lengths relevant to the detection of mutation. An oligonucleotide with damaged sites was inserted at the SmaI restriction site of pUC18. Misrepair of the damage leads to ‘mutant’ fragment of 1755 bp length, whereas successful repair results in 1352 and 403 bp fragments after digestion with BsmAI.
Statistical significance of the effects of clustered damage sites relative to that of single lesions or of interlesion distances was determined by t-test. To determine whether mutation frequencies differed between strains, analysis of variance was carried out.
Sequence analysis of plasmid DNA
After transformation (as above), the bacteria were incubated for 1 h at 37°C in 1 ml SOC. An aliquot of 0.5 µl was plated on to LB–ampicillin (100 µg/ml) agar and the bacteria were incubated overnight at 37°C. Single colonies were picked at random and used to inoculate separate 2 ml aliquots of LB–ampicillin (100 µg/ml) broth and grown at 37°C overnight. The plasmid DNA retrieved from the bacteria was sequenced using an Applied Biosystems Big Dye sequencing kit using the following forward and reverse primers 5′-CTTCGCTATTACGCCAGCTG-3′ and 5′-GGCACGACAGGTTTCCCGACTGGA-3′, respectively. These primers amplify sequence across the site of damage in the cloned oligonucleotide. The sequencing data were analysed using the Genetyx Mac software (ver. 10).
RESULTS
We developed previously a plasmid-based assay to measure the frequency of mutation induced by bistranded clustered damage sites containing two synthetic lesions at varying distances relative to each other (18). In the present study we have used this assay to test the mutability of clustered damage sites consisting primarily of 8-oxoG and DHT. The 8-oxoG lesion was placed within a BsmAI restriction site and the DHT lesion was positioned at sites 1, 3 or 5 bp away on the complementary strand in both orientations (see Table 1, where these positions are termed negative when DHT is placed 3′ to 8-oxoG, and positive when DHT is 5′ to 8-oxoG). Damaged DNA was transfected into wild-type or glycosylase-deficient strains (fpg, mutY, nth, fpg mutY or fpg mutY nth) of E.coli, and mutation was assessed by the inability to cut at the BsmAI site following plasmid rescue.
Close proximity of DHT and 8-oxoG leads to high levels of mutation
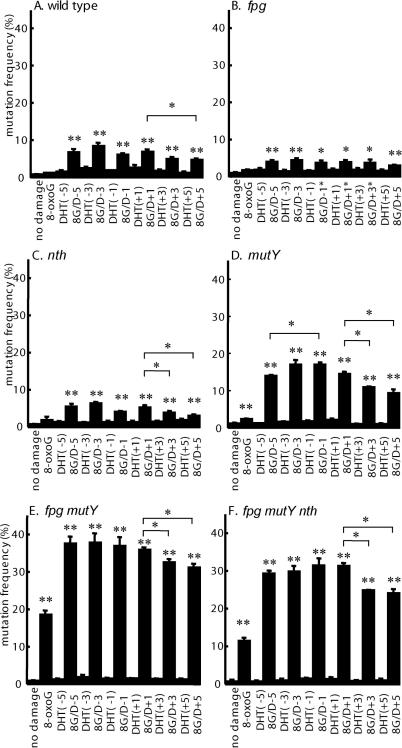
A representative gel image of the mutation analysis in the fpg mutY double mutant strain is shown in Figure 2. As expected, DHT alone at each position in the oligonucleotide gives mutation frequencies that are always close to background frequencies in all strains (Figure 3; DHT−1, DHT−3, DHT−5, DHT+1, DHT+3, DHT+5). The 8-oxoG lesion alone also gives relatively low frequencies of mutation in the single glycosylase-deficient strains (Figure 3B–D), but in contrast to DHT, a substantial mutagenic effect was seen in multiple glycosylase-deficient strains (Figure 3E and F). However, in all strains, clustered lesions lead to higher mutation frequencies than those for a single 8-oxoG lesion (all differences significant at the 1% level, except for the clustered damage containing DHT at positions −1, +1 and +3 in the fpg strain where the differences are significant at the 5% level). The mutability of clustered damage in the nth or fpg strains was found to be slightly less than that in wild type (differences significant at the 1% level) (compare Figure 3A–C). Relative to results with the wild-type strain, a marked increase of mutation frequency was observed when clustered damage was introduced into the mutY strain (Figure 3D). Even higher frequencies were observed in the fpg mutY double mutant strain, where the frequencies increase up to 35–40% (Figure 3E), but overall mutation frequencies in the fpg mutY nth triple mutant strain were lower than in the double mutant (difference significant at the 1% level; Figure 3F).
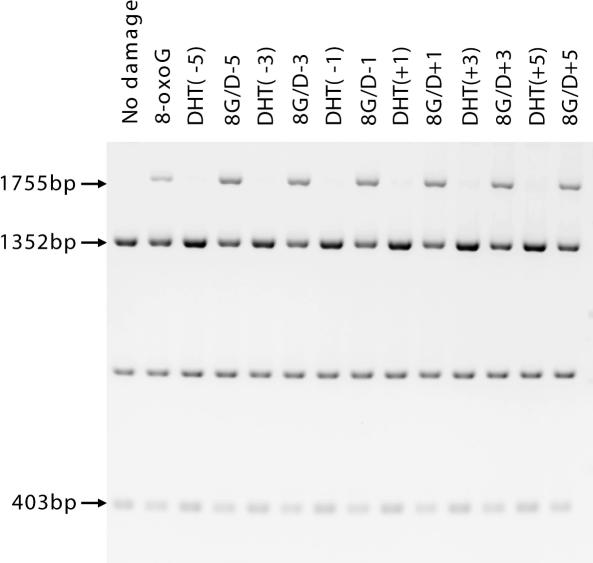
Figure 2.
Gel image of BsmAI restricted plasmid DNA processed in fpg mutY strain of E.coli. The types of damage introduced in the cell are indicated above each lane. The intensity of the 1755 bp band indicates the extent of mutation.
Figure 3.
Mutation frequencies (±SE) of clustered damage transformed into (A) wild type, (B) fpg strain, (C) nth strain, (D) mutY strain, (E) fpg mutY strain or (F) fpg mutY nth strain of E.coli. Types of damage are shown along the x-axis. The differences between a single 8-oxoG lesion and no damage, or between each type of clustered damage and a single 8-oxoG are shown where significant at the 5% (*) or at the 1% (**) level. Interlesion distance and lesion orientation also produce some significant differences at the 5% level (* above pairs of bars).
Interlesion distance and lesion orientation affect mutation frequencies
The mutation frequency decreases with increasing separation of DHT from 8-oxoG for the +orientation, but corresponding decreases were not clearly observed for the −orientation (Figure 3). The decrease is significant at the 5% level when the +5 cluster (8G/D+5) was compared to the +1 cluster (8G/D+1) in every strain, except for the fpg strain. Similarly, the cluster containing DHT at position +3 (8G/D+3) is less effective at inducing mutation than when at position +1 of the cluster (8G/D+1) in the nth, mutY, fpg mutY and fpg mutY nth strains (difference significant at 5% level). The only clear decrease for the −orientation was seen when the −5 cluster (8G/D−5) was compared to the −1 cluster (8G/D−1) in the mutY strain (difference significant at the 5% level).
Clustered base lesions can lead to incomplete mutations
We showed previously that bistranded clustered damage initially consisting of an AP site opposed 8-oxoG gives rise to ‘complete’ mutations after bacterial processing, i.e. only the mutated plasmid DNA strand gives rise to progeny (18). However, sequencing of plasmid DNA retrieved from individual bacterial colonies derived from clustered DHT/8-oxoG transfections revealed that unmutated and mutated sequences were often simultaneously present in an individual clone. To confirm that the majority of the mutations were ‘incomplete’, plasmids were isolated from ∼100 individual clones after processing of the single 8-oxoG lesion or clustered DHT/8-oxoG lesions in double or triple mutant strains, and analysed on gels after cutting with BsmAI (where the proportion of cut:uncut plasmid reveals the degree of ‘incompleteness’). In each lesion/strain combination, the distribution was found to vary continuously from no mutation (completely repaired) to 100% mutation (complete mutation) (Supplementary Figure 1). Adding together the mutation frequencies estimated from individual clones gave a value similar to the overall average frequency determined from the gel analysis using bulk plasmids (Figure 3).
Sequencing of the plasmids from each clone showed that the 8-oxoG lesion alone generated predominantly G:C to T:A transversion in the fpg mutY strain, consistent with previous findings (23,30). Similarly, the main mutation type arising from clustered damage with DHT positioned at either −1 or −5 on the opposite strand to 8-oxoG in fpg mutY or fpg mutY nth strains is also G:C to T:A transversion at the site of 8-oxoG. However, mutations such as 1 bp and several base pair deletions were detected not as mixed populations but as pure clones (Table 2).
Table 2.
Type of mutation induced by the DHT/8-oxoG cluster
| Type of damage | Strain | No. of clones sequenced | Type of mutation | Frequencya (%) |
|---|---|---|---|---|
| 8-Oxog | fpg mutY | 92 | G→Tb | 19.2 |
| G→deletionb | 3.2 | |||
| 22.4 (total) | ||||
| 8G/D−5 | fpg mutY | 95 | G→Tb | 28.1 |
| G→deletionb | 3.1 | |||
| 15 bp deletionc | 1.0 | |||
| 32.2(total) | ||||
| 8G/D−1 | fpg mutY | 95 | G→Tb | 35.9 |
| G→deletionb | 2.1 | |||
| T→insertiond | 0.7 | |||
| AGAG→GTc | 1.0 | |||
| 39.7(total) | ||||
| 8G/D−1 | fpg mutY nth | 98 | G→Tb | 28.2 |
| G→deletionb | 2.0 | |||
| G→insertione | 0.9 | |||
| 31.1 (total) |
aFrequency of each type of mutation was calculated by first determining the type of mutation from the sequence profile of the clone and then averaging the frequencies detected from the restriction analysis of the plasmid.
bPosition of the mutation was at the site of 8-oxoG (shown as bold G)
csequence includes the 8-oxoG site.
dT inserted at the next base 3′ to the site of 8-oxoG.
eG inserted at the next base either 5′ or 3′ to the site of 8-oxoG.
Mutations induced by an 8-oxoG/8-oxoG clustered damage site
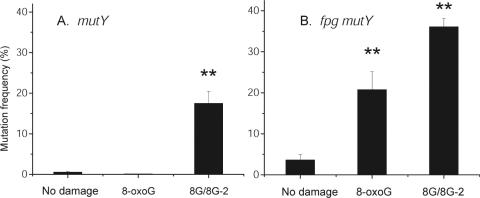
The occurrence of incomplete mutations may serve as an indication of the processing of the DHT/8-oxoG cluster, by removal of one lesion to form an intermediate with one intact lesion and an abasic site or SSB (Discussion). If this is true, then clustered damage sites containing different combinations of excisable base lesions should also yield incomplete mutations at frequencies similar to the DHT/8-oxoG cluster. As a test of this prediction, we analysed mutation in an oligonucleotide with 8-oxoG opposing another 8-oxoG at the −2 position (Table 1). Plasmids carrying this oligonucleotide were transfected into the single and double mutant strains with the highest mutation frequencies, namely mutY and fpg mutY. The frequency of mutation in the 8-oxoG/8-oxoG cluster is significantly higher (at the 5% level) than that of the single 8-oxoG lesion (Figure 4) in both strains as would be predicted from the preceding data and studies published previously (17). It is also seen that the maximum mutation frequency found for this cluster is very similar to thatfound for the DHT/8-oxoG cluster in these mutant strains (compare Figures 3 and 4). Sequence analysis of the mutations arising in this oligonucleotide showed again that the main mutation type was the G:C to T:A transversion, with no evidence of deletions (data not shown). Also, as for the DHT/8-oxoG cluster, most of the recovered 8-oxoG/8-oxoG plasmids showed incomplete mutation (Supplementary Figure 1), confirming that different types of lesion opposing 8-oxoG within a bistranded clustered damage site can lead to this outcome.
Figure 4.
Mutation frequencies (±SE) of 8-oxoG/8-oxoG damage transformed into the mutY and fpg mutY strains of E.coli. Types of damage are shown along the x-axis (8G/8G-2 = clustered 8-oxoG/8-oxoG lesions at the −2 spacing). Statistical significance at the 1% (**) level is indicated.
DISCUSSION
The effect of damage clustering on lesion excision
Although DHT alone is neither a replication block nor a mutagenic lesion (24,25), as confirmed here, we have shown that the mutagenic potential of 8-oxoG is enhanced when present within a clustered damage site containing DHT on the complementary strand. This result suggests that the presence of DHT has interfered with the repair of 8-oxoG, despite previous in vitro experimental data indicating that DHT does not have a significant influence on the excision efficiency of 8-oxoG by Fpg (11). We consider that our data support the sequential removal of these lesions, with DHT removed first to give an AP site opposing the 8-oxoG lesion. DHT may be removed by Nth, although there is at least one other glycosylase (endonuclease VIII; Nei) with overlapping substrate preferences (31). The fact that more than one glycosylase removes DHT lesions may increase the probability that DHT is removed first from this clustered damage site. The AP site can also be converted rapidly to an SSB, possibly by the associated AP lyase activity of Nth (32–34) or Nei (35), and the SSB can significantly retard the excision of an adjacent 8-oxoG lesion by Fpg (11). Transfection of bacteria with an AP/8-oxoG clustered damage substrate can be highly mutagenic, as shown in our previous studies (18) where we suggested that the presence of the AP site (or SSB) retards the excision of 8-oxoG so that it persists into plasmid replication. Most strikingly, in the present study we see that the frequencies of mutation are very similar for the DHT/8-oxoG cluster (Figure 3) and for the 8-oxoG/8-oxoG cluster (Figure 4), as well as to those reported previously for the AP/8-oxoG cluster (18) in the same bacterial strains, suggesting that the same key intermediate (AP/8-oxoG or SSB/8-oxoG) is involved.
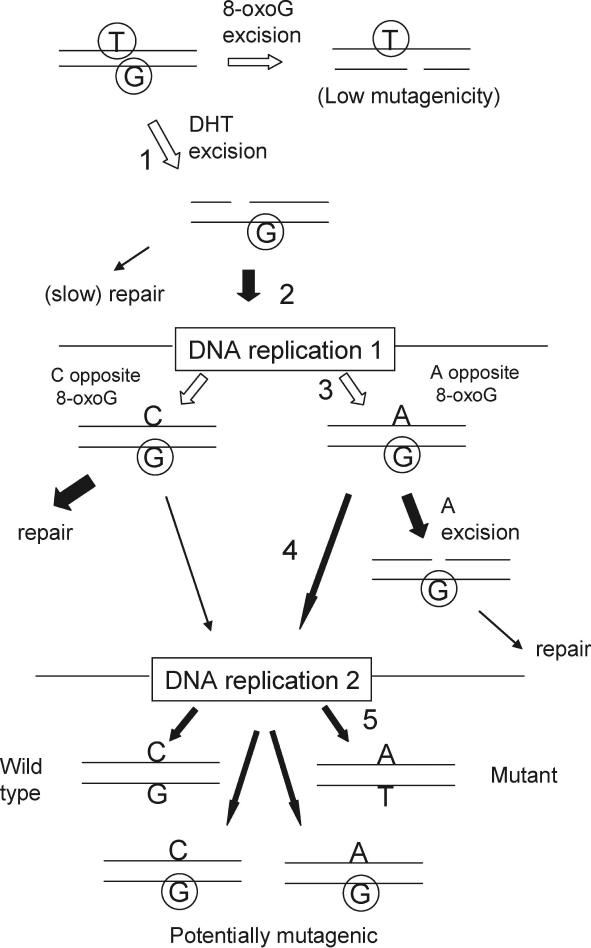
Based upon these considerations, in Figure 5 we show the most probable pathway for the DHT/8-oxoG clustered damage site leading to mutation. The removal of DHT (Step 1) leads to an intermediate which may not be repaired before undergoing replication (Step 2), when 8-oxoG may be paired with adenine (Step 3). In the absence of MutY, the adenine:8-oxoG pairing may persist into another replication cycle (Step 4) when the adenine can pair with thymine to lead to a transversion mutation (Step 5).
Figure 5.
Pathways of mutation fixation for the DHT/8-oxoG cluster. The damaged DNA is depicted at the top left, followed by the processing of the damage through two replication cycles.  represents 8-oxoG and
represents 8-oxoG and  represents DHT. Open arrows indicate alternative pathways for the damaged DNA. For simplicity, only the strand carrying the 8-oxoG is shown at the first replication (the other strand may be lost or replicated inefficiently, depending on lesion processing; see text). The major mutagenic pathway is numbered in Steps 1–5, where an A is paired with 8-oxoG at the first replication, and if this is not removed it will be paired with T at the second replication (GC to TA transversion).
represents DHT. Open arrows indicate alternative pathways for the damaged DNA. For simplicity, only the strand carrying the 8-oxoG is shown at the first replication (the other strand may be lost or replicated inefficiently, depending on lesion processing; see text). The major mutagenic pathway is numbered in Steps 1–5, where an A is paired with 8-oxoG at the first replication, and if this is not removed it will be paired with T at the second replication (GC to TA transversion).
Although we consider that the evidence points primarily to the removal of DHT first from the clustered DHT/8-oxoG site, we also considered what the mutagenic outcome would be if the 8-oxoG lesion was removed first (Figure 5, Step 1). To examine this we created a DHT/SSB clustered damage site with DHT at the (−1) position and measured its mutagenicity in the wild-type and glycosylase-deficient strains. We found that all strains (wild-type, single, double and triple mutant) gave approximately the same mutation frequency (range 5.7–6.5% in three independent experiments; data not shown), suggesting that this intermediate does not contribute to the high mutation frequencies found with the DHT/8-oxoG cluster (Figure 3).
Additionally, if two closely spaced base lesions were excised simultaneously, DSBs could be formed leading to reduced transformation efficiency and a high frequency of DNA deletions. However, if the processing of one of the lesions leads to the retardation of excision of the other, and repair occurs sequentially, DSB formation would be avoided [consistent with data from the action of purified base excision proteins on similar DNA substrates (11)]. Our transformation and sequencing data with opposed DHT and 8-oxoG lesions clearly support the sequential repair model, in agreement with recent results with 8-oxoG/8-oxoG (19) and AP/8-oxoG (18) clustered lesions.
Effect of interlesion separation on mutation frequency
The effect of interlesion position is much more pronounced with DHT placed in the 5′ direction relative to 8-oxoG (+orientation; Figure 3), as shown previously for other 8-oxoG-containing clustered damage sites when the second lesion (AP site or another 8-oxoG) is positioned 5–6 bp away (18,19). One explanation for this effect of 5′ interlesion distance may lie in the nature of the 8-oxoG repair process: Lomax et al. (14,15) found that cellular repair involves the resynthesis of several nucleotides (long-patch repair) when an SSB and to a lesser extent an AP site is placed in the −orientation relative to 8-oxoG, whereas single-nucleotide replacement (short-patch repair) occurs when such lesions are located in the +orientation relative to 8-oxoG. With long-patch repair, as long as the repair patch size is larger than the distance of separation, a nucleotide will always be inserted opposite 8-oxoG by repair synthesis (Figure 5) and the mutation frequency will be independent of the separation distance of lesions. With short-patch repair, the most closely proximal lesions will lead to the strongest inhibition of 8-oxoG excision and thus will result in a greater chance of incorrect repair and mutagenesis.
Importance of different glycosylases for the repair of clustered damage
We have addressed the question of which glycosylase is most important in the initial stages of repair of clustered damage containing 8-oxoG and DHT (Figure 5, Step 1). However, we have found that lack of MutY, but not Fpg or Nth, leads to a marked increase of mutation frequency compared to that in wild type. MutY alleviates the induction of mutation after plasmid DNA replication through excision of adenine opposed to 8-oxoG (23); its importance is underscored by the recent finding that biallelic loss of the human MutY homologue (MYH) leads to a high risk of colorectal cancer [the MYH polyposis syndrome; (36,37)]. The 8-oxoG:adenine base pair can arise from the presence of 8-oxoG at replication due to retardation or loss of its excision in the cluster (Figure 5, Step 3), but will be readily repairable since after replication it is no longer part of a cluster. As noted above, the action of Fpg to cleave 8-oxoG is most likely retarded by the initial processing of the DHT lesion to an AP site/SSB on the other strand, giving the potential for a marked increase in mutagenesis by clustered damage in the mutY strain.
It is perhaps surprising that Fpg deficiency gives a slightly lower mutation frequency with clustered DHT/8-oxoG damage than that for wild-type cells (Figure 3A and B), although we reported recently a similar effect with another type of clustered damage [AP/8-oxoG; (18)]. A possible explanation for this is the reported inhibition of the in vitro activity of MutY by Fpg in adenine excision from 8-oxoG:A (38), so that paradoxically the absence of Fpg would increase the chance of correct post-replication repair. However, we find that strains lacking both Fpg and MutY (fpg mutY and fpg mutY nth) show increases in mutation frequency with both 8-oxoG alone and clustered damage, relative to the single mutY strain (Figure 3D–F). These results suggest that in our experiments Fpg is important in two contexts: first, with single 8-oxoG sites, where there is no inhibition by adjacent lesion processing, and second, after replication of a clustered damage site where a persisting 8-oxoG lesion will become separated from the other lesion. Although the clustered DHT/8-oxoG damage sites in the multiple mutant strains still give a larger mutation frequency increase than 8-oxoG alone (two to three times), the relative increase is less than that in the single mutY strain (seven times). These results show that more of the lesions are being fixed into mutations when there are multiple repair-protein deficiencies, but that the relative effectiveness of lesion clustering in leading to mutation becomes less important when repair is severely compromised.
The loss of Nth also leads to a significant reduction in mutations arising from the clustered damage site, in both the nth strain (relative to wild type) and the triple mutant fpg mutY nth strain (relative to the double mutant fpg mutY) (Figure 3). It has been shown previously that normally Nth protects against the formation of spontaneous mutations [i.e. compared to wild-type bacteria, the nth strain shows a 3-fold increase in mutations arising from randomly occurring lesions (39)]. This highlights the difference between the mutagenic consequences of processing clustered damage relative to those from single lesions. As we have proposed, the processing of DHT to AP/SSB by Nth in a clustered damage site is likely to cause the retardation of 8-oxoG processing, and thereby increase the mutation frequency. Although these data suggest that Nth has a major role in determining the mutagenic consequences of clustered damage, this may not be the complete picture: as noted above, endonuclease VIII (Nei) has an overlapping substrate specificity with Nth. If Nei is able to remove DHT in the absence of Nth, it should also lead to mutagenic intermediates (AP/8-oxoG or SSB/8-oxoG). Data from knockout strains suggest that Nth and Nei back each other up in combating radiation damage, but in the present context there are other factors to take into account. First, the levels of Nth present in bacterial cells are much higher than those of Nei (ratio ∼20:1) (40). Second, while bacteria lacking Nth have a 3-fold increase in spontaneous mutation frequency, in the same assay those lacking Nei were found to have a decreased mutation frequency (0.56-fold) suggesting a different role for these two glycosylases in mutation formation (39). In the absence of more direct evidence, we conclude that Nth is likely to be more important than Nei in the processing of potentially mutagenic damage in our experiments.
In our study, the fpg mutY nth strain also showed a reduced mutation frequency at a single 8-oxoG lesion, relative to that in the fpg mutY strain, and we do not at present have an explanation for this result. Although Nth binds to base pairs containing 8-oxoG, so that it may interfere with the repair activity of other glycosylases, this activity preferentially affects 8-oxoG:G and not 8-oxoG:C base pairs (41).
Incomplete mutation induced by 8-oxoG alone and in damage clusters
In principle potentially mutagenic damage on one strand of plasmid DNA should give rise to both mutant and non-mutant progeny, providing that damage survives until DNA replication. The proportion of each plasmid DNA strand represented in the final progeny will depend on the efficiency with which the two strands are replicated, and on the probability of conversion of the damage to a mutation. However, we showed previously that certain types of clustered DNA damage can lead to ‘complete’ mutations (18), where single cell clones have a pure mutant genotype [as described in early bacterial and yeast experiments; (42–45)]. The analysis of individual clones carrying plasmids initially containing 8-oxoG alone or clustered DHT/8-oxoG lesions in fpg mutY or fpg mutY nth strains showed that most mutations were ‘incomplete’. Furthermore, these clones showed a broad distribution of mutation frequencies (Supplementary Figure 1). This might be expected when 8-oxoG remains unrepaired, from the stochastic incorporation of adenine or cytosine at each round of plasmid replication (Figure 5, Step 2). It should be noted that the replicative polymerase in E.coli, Pol III, has a very strong preference for putting adenine opposite 8-oxoG (46). However, cytosine is also incorporated opposite 8-oxoG by DNA polymerase in our plasmid assay, because we observed that ∼30% of amplified plasmids were wild type (G:C at the 8-oxoG site) even when fpg mutY cells were initially transformed with plasmids harbouring the 8-oxoG:adenine pair (data not shown).
To explain mutation frequencies of >50%, especially with clustered damage sites, the strand harbouring cytosine opposite 8-oxoG could be lost or its rate of amplification could be reduced so that the 8-oxoG-carrying strand serves as the major replication template (Figure 5, Step 2). Strand loss leading to a reduced probability of progeny plasmids derived from the strand harbouring specific forms of DNA damage, such as DNA adducts N-2-acetylaminofluorene and 1,N6-ethenodeoxyadenosine, has indeed been demonstrated previously (47,48).
Our observations emphasize the importance of the type of lesions, interlesion distance and relative orientation of lesions within a cluster for the effective processing of clustered damage sites in the cell. We find that among the glycosylases that attempt repair of clustered damage, MutY has the most important anti-mutagenic role, consistent with the involvement of its human homologue in preventing cancer. Our data highlight the complex interaction of damage excision and replication underlying the mutagenic potential of clustered damage sites, even when a component lesion is non-mutagenic, and show that a detailed description of DNA metabolism following damage is critical for understanding the mechanisms of mutagenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to the Japan Atomic Energy Research Institute for a study grant, and to the Royal Society for a short-term visit grant, to N.S.; to the Medical Research Council for a studentship to C.P.; and to the European Commission (CLUSTOXDNA, MCRTN-CT-2003-505086) for partial support of this study. Funding to pay the Open Access publication charges for this article was provided by Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ward J.F. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 2.Goodhead D.T. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 3.Nikjoo H., O'Neill P., Wilson W.E., Goodhead D.T. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Goodhead D.T., Thacker J., Cox R. Weiss Lecture. Effects of radiations of different qualities on cells: molecular mechanisms of damage and repair. Int. J. Radiat. Biol. 1993;63:543–556. doi: 10.1080/09553009314450721. [DOI] [PubMed] [Google Scholar]
- 5.Blakely E.A., Kronenberg A. Heavy-ion radiobiology: new approaches to delineate mechanisms underlying enhanced biological effectiveness. Radiat Res. 1998;150:S126–S145. [PubMed] [Google Scholar]
- 6.Sutherland B.M., Bennett P.V., Sidorkina O., Laval J. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland B.M., Bennett P.V., Sutherland J.C., Laval J. Clustered DNA damages induced by X rays in human cells. Radiat Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Gulston M., Fulford J., Jenner T., de Lara C., O'Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furlong E.A., Jorgensen T.J., Henner W.D. Production of dihydrothymidine stereoisomers in DNA by gamma-irradiation. Biochemistry. 1986;25:4344–4349. doi: 10.1021/bi00363a025. [DOI] [PubMed] [Google Scholar]
- 10.Dawidzik J.B., Budzinski E.E., Patrzyc H.B., Cheng H.C., Iijima H., Alderfer J.L., Tabaczynski W.A., Wallace J.C., Box H.C. Dihydrothymine lesion in X-irradiated DNA: characterization at the molecular level and detection in cells. Int. J. Radiat. Biol. 2004;80:355–361. doi: 10.1080/09553000410001695877. [DOI] [PubMed] [Google Scholar]
- 11.David-Cordonnier M.H., Laval J., O'Neill P. Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry. 2001;40:5738–5746. doi: 10.1021/bi002605d. [DOI] [PubMed] [Google Scholar]
- 12.David-Cordonnier M.H., Laval J., O'Neill P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- 13.Harrison L., Hatahet Z., Wallace S.S. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 14.Lomax M.E., Cunniffe S., O'Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 15.Lomax M.E., Cunniffe S., O'Neill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair (Amst.) 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Eot-Houllier G., Eon-Marchais S., Gasparutto D., Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malyarchuk S., Youngblood R., Landry A.M., Quillin E., Harrison L. The mutation frequency of 8-oxo- 7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair (Amst.) 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 18.Pearson C.G., Shikazono N., Thacker J., O'Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malyarchuk S., Brame K.L., Youngblood R., Shi R., Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza D.I., Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson C.G. UK: University of Oxford; 2005. Investigating the mutagenic potential of clustered DNA damage in E.coli. D.Phil. thesis. [Google Scholar]
- 22.Harrison L., Brame K.L., Geltz L.E., Landry A.M. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair (Amst.) 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaels M.L., Cruz C., Grollman A.P., Miller J.H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans J., Maccabee M., Hatahet Z., Courcelle J., Bockrath R., Ide H., Wallace S. Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 25.Ide H., Petrullo L.A., Hatahet Z., Wallace S.S. Processing of DNA base damage by DNA polymerases. Dihydrothymine and beta-ureidoisobutyric acid as models for instructive and noninstructive lesions. J. Biol. Chem. 1991;266:1469–1477. [PubMed] [Google Scholar]
- 26.Dizdaroglu M., Laval J., Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 27.Wallace S.S. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150:S60–79. [PubMed] [Google Scholar]
- 28.D'Ham C., Romieu A., Jaquinod M., Gasparutto D., Cadet J. Excision of 5,6-dihydroxy-5,6-dihydrothymine, 5,6-dihydrothymine, and 5-hydroxycytosine from defined sequence oligonucleotides by Escherichia coli endonuclease III and Fpg proteins: kinetic and mechanistic aspects. Biochemistry. 1999;38:3335–3344. doi: 10.1021/bi981982b. [DOI] [PubMed] [Google Scholar]
- 29.Cupples C.G., Miller J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grollman A.P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 31.Wallace S.S., Bandaru V., Kathe S.D., Bond J.P. The enigma of endonuclease VIII. DNA Repair (Amst.) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 32.Bailly V., Verly W.G. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., Linn S. The mechanisms of action of E.coli endonuclease III and T4 UV endonuclease (endonuclease V) at AP sites. Nucleic Acids Res. 1988;16:1135–1141. doi: 10.1093/nar/16.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhry M.A., Weinfeld M. The action of Escherichia coli endonuclease III on multiply damaged sites in DNA. J. Mol. Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- 35.Jiang D., Hatahet Z., Melamede R.J., Kow Y.W., Wallace S.S. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 36.Lipton L., Tomlinson I. The multiple colorectal adenoma phenotype and MYH, a base excision repair gene. Clin. Gastroenterol. Hepatol. 2004;2:633–638. doi: 10.1016/s1542-3565(04)00286-1. [DOI] [PubMed] [Google Scholar]
- 37.Jo W.S., Chung D.C. Genetics of hereditary colorectal cancer. Semin. Oncol. 2005;32:11–23. doi: 10.1053/j.seminoncol.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 38.Hazra T.K., Hill J.W., Izumi T., Mitra S. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:193–205. doi: 10.1016/s0079-6603(01)68100-5. [DOI] [PubMed] [Google Scholar]
- 39.Jiang D., Hatahet Z., Blaisdell J.O., Melamede R.J., Wallace S.S. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melamede R.J., Hatahet Z., Kow Y.W., Ide H., Wallace S.S. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto Y., Zhang Q.M., Takao M., Yasui A., Yonei S. Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holliday R. Mutation and replication in Ustilago maydis. Genet. Res. 1962;3:472–486. [Google Scholar]
- 43.Kubitschek H.E. Mutation without segregation. Proc. Natl Acad. Sci. USA. 1964;52:1374–1381. doi: 10.1073/pnas.52.6.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munson R.J., Bridges B.A. Segregation of radiation-induced mutations in Escherichia coli. Nature. 1964;203:270–273. doi: 10.1038/203270a0. [DOI] [PubMed] [Google Scholar]
- 45.Witkin E.M., Sicurella C. Pure clones of lactose-negative mutants obtained in Escherichia coli after treatment with 5-bromouracil. J. Mol. Biol. 1964;8:610–613. doi: 10.1016/s0022-2836(64)80017-6. [DOI] [PubMed] [Google Scholar]
- 46.Moriya M., Grollman A.P. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C→T:a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol. Gen. Genet. 1993;239:72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 47.Koffel-Schwartz N., Maenhaut-Michel G., Fuchs R.P. Specific strand loss in N-2-acetylaminofluorene-modified DNA. J. Mol. Biol. 1987;193:651–659. doi: 10.1016/0022-2836(87)90348-2. [DOI] [PubMed] [Google Scholar]
- 48.Pandya G.A., Yang I.Y., Grollman A.P., Moriya M. Escherichia coli responses to a single DNA adduct. J. Bacteriol. 2000;182:6598–6604. doi: 10.1128/jb.182.23.6598-6604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]