Abstract
Lipoarabinomannan (LAM), one of the few known bacterial glycosylphosphoinositides (GPIs), occurs in various structural forms in Mycobacterium species. It has been implicated in key aspects of the physiology of Mycobacterium tuberculosis and the immunology and pathogenesis of tuberculosis. Yet, little is known of the biosynthesis of LAM. A bioinformatics approach identified putative integral membrane proteins, MSMEG4250 in Mycobacterium smegmatis and Rv2181 in M. tuberculosis, with 10 predicted transmembrane domains and a glycosyltransferase (GT) motif (DID), features that are common to eukaryotic mannosyltransferases (ManTs) of the GT-C superfamily that rely on polyprenyl-linked rather than nucleotide-linked sugar donors. Inactivation of M. smegmatis MSMEG4250 by allelic exchange resulted in altered growth and inability to synthesize lipomannan (LM) but accumulation of a previously uncharacterized, truncated LAM. MALDI-TOF/MS and NMR indicated a structure lower in molecular weight than the native molecule, a preponderance of 6-linked Manp residues, and the absence of 2,6-linked and terminal Manp. Complementation of the mutant with the corresponding ortholog of M. tuberculosis H37Rv restored normal LM/LAM synthesis. The data suggest that MSMEG4250 and Rv2181 are ManTs that are responsible for the addition of α(1→2) branches to the mannan core of LM/LAM and that arrest of this branching in the mutant deters formation of native LAM. The results allow for the presentation of a unique model of LM and LAM biosynthesis. The generation of mutants defective in the synthesis of LM/LAM will help define the role of these GPIs in the immunology and pathogenesis of mycobacterial infections and physiology of the organism.
Keywords: mutants, Mycobacterium tuberculosis, glycosyltransferase
Mycobacterial diseases, such as tuberculosis and leprosy, remain serious human public health problems. One critical feature that contributes to the particular pathogenicity and physiology of mycobacteria is the unique, highly hydrophobic and impermeable cell wall (1). Beyond the cytoplasmic membrane, the cell wall of Mycobacterium tuberculosis consists of a core of mycolic acids, arabinogalactan and peptidoglycan, providing the template for insertion of products such as the phthiocerol-containing lipids, the trehalose mycolates, phosphatidylinositol mannosides (PIMs), and their more glycosylated derivatives, lipomannan (LM) and lipoarabinomannan (LAM) (2), all of which contribute to the particular physiology and disease induction capacity of Mycobacterium species. LAM, in its various forms, including those with mannose (Man) “caps” (ManLAM), has been implicated in many of the key aspects of the pathogenesis of tuberculosis and leprosy, such as induction of phagocytosis, phagosomal alteration and inhibition of fusion with lysosomes, and induction of innate, humoral, and acquired T cell-mediated immunity (3, 4). However, all of these studies (often conflicting) were conducted with the isolated molecule. Mutants devoid of LAM are crucial to fully resolve its role in disease pathogenesis and bacterial physiology.
Although structurally well defined (5), the underlying enzymology and genetics of LAM biosynthesis are unknown. It has been believed, although not empirically demonstrated, that both LM and LAM have their origins in the PIMs, because all contain a phosphatidylinositol (PI) moiety (5, 6). The first step in PIM synthesis involves the transfer of a Manp residue to the 2 position of the myo-inositol ring of PI to form PIM1, catalyzed by PimA (7, 8). Biosynthesis proceeds by means of the sequential addition of Manp residues to PIM1 or its acylated counterpart (AcPIM1),§ catalyzed in part by PimB and PimC, to produce PIM species having from two (PIM2) to three (PIM3) Manp residues (9, 10). The Manp units at position 6 of the inositol of PIM3 are further elongated with Manp to generate higher PIMs (PIM4, PIM5, and PIM6) (11, 12). However, the mannosyltransferases (ManTs) involved in these biosynthetic steps have not been identified. PIM6 is likely a “dead-end” product, because it contains 2-linked Manp, a combination not found in the mannan core of LM/LAM (13); PIM4 is the likely precursor for the subsequent extension of the mannan chain, giving rise to “linear LM” (12), which is further mannosylated at the C-2 positions, resulting in mature, branched LM. LAM itself retains the PI end and mannan backbone of LM; the Araf (arabinofuranose) residues originate in decaprenyl-P-arabinofuranose (C50-P-Araf), and addition is partially catalyzed by EmbC (14).
The only well characterized glycosyltransferases (GTs) implicated in PIM/LM/LAM biosynthesis are the three initial ManTs, PimA, PimB, and PimC, and EmbC (8–10, 14). Apparently, the ManTs that use GDP-Man as sugar donors occur on the cytoplasmic face of the plasma membrane (15), whereas those responsible for further elongation, requiring C50-P-Man as the sugar donor, are likely located in the cytoplasmic membrane or extracytoplasmic space (12, 15, 16). The lumen-associated ManTs of the eukaryotic protein glycosylation pathway, relying on polyprenyl-linked sugar as donors and members of the GT-C superfamily of enzymes for glycosylation, are also invariably integral membrane proteins, sharing similar hydropathy plots and the presence of a conserved motif in an extracytoplasmic loop (17). Recent iterative BLAST searches of sequence databases advanced a list of 11 ORFs from the genome of M. tuberculosis with multiple transmembrane domains (17) (Table 2, which is published as supporting information on the PNAS web site). This development prompted us to examine these putative GTs through bioinformatics-informed mutagenesis as being responsible for the latter steps in LM/LAM biosynthesis.
Results
Sequence of MSMEG4250 Suggests a Unique Family of GTs.
Of the 11 ORFs in the M. tuberculosis genome that were identified as putative polyprenyl-dependent GT-Cs, 4 (Rv0051, Rv1159, Rv2181, and Rv2673) were predicted to be homologous to the eukaryotic PIG-M protein, a ManT that transfers the first Man to the glycosylphosphoinositide anchor of human cell surface glycoproteins (18). Rv2181 (MSMEG4250) is the only one with a DID motif, which is known to be critical for the function of PIG-M (18). MSMEG4250 and PIG-M are of similar size and contain 10 predicted transmembrane-spanning domains with the DID motif located in the first extracytoplasmic loop and both the N and C termini within the cytoplasm, all of which are indicative of analogous functions. Moreover, a homologous ORF is present in the known genomes of other actinomycetes, all with the ability to synthesize PIMs and related products (19).
Construction of Mycobacterium smegmatis ΔMSMEG4250 by Allelic Exchange and Phenotypic Analysis.
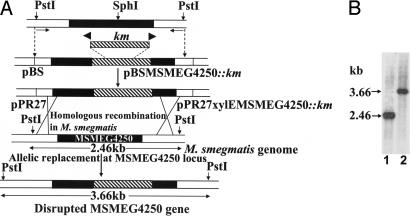
The ortholog of MSMEG4250 in M. tuberculosis H37Rv is Rv2181 and has been reported to be nonessential by saturated transposon mutagenesis experiments (20). A knockout mutant of MSMEG4250 in M. smegmatis was successfully generated by homologous recombination by using the thermosensitive vector pPR27xylE (Fig. 1A). Transformation of M. smegmatis with the vector carrying a kanamycin (Km) disrupted copy of the gene and selection at 42°C for Km resistance (Kmr) and sucrose resistance (Sucr) led to the identification of 17% white colonies when sprayed with catechol, suggesting that allelic exchange had occurred. Further analysis by Southern blot of 9 Kmr-Sucr-XylE− colonies revealed that all had undergone gene replacement at the MSMEG4250 locus (Fig. 1B). ΔMSMEG4250 grew normally at 30°C and slowly at 37°C but failed to grow at 42°C in Sauton medium; however, it grew normally on LB-Km plates and in LB broth at 37°C. The WT and mutant strains exhibited identical sensitivity to isoniazid, chloramphenicol, novobiocin, streptomycin, ethambutol, and erythromycin (minimum inhibitory concentration values of 5, 10, 2.5, 2.5, 1, and 2.5 μg/ml, respectively). Clearly, MSMEG4250 is not essential in M. smegmatis but is required for growth at elevated temperatures. Electron microscopy analyses of the mutant strain as compared with the WT did not reveal any marked ultrastructural changes, in accord with the observation that a LAM-like deficient mutant of Corynebacterium did not display any altered ultrastructural morphology (19).
Fig. 1.
Generation of M. smegmatis ΔMSMEG4250 by targeted disruption. (A) Schematic representation of the genetic structures obtained during the construction of the mutation in MSMEG4250 of M. smegmatis. Black boxes indicate coding sequence of MSMEG4250. Hatched regions represent the intragenic DNA fragment replaced with a Km cassette from pUC4K. The locations of primers for PCR are indicated by small arrows. Homologous recombination and selection for Km and sucrose resistance and xylE resulted in colonies with a disrupted MSMEG4250, which remained white upon spraying with catechol (XylE−). (B) Southern blot hybridization was performed on genomic DNA prepared from M. smegmatis mc2155 cells (lane 1) and the candidate mutant colony (lane 2). DNA was restricted with PstI and probed with the PCR product generated with the primers used in the initial step.
Effects of MSMEG4250 Inactivation on LM/LAM Synthesis.
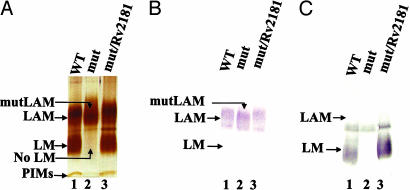
The similarities between ΔMSMEG4250 and PIG-M, an enzyme using only a polyprenyl-linked sugar donor and a PI-anchored acceptor, dictated an examination of the status of PIM, LM, and LAM biosynthesis in MSMEG4250. Comparison of CHCl3/CH3OH (2:1) and CHCl3/CH3OH/H2O (10:10:3) soluble lipids demonstrated no obvious differences in the profiles of the PIM family when analyzed by high-performance TLC or 2D TLC; WT and ΔMSMEG4250 expressed similar levels of AcPIM2, Ac2PIM2, AcPIM6, and Ac2PIM6 (8, 16) (data not shown). However, analysis of the phenol-extracted LM/LAM fraction by SDS/PAGE revealed the complete absence of LM in the mutant but the presence of a product slightly different in mobility from WT LAM named mutLAM (Fig. 2A). The mutLAM reacted strongly to monoclonal antibody CS-35 (Fig. 2B), which is known to recognize only the arabinan component of WT LAM (21). Con A reacts avidly to WT LM and some minor glycoproteins (Fig. 2C). However, the lack of reactivity in the LM region of the gel in the case of ΔMSMEG4250 further points to its absence.
Fig. 2.
Analysis of LM/LAM from M. smegmatis mc2155 (WT), ΔMSMEG4250 (mut), and ΔMSMEG4250/Rv2181 (mut/Rv2181) cells. LM/LAM was extracted from cells of WT (lanes 1), ΔMSMEG4250 (lanes 2), and ΔMSMEG4250/Rv2181 (lanes 3) by the phenol procedure, separated on a 10–20% Tricine gel, and visualized by periodic acid/Schiff reagent staining (A), Western blot with monoclonal antibody CS-35 (B), and Con A (C).
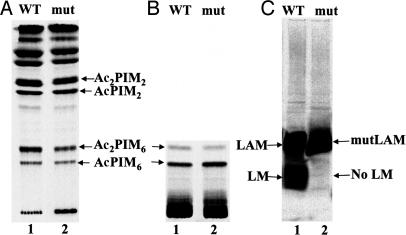
Results from examination of 14C-labeled PIMs and lipoglycans from M. smegmatis WT and ΔMSMEG4250 grown in the presence of [14C]glucose was in full conformity with the chemical analysis; namely, the CHCl3/CH3OH (2:1) and CHCl3/CH3OH/H2O (10:10:3) extracts, when examined by high-performance TLC (Fig. 3A and B) or 2D TLC, revealed no significant changes in PIM composition, whereas [14C]LM was not evident while the 14C-labeled LAM-like molecule was present (Fig. 3C). TFA hydrolysis revealed the presence of arabinose and Man as the predominant sugars, confirming the LAM-like nature of mutLAM.
Fig. 3.
Analysis of 14C-labeled CHCl3/CH3OH (2:1) (A) and CHCl3/CH3OH/H2O (10:10:3) (B) and LM/LAM fraction of M. smegmatis mc2155 (WT) (lanes 1) and ΔMSMEG4250 (mut) (lanes 2) cells (C). Cells were grown to an OD600 of 0.4 and then incubated in the presence of 1 μCi/ml [14C]glucose for an additional 8 h. Cells were extracted with CHCl3/CH3OH (2:1) and CHCl3/CH3OH/H2O (10:10:3) and analyzed by high-performance TLC. LM/LAM was extracted and analyzed by Tricine gel electrophoresis and subsequent blotting to a nitrocellulose membrane for autoradiography.
Taken together, these data point to a block in LM synthesis, but not in LAM synthesis, which is surprising in light of the dogma that LM is an intermediate on the pathway to LAM (12). Obviously, the putative ManT, MSMEG4250, plays a key role in the biosynthesis of mature, branched LM, an event, which, when incapacitated, apparently does not affect arabinosylation. These observations raise the possibility that the synthesis of LM and LAM proceeds independently after a bifurcation in the basic pathway emanating from the PIMs.
Structural Characterization of mutLAM.
MALDI-TOF/MS analysis of WT LAM showed a broad unresolved peak centered at ≈19 kDa. However, mutLAM showed a sharper peak at an average molecular mass of 14 kDa, indicative of a smaller size (Fig. 7, which is published as supporting information on the PNAS web site). Apparently, the mutLAM is a truncated form of WT LAM, incomplete in some respects within the arabinan or mannan components.
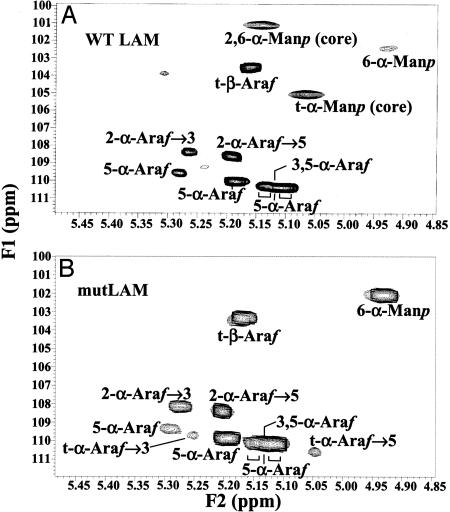
Per-O-methylation analysis of mutLAM demonstrated amounts of t-Araf, 5-linked Araf, and 3,5-linked Araf similar to those in WT LAM, indicative of little to no alteration in the nature of the arabinan component (Fig. 4). However, there was an increase in the proportion of 6-linked Manp, no evident 2,6-linked Manp, and a decrease in terminal t-Manp residues; these traces of t-Manp can be attributed to the Manp linked to the 2 position of inositol or to the t-Manp at the end of the mannan core. Analysis of partially methylated alditol acetate derivatives of WT and mutLAM demonstrated that the mole percentage decrease in 2,6-linked Manp and t-Manp in the latter was approximately equivalent to the increase in 6-linked Manp (Table 3, which is published as supporting information on the PNAS web site). The results indicate that the mannan domain of mutLAM consists only of a linear α(1→6) Manp backbone devoid of the normal α(1→2) Manp branches and that MSMEG4250 is apparently a ManT that is responsible for the addition of α(1→2) Manp branches to the linear mannan core on the biosynthetic pathway to mature LAM.
Fig. 4.
Glycosyl linkage analysis of per-O-methylated WT LAM and mutLAM prepared from M. smegmatis (WT) (A) and ΔMSMEG4250 (B). Samples of purified LAM and mutLAM were prepared as described in Materials and Methods, per-O-methylated, hydrolyzed, and reduced, and the per-O-acetylated glycosyl derivatives were analyzed by GC/MS. The arrow indicates the absence of 2,6-linked Man.
2D 1H13C heteronuclear single quantum correlation (HSQC) spectroscopy NMR was applied to mutLAM, and the 1H and 13C resonances of the different spin systems were attributed as described in refs. 22 and 23 (Table 1). WT M. smegmatis LAM (Fig. 5A) showed no discrepancy from the published spectrum of M. smegmatis LAM (22). The overall arabinosylation pattern was retained in mutLAM except for the presence of small amounts t-α-Araf, which arises from variability in the extent of β(1→2) Araf termini of the arabinan in various strains of mycobacteria (23) (Fig. 5B; see also Fig. 8, which is published as supporting information on the PNAS web site). However, two of the signals attributed to t-Manp and 2,6-linked Manp in the WT LAM are missing in the mutLAM (Fig. 5). Thus, the data from 1H13C HSQC spectra provide further evidence that mutLAM contained no evident 2,6-linked Manp or t-Manp units. This observation was reinforced by the fact that the volume intensity of 6-Manp was higher for mutLAM (Table 1). Apparently, disruption of MSMEG4250 results in the complete loss of the branched Manp units normally attached to C-2 of the α(1→6) mannan backbone of LAM, resulting in the absence of the conventional, mature, branched LM but retention, in the form of mutLAM, of the normal arrangement of arabinan motifs on an abnormal LM template, itself induced by the mutant. These observations allow for the presentation of a previously uncharacterized pathway for the bifurcation of LM and LAM synthesis (Fig. 6).
Table 1.
1H13C NMR chemical shift assignment based on 2D HSQC of LAM and mutLAM from WT and ΔMSMEG4250 cells
| Glycosyl residues | WT LAM |
mutLAM |
||||
|---|---|---|---|---|---|---|
| 13C | 1H | Volume | 13C | 1H | Volume | |
| 5-α-Araf | 110.0 | 5.18 | 83 | 109.8 | 5.19 | 95 |
| 5-α-Araf | 109.6 | 5.28 | 15 | 109.3 | 5.30 | 12 |
| 3,5-α-Araf | 110.3 | 5.13 | 347 | 110.1 | 5.11 | 392 |
| 2-α-Araf→5 | 108.6 | 5.19 | 75 | 108.4 | 5.20 | 79 |
| 2-α-Araf→3 | 108.4 | 5.26 | 19 | 108.1 | 5.27 | 25 |
| t-α-Araf→5 | — | — | — | 110.6 | 5.04 | 6 |
| t-α-Araf→3 | — | — | — | 109.7 | 5.25 | 4 |
| t-β-Araf | 103.5 | 5.16 | 100 | 103.3 | 5.17 | 100 |
| t-α-Manp | 105.1 | 5.07 | 37 | — | — | — |
| 2,6-α-Manp | 101.1 | 5.14 | 28 | — | — | — |
| 6-α-Manp | 102.4 | 4.93 | 8 | 102.0 | 4.92 | 93 |
Signal volumes were integrated and normalized to t-β-Araf, which was taken as the reference signal. Chemical shifts were measured in 2H2O at 500 MHz and 295 K. —, not detected.
Fig. 5.
Two-dimensional 1H13C HSQC spectra of purified WT LAM of M. smegmatis (A) and mutLAM of ΔMSMEG4250 (B). Only the expanded anomeric regions are shown.
Fig. 6.
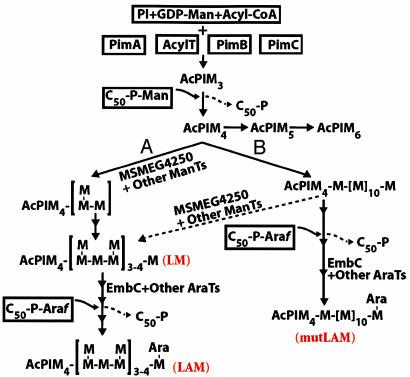
Proposed biosynthetic pathway for mycobacterial LM/LAM. AcPIM4 is proposed as the branch point on the pathways leading to AcPIM6, LM, and LAM. Normally it is thought that linear mannan backbone (pathway B) is substituted at C-2 by Manp residues (dotted arrow) to form the native branched LM, which is subsequently arabinosylated to form mature LAM. However, in the absence of the α(1→2) branching enzyme, MSMEG4250, linear mannan formed from pathway A or B can be directly arabinosylated to form mutLAM.
Complementation of ΔMSMEG4250 with Rv2181 Restores LM and LAM Synthesis.
The degree of identity between the amino acid sequence of MSMEG4250 and its ortholog, Rv2181, of M. tuberculosis H37Rv is 53%. To obtain evidence that Rv2181 is also a functional homolog of MSMEG4250, it was placed under the control of the hsp60 promoter in the mycobacterial expression vector pVV16 and transformed into ΔMSMEG4250. Analysis of the LM/LAM population by SDS/PAGE and immunoblotting demonstrated restoration of LM and LAM content (Fig. 2A), whereas cells transformed with the vector alone had the same phenotype as ΔMSMEG4250. The reactivity of these molecules to monoclonal antibody CS-35 (Fig. 2B) and Con A (Fig. 2C) showed patterns consistent with those of native LAM and LM. The results suggest that Rv2181 is the functional equivalent of MSMEG4250 and is sufficient in restoring biosynthesis of native LM and LAM in M. smegmatis ΔMSMEG4250.
Discussion
The implied biosynthetic sequence of PI advancing to the PIMs, LM, and, finally, LAM is supported by structural relationships and some recent biochemical and genetic studies (8–10, 12, 13). PimA, PimB, and PimC, mediating the early steps in PIM synthesis (Fig. 6), are the only well characterized enzymes in the series. These enzymes use GDP-Man as the sugar donor and clearly belong to family 4 of the CAZy classification of GTs and are of the GT-A superfamily (http://afmb.cnrs-mrs.fr/CAZY). There is now consensus (16, 24) that, beyond PIM4, the pathway diverges, on the one hand toward PIM6 and, on the other, toward LM/LAM synthesis (Fig. 6).
Little is known of the nature of the ManTs or arabinosyltransferases or of the intermediate products that are responsible for the synthesis of the more extended PIMs, LM, and LAM. EmbC is the only known GT implicated in the synthesis of the arabinan of LAM, and it may have ancillary functions, such as in conjoint transport (14). A search for previously unidentified genes by using an initial bioinformatics approach identified MSMEG4250 as a putative polyprenyl-linked sugar-dependent ManT, and sequence analysis indicated a distinctiveness from all ManTs described to date, including those responsible for the initial steps in PIM synthesis. In particular, in contrast to the GDP-Man-requiring enzymes, MSMEG4250 shares structural similarities with proteins belonging to the recently identified GT-C superfamily, members of which uniformly use polyprenyl-linked sugar donors (17). Also, the presence of GT-C-like predicted membrane topology and a putative GT motif on the extracytoplasmic face of the cytoplasmic membrane, and the overall similarity to PIG-M with its specificity for glycosylation of glycosylphosphoinositide anchors, implied that MSMEG4250 may catalyze the synthesis of mature LM/LAM, perhaps on the external face of the plasma membrane. This scenario was also consistent with the inverting mechanism of GT catalysis leading to a product in the α-configuration.
The overall evidence that MSMEG4250 is responsible for the α(1→2) Manp branching of mature LM and LAM is generally persuasive. First, disruption of this gene by targeted replacement blocked the synthesis of mature, branched LM and resulted in the accumulation of a product shown by MALDI-TOF/MS to be lower in molecular weight than WT LAM and devoid of 2,6-linked Manp and terminal Manp residues. NMR analysis, in particular, was unequivocal in demonstrating the complete loss of t-Manp and 2,6-linked Manp residues from mutLAM, with little alteration in the known arabinan motifs (23), suggesting that a linear α(1→6) mannan devoid of any α(1→2) Manp branching is capable of being arabinosylated in a normal fashion. Second, a key factor in attributing these particular functions to this enzyme is the evidence that complementation of MSMEG4250 with a plasmid containing only Rv2181 of M. tuberculosis restored the production of mature LM and LAM. Third, apparently, this form of branching activity is selective for the α(1→6)-linked mannan, because synthesis of PIM6, as for all PIMs, was not affected in the mutant, demonstrating that MSMEG4250 is not responsible for addition of the α(1→2) Manp unit to PIM4 leading to PIM5 and PIM6.
In the absence of mannan side branches, LM is likely either degraded by an endomannosidase (25) or rapidly arabinosylated to form the mutLAM. Linear LM, as a precursor, may be transient; such a product has been recognized but only through metabolic labeling of cell-free extracts (12). Thus, the absence of native branched LM and accumulation of mutLAM puts paid to the longstanding notion that mature, branched LM is an obligate precursor of LAM. In general, the data support the hypothesis that disruption of the branching ManT, MSMEG4250, leads not only to the absence of mature, branched LM but to a redirection of its precursor to the normal arabinosylation mechanism with the resultant synthesis of mutLAM.
A proposed pathway that takes into account the potential role of MSMEG4250 and the accumulation of the observed products is presented in Fig. 6. At least two [α(1→6) and α(1→2)] ManT enzyme activities are required for formation of mature LM. LM contains ≈20–25 α(1→6)-linked Manp units, of which 8–9 are decorated with single Manp residues by means of α(1→2) linkages (26). The spacing between the branches, and therefore whether there is a uniformity hinting of “block” addition, is not known. Theoretically, a Wzy-dependent pathway, such as for the synthesis of Gram-negative LPSs (27), may apply. At the periplasmic face of the cytoplasmic membrane, decaprenyl-P-linked mannoligosaccharides may arise by chain extension at the reducing end terminus, with the nascent polymer then being transferred from its C50-P carrier to the nonreducing end of the PIMs. Under this scenario, a repeating unit of 1,6 Manp branched at the C-2 position could be synthesized on the C50-P carrier. It is also possible that a processive GT synthesizes an α(1→6)-linked mannooligosaccharide built on a C50-P anchor, which is then transferred to the nonreducing end of PIM intermediates, followed by branching. However, lipid-linked oligomannosides have not been reported in mycobacteria. Invoking a simpler synthetic pathway that is more in accord with the existing sparse data involving only one acceptor (AcPIM4) and addition of sugars at the nonreducing end (Fig. 6), it is not known whether α(1→6) and α(1→2) branching ManT activities work in concert [i.e., by addition of one or two α(1→6) Manp units, followed by α(1→2) Manp, before further elongation (pathway A in Fig. 6)] or whether the α(1→2) Manp residues are added after the α(1→6) mannan is synthesized (pathway B in Fig. 6).
Clearly, MSMEG4250 and Rv2181 are involved in the formation of the proper, fully branched LM and LAM. This observation paves the way for future studies with the recombinant enzyme and appropriate acceptors to identify the individual steps that are responsible for the latter stages of LM/LAM biosynthesis and their regulation.
ManLAM is known to be antiinflammatory, whereas PIM2, PIM6, and LM are Toll-like receptor 2-dependent proinflammatory molecules (28, 29). The molecular basis of proinflammatory versus antiinflammatory activities is not clear. It has been hypothesized that the ratio of LM/LAM might be a key parameter influencing the net immune response against mycobacteria and the outcome of infection (28, 29). However, all of these findings were derived from studies using purified PIMs, LM, or LAM with cellular models. The relevance and individual contribution of these molecules in mycobacterial infections remain to be determined. Therefore, the availability of defined mutants deficient in some aspects of PIM, LM, or LAM synthesis in M. tuberculosis should enable a precise measurement of the contribution of these molecules to the immunopathogenesis of tuberculosis and suggest innovative therapeutic strategies for the treatment of tuberculosis.
Materials and Methods
Bacterial Strains and Growth Conditions.
Escherichia coli DH5α was grown in LB broth at 37°C. M. smegmatis mc2155 cells and the mutant were grown either in 7H9 broth containing oleic acid, albumin, dextran, and 0.05% Tween 80 or LB broth. Antibiotics were applied as follows: 50 μg/ml Km, 50 μg/ml hygromycin, and 100 μg/ml ampicillin. Growth curves were performed by using defined Sauton medium supplemented with 1% ZnSO4 (30). Electrocompetent (8) M. smegmatis mc2155 (WT) and ΔMSMEG4250 cells were transformed by using a Gene Pulser unit (Bio-Rad, Hercules, CA). Sucrose was added at a final concentration of 10% to select for allelic exchange mutants. DNA sequence information from finished and unfinished genomes was obtained from TIGR (www.tigr.org) and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Amino acid sequences were aligned with ClustalW, and similarity searches were performed by SIM alignment. Secondary structure, hydrophobicity, and transmembrane topology was predicted by the Transmembrane Hidden Markov Model (TMHMM 2.0). Minimal inhibitory concentrations were determined by 2-fold serial broth dilution. The inoculum was 5 × 104 cells per tube, and the results were read after 48-h incubation. Samples for electron microscopy were prepared and analyzed as described in ref. 31.
Construction of M. smegmatis ΔMSMEG4250 and Cloning of Rv2181.
Deletion of the M. smegmatis MSMEG4250 gene was performed as described in ref. 32. MSMEG4250 and its flanking regions, including two natural PstI sites, were amplified by PCR with the forward 5′-AACCTCGGCATCCCGGTG-3′ and reverse 5′-GCAAGACGGGCCTGGCC-3′ primers. The 2.5-kb PCR fragment was subcloned into pBS(KS−). The resulting plasmid pBS-MSMEG4250 was digested with SphI, blunt-ended with T4 DNA polymerase, ligated with a Km resistance (Kmr) gene excised from pUC4K (Amersham Biosciences, Piscataway, NJ) by PstI digestion, and blunt-ended. A 3.7-kb fragment of MSMEG4250::Km was then excised from the resulting plasmid with PstI, blunt-ended, and inserted into the XbaI-digested, blunt-ended site of pPR27-xylE, the vector used to achieve allelic replacement at the MSMEG4250 locus. Allelic replacement was confirmed by Southern hybridization as described in ref. 32. The blots were hybridized with a 2.5-kb 32P-labeled DNA probe generated by digestion of the PCR fragment subcloned in the pBS(KS−) vector as described above. To produce the complementation plasmid, pVV16Rv2181, the full-length Rv2181 gene, was amplified by PCR from M. tuberculosis H37Rv genomic DNA by using the primers 5′-TATAACATATGAGTGCATGGCGGGC-3′ and 5′-TATAAAAGCTTGCTGGCCGTCGGCGCC-3′, which contained a NdeI and HindIII restriction site (italicized), respectively, to enable direct cloning into the expression vector pVV16, a derivative of pMV261 (33) harboring Km- and hygromycin (Hyg)-resistant markers and the hsp60 promoter. Plasmid pVV16Rv2181 was transferred by electroporation into ΔMSMEG4250, and transformants were selected on plates containing Km and Hyg.
Analysis of PIMs, LM, and LAM.
In vivo radiolabeling of M. smegmatis mc2155 and ΔMSMEG4250 cells was performed in LB supplemented with 0.05% Tween 80, with or without Km, as appropriate. Subsequently, 1 μCi/ml (1 Ci = 37 GBq) [14C]glucose (50–62 mCi/mmol, Amersham Biosciences) was added to the cultures at A600 = 0.4. Cells were harvested by centrifugation after 8 h and washed once with PBS. Lipids were obtained by two consecutive overnight extractions with CHCl3/CH3OH (2:1), followed by centrifugation. The resulting insoluble pellet was further extracted with CHCl3/CH3OH/H2O (10:10:3). These extracts were dried and applied to aluminum-backed high-performance TLC silica gel sheets (Merck, Darmstadt, Germany). The solvent system used was as described in ref. 16. The final insoluble pellet was extracted with an equal volume of water and PBS-saturated phenol at 80°C for 2 h (14). The resulting aqueous layer containing LAM, LM, and the polar PIMs was dialyzed and analyzed on commercial 10–20% gradient Tricine SDS-polyacrylamide gels (Invitrogen, Carlsbad, CA). Blotting to nitrocellulose membrane was performed at 50 V for 1 h. Immunodetection was performed with monoclonal antibody CS-35 (21) and Con A conjugated to peroxidase (Sigma, St. Louis, MO) (34). The membranes probed with Con A-peroxidase were subsequently developed with the 4-chloro-1-naphthol/3,3′-diaminobenzidine tetrahydrochloride substrate kit (Pierce, Rockford, IL). Sugar composition of 14C-labeled LAM was determined as described in ref. 35.
Extraction of LAM from WT and ΔMSMEG4250 for Structural Characterization.
M. smegmatis mc2155 and ΔMSMEG4250 cells were harvested in late logarithmic phase from 12 liters of liquid medium and lyophilized. Dried cells were delipidated with organic solvents and freeze-thawed before disruption by sonication (35). The resulting suspension was refluxed in 50% ethanol several times; extracts were combined, evaporated, and digested with proteinase K (Invitrogen), followed by dialysis and subjection to a two-step protocol involving hydrophobic and gel exclusion chromatography (35). These LAM- and LM-containing fractions were monitored on Tricine gels stained with periodic acid/Schiff reagent (36), pooled, and dialyzed.
MALDI-TOF/MS.
The matrix used was 2,5-dihydroxybenzoic acid at a concentration of 10 mg/ml in a mixture of water/acetonitrile [1:1 (vol/vol)] containing 0.1% TFA and 10 μg of LAM mixed with 1.0 μl of the matrix solution. Analyses were performed on an Ultraflex MALDI-TOF/TOF instrument (Bruker, Bremen, Germany) using reflector mode detection. Mass spectra were recorded in the negative mode for the underivatized sample on a 30-ns time delay with a grid voltage of 94% and full accelerating voltage (25 kV). The mass spectra were mass assigned through external calibration.
Glycosidic Linkage Analysis.
To determine linkage patterns, samples were per-O-methylated (37); alditol acetates were prepared as described in ref. 38, solubilized in CHCl3, and analyzed by GC/MS (Thermoquest GCQ Plus, Thermo Electron, Austin, TX).
NMR Spectroscopy.
Spectra were acquired after several lyophilizations in D2O at a concentration of 4–5 mg per 0.6 ml of 100% D2O. Two-dimensional 1H13C HSQC and HSQC-total correlation spectroscopy NMR spectra were acquired on an Inova 500-MHz NMR spectrometer (Varian, Palo Alto, CA) by using the supplied Varian pulse sequences. The HSQC data were acquired as described in ref. 39.
Supplementary Material
Acknowledgments
We thank Christopher D. Rithner and Jian Zhang for NMR, Michael S. Scherman for electron microscopy, and Jessica Prenni for MALDI-TOF/MS analysis. This work was supported by National Institute of Allergy and Infectious Diseases/National Institutes of Health Grant AI-18357.
Abbreviations
- LM
lipomannan
- LAM
lipoarabinomannan
- PIM
phosphatidylinositol mannoside
- GT
glycosyltransferase
- ManT
mannosyltransferase
- Araf
arabinofuranose
- Man
mannose
- PI
phosphatidylinositol
- HSQC
heteronuclear single quantum correlation
- Km
kanamycin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
AcPIM or Ac2PIM is used to define the number of acyl groups, in addition to those attached to glycerol, esterified either to the 6-position of the Manp on carbon-2 of myo-inositol, or directly to the 3-position of myo-inositol.
References
- 1.Brennan PJ, Nikaido H. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Crick DC, Brennan PJ, McNeil MR. In: Tuberculosis. Rom WN, Garay SM, editors. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 115–134. [Google Scholar]
- 3.Nigou J, Gilleron M, Rojas M, Garcia LF, Thurnher M, Puzo G. Microbes Infect. 2002;4:945–953. doi: 10.1016/s1286-4579(02)01621-0. [DOI] [PubMed] [Google Scholar]
- 4.Briken V, Porcelli SA, Besra GS, Kremer L. Mol Microbiol. 2004;53:391–403. doi: 10.1111/j.1365-2958.2004.04183.x. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee D, Hunter SW, McNeil M, Brennan PJ. J Biol Chem. 1992;267:6228–6233. [PubMed] [Google Scholar]
- 6.Hunter SW, Brennan PJ. J Biol Chem. 1990;265:9272–9279. [PubMed] [Google Scholar]
- 7.Hill DL, Ballou CE. J Biol Chem. 1966;241:895–902. [PubMed] [Google Scholar]
- 8.Kordulakova J, Gilleron M, Mikusova K, Puzo G, Brennan PJ, Gicquel B, Jackson M. J Biol Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- 9.Schaeffer ML, Khoo KH, Besra GS, Chatterjee D, Brennan PJ, Belisle JT, Inamine JM. J Biol Chem. 1999;274:31625–31631. doi: 10.1074/jbc.274.44.31625. [DOI] [PubMed] [Google Scholar]
- 10.Kremer L, Gurcha SS, Bifani P, Hitchen PG, Baulard A, Morris HR, Dell A, Brennan PJ, Besra GS. Biochem J. 2002;363:437–447. doi: 10.1042/0264-6021:3630437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan P, Ballou CE. J Biol Chem. 1967;242:3046–3056. [PubMed] [Google Scholar]
- 12.Besra GS, Morehouse CB, Rittner CM, Waechter CJ, Brennan PJ. J Biol Chem. 1997;272:18460–18466. doi: 10.1074/jbc.272.29.18460. [DOI] [PubMed] [Google Scholar]
- 13.Khoo KH, Dell A, Morris HR, Brennan PJ, Chatterjee D. Glycobiology. 1995;5:117–127. doi: 10.1093/glycob/5.1.117. [DOI] [PubMed] [Google Scholar]
- 14.Zhang N, Torrelles JB, McNeil MR, Escuyer VE, Khoo KH, Brennan PJ, Chatterjee D. Mol Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
- 15.Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. J Biol Chem. 2005;280:21645–21652. doi: 10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- 16.Morita YS, Patterson JH, Billman-Jacobe H, McConville MJ. Biochem J. 2004;378:589–597. doi: 10.1042/BJ20031372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Mushegian A. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda Y, Watanabe R, Harris CL, Hong Y, Ohishi K, Kinoshita K, Kinoshita T. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson KJ, Eggeling L, Maughan WN, Krumbach K, Gurcha SS, Nigou J, Puzo G, Sahm H, Besra GS. J Biol Chem. 2003;278:40842–40850. doi: 10.1074/jbc.M307988200. [DOI] [PubMed] [Google Scholar]
- 20.Sassetti CM, Boyd DH, Rubin EJ. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 21.Kaur D, Lowary TL, Vissa VD, Crick DC, Brennan PJ. Microbiology. 2002;148:3049–3057. doi: 10.1099/00221287-148-10-3049. [DOI] [PubMed] [Google Scholar]
- 22.Gilleron M, Himoudi N, Adam O, Constant P, Venisse A, Riviere M, Puzo G. J Biol Chem. 1997;272:117–124. doi: 10.1074/jbc.272.1.117. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Berg S, Lee A, Spencer JS, Zhang J, Vissa V, McNeil MR, Khoo KH, Chatterjee D. J Biol Chem. 2006;281:19512–19526. doi: 10.1074/jbc.M513846200. [DOI] [PubMed] [Google Scholar]
- 24.Kovacevic S, Anderson D, Morita YS, Patterson J, Haites R, McMillan BN, Coppel R, McConville MJ, Billman-Jacobe H. J Biol Chem. 2006;281:9011–9017. doi: 10.1074/jbc.M511709200. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama K, Ballou CE. J Biol Chem. 1989;264:21621–21628. [PubMed] [Google Scholar]
- 26.Khoo KH, Douglas E, Azadi P, Inamine JM, Besra GS, Mikusova K, Brennan PJ, Chatterjee D. J Biol Chem. 1996;271:28682–28690. doi: 10.1074/jbc.271.45.28682. [DOI] [PubMed] [Google Scholar]
- 27.Raetz CR, Whitfield C. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilleron M, Quesniaux VF, Puzo G. J Biol Chem. 2003;278:29880–29889. doi: 10.1074/jbc.M303446200. [DOI] [PubMed] [Google Scholar]
- 29.Quesniaux VJ, Nicolle DM, Torres D, Kremer L, Guerardel Y, Nigou J, Puzo G, Erard F, Ryffel B. J Immunol. 2004;172:4425–4434. doi: 10.4049/jimmunol.172.7.4425. [DOI] [PubMed] [Google Scholar]
- 30.Atlas RM. In: Handbook of Microbial Media. 2nd Ed. Parks LC, editor. Boca Raton, FL: CRC; 1997. pp. 1225–1226. [Google Scholar]
- 31.Escuyer VE, Lety MA, Torrelles JB, Khoo KH, Tang JB, Rithner CD, Frehel C, McNeil MR, Brennan PJ, Chatterjee D. J Biol Chem. 2001;276:48854–48862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- 32.Jackson M, Crick DC, Brennan PJ. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- 33.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, et al. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 34.Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. J Bacteriol. 1996;178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg S, Starbuck J, Torrelles JB, Vissa VD, Crick DC, Chatterjee D, Brennan PJ. J Biol Chem. 2005;280:5651–5663. doi: 10.1074/jbc.M411418200. [DOI] [PubMed] [Google Scholar]
- 36.Prinzis S, Chatterjee D, Brennan PJ. J Gen Microbiol. 1993;139:2649–2658. doi: 10.1099/00221287-139-11-2649. [DOI] [PubMed] [Google Scholar]
- 37.Dell A, Reason AJ, Khoo KH, Panico M, McDowell RA, Morris HR. Methods Enzymol. 1994;230:108–132. doi: 10.1016/0076-6879(94)30010-0. [DOI] [PubMed] [Google Scholar]
- 38.McNeil M, Chatterjee D, Hunter SW, Brennan PJ. Methods Enzymol. 1989;179:215–242. doi: 10.1016/0076-6879(89)79123-0. [DOI] [PubMed] [Google Scholar]
- 39.Torrelles JB, Khoo KH, Sieling PA, Modlin RL, Zhang N, Marques AM, Treumann A, Rithner CD, Brennan PJ, Chatterjee D. J Biol Chem. 2004;279:41227–41239. doi: 10.1074/jbc.M405180200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.