Abstract
RNAi is proving to be a powerful experimental tool for the functional annotation of mammalian genomes. The full potential of this technology will be realized through development of approaches permitting regulated manipulation of endogenous gene expression with coordinated reexpression of exogenous transgenes. We describe the development of a lentiviral vector platform, pSLIK (single lentivector for inducible knockdown), which permits tetracycline-regulated expression of microRNA-like short hairpin RNAs from a single viral infection of any naïve cell system. In mouse embryonic fibroblasts, the pSLIK platform was used to conditionally deplete the expression of the heterotrimeric G proteins Gα12 and Gα13 both singly and in combination, demonstrating the Gα13 dependence of serum response element-mediated transcription. In RAW264.7 macrophages, regulated knockdown of Gβ2 correlated with a reduced Ca2+ response to C5a. Insertion of a GFP transgene upstream of the Gβ2 microRNA-like short hairpin RNA allowed concomitant reexpression of a heterologous mRNA during tetracycline-dependent target gene knockdown, significantly enhancing the experimental applicability of the pSLIK system.
Keywords: G protein, tetracycline
The discovery of the RNAi pathway in mammalian cells heralded a new era in the analysis of mammalian gene function (1, 2). The sequence specificity of RNAi permits inhibition of endogenous gene expression by introduction of gene-specific dsRNA into cells. Stable expression of such dsRNA in the form of a short hairpin RNA (shRNA) expressed from an RNA polymerase (pol) III promoter is a useful and widely used approach for the application of RNAi (3–5). However, long-term suppression using pol III shRNAs can be problematic (6), and options for conditional expression are limited. For RNAi-based gene therapy applications, an ideal platform would constitute a regulatable shRNA exhibiting high efficacy when expressed at single copy in the genome.
The design of shRNA transcripts as primary microRNA (miR) mimics (miR-shRNAs) driven by pol II promoters significantly expands the possibilities for conditional RNAi in mammalian cells (7–9). Accordingly, tetracycline (Tet)-inducible expression of miR-shRNAs has recently been described (10, 11), but these studies either were limited to cell systems with constitutive expression of the Tet-transactivating components or required cumbersome multivirus infection. The development of a flexible single vector configuration for regulatable knockdown of endogenous genes with miR-shRNAs, combined with reexpression of a transgene, would provide a valuable experimental tool, with wide applicability in mammalian cell systems.
In this study we describe the development of a flexible lentiviral vector platform supporting constitutive expression of a Tet-transactivating component with a selection marker and coordinated conditional expression of miR-shRNA(s) targeting single or multiple gene products. We also demonstrate an additional powerful feature of this system by combining miR-shRNA and cDNA expression from the conditional promoter, which increases the flexibility of this system for functional genomic and gene therapy applications.
Results
Characterization of Single Lentiviral Vectors Expressing All of the Required Components for Tet-Regulated pol II Promoter-Driven RNAi.
Self-inactivating lentivirus has been established as an efficient vehicle for introducing exogenous expression cassettes into a wide range of cell lines, primary cells, and transgenic animals (10, 12–19) and has become the favored approach for stable expression of shRNAs in intractable cells (12–15). We hypothesized that packaging of all of the necessary components for Tet-regulated expression of miR-shRNAs in a lentiviral vector would permit robust drug-inducible RNAi in mammalian cells transduced at single copy. To increase the flexibility of such a system, we adopted a recombination-based cloning strategy that would permit preliminary identification of potent miR-shRNAs for genes of interest in a gateway entry vector, such that validated sequences could be easily shuttled to multiple viral expression platforms (Fig. 1a). We used the previously described approach of embedding gene-specific shRNAs in the primary transcript of human miR30 (7, 9), and we devised a simplified cloning strategy to create miR-shRNA entry clones (Fig. 5 a and b, which is published as supporting information on the PNAS web site). Potent miR-shRNAs were identified for the heterotrimeric G proteins Gα12 and Gα13 (Fig. 5d) and subcloned to lentiviral vectors containing all of the required components for Tet-ON and Tet-OFF conditional expression with constitutive coexpression of the fluorescent protein Venus (Fig. 6, which is published as supporting information on the PNAS web site). We infected a mouse embryonic fibroblast (MEF) cell line with the Tet-ON and Tet-OFF lentiviruses at low copy [multiplicity of infection (MOI) < 1], and, after selection of Venus-expressing cells by FACS, miR-shRNA expression was induced by doxycycline (DOX) withdrawal (Tet-OFF) or addition (Tet-ON). We observed relatively poor induction of Gα12 and Gα13 knockdown with the Tet-OFF system, but the Tet-ON vectors gave extremely potent conditional knockdown of the endogenous G proteins (Fig. 6). The superior performance of the Tet-ON vector may be attributed to our use of a third-generation version of the reverse Tet transactivator (rtTA) (S12G F86Y A209T = rtTA3), which has been shown to have improved DOX sensitivity and activity (20, 21). We named this vector system pSLIK (single lentivector for inducible knockdown) and created versions with alternative markers named pSLIK-Venus and pSLIK-Neo (Fig. 1a).
Fig. 1.
Characteristics of the pSLIK single vector system for conditional RNAi. (a) Schematics of the pSLIK-Venus and pSLIK-Neo vector platforms. Validated miR-shRNAs in the pEN_TmiRc2 vector are subcloned by site-specific recombination to either pSLIK vector. (b) Schematic showing predicted constitutive (black) and DOX-induced (green) transcripts from the pSLIK lentivirus. (c) FACS analysis of Venus expression in pSLIK-Venus transduced MEFs in the absence (−) and presence (+) of DOX (1 μg/ml). The presence of DOX induces a 10-fold increase in the Venus mean fluorescence intensity. (d) DOX-dependent increase in rtTA3 and Neo transcript expression in pSLIK-Neo transduced RAW264.7 cells. Neo selection with G418 was withdrawn from cells during DOX induction. mRNA levels were assessed by quantitative RT-PCR and normalized to the expression level in the absence of DOX (1 μg/ml). (e) Kinetics of conditional Gα12 knockdown in MEFs. Western blot analysis of Gα12 protein levels in pSLIK-Venus-Gα12 transduced MEFs cultured in the presence of 1 μg/ml DOX for 8 days (DOX addition). (f) Gα12 knockdown is readily reversed during continued culture for 8 days in the absence of DOX. (g) DOX dose–response in pSLIK transduced MEFs. Potent Gα12 target protein knockdown is induced by 100 ng/ml DOX.
We noted an interesting property of pSLIK-Venus transduced cells where the fluorescence intensity of Venus increased 10-fold in response to DOX (Fig. 1c). We hypothesized that this increase could be attributed to additional expression of Venus from the DOX-induced transcript (Fig. 1b). To test this hypothesis, we assessed the DOX-dependent expression of both rtTA3 and neomycin (Neo) mRNA by quantitative RT-PCR from cells transduced with a pSLIK-Neo lentivirus, and we observed >10-fold increases in mRNA levels of both transcripts in the presence of DOX (Fig. 1d). We propose that the addition of DOX initiates a positive feedback loop where rtTA3 can increase its own expression from the Tet response element (TRE) promoter, which may also induce higher levels of conditional miR-shRNA expression. Comparable autoregulatory increase in rtTA expression has been observed previously (21).
MEF cells infected at MOI <1 with a pSLIK-Venus-Gα12 virus showed rapid RNAi kinetics, with significant target knockdown (>90%) observed after 2 days of DOX treatment (Fig. 1e). Knockdown was stable for >1 week, after which knockdown was readily reversed upon DOX removal (Fig. 1f). Dose–response analysis showed significant Gα12 knockdown at 100 ng/ml DOX (Fig. 1g). These data demonstrate that, using the pSLIK lentiviral vector, we can introduce a tightly regulated miR-shRNA expression system for conditional knockdown of endogenous mammalian genes from a single viral infection.
pSLIK Lentivectors Can Mediate the Conditional Knockdown of Multiple Genes.
It has been demonstrated that certain cellular responses involving the Gα12 family can be mediated by either Gα12 or Gα13 (22–24). It would therefore be desirable to develop an approach that would permit conditional knockdown of both genes simultaneously. Because the Drosha ribonuclease often processes primary miRs from within a larger transcript (25) and previous studies have shown that multiple mature miRs can be produced from a single transcript (7), we hypothesized that potent miR-shRNAs targeting Gα12 and Gα13 (Fig. 2a) could be expressed in tandem to permit depletion of both targets simultaneously. We devised a cloning strategy to concatenate miR-shRNAs targeting different genes in a gateway entry vector followed by subcloning to pSLIK-Venus (Fig. 2b). MEF cells were transduced at MOI <1 with pSLIK-Venus lentiviruses containing miR-shRNAs targeting Gα12 and Gα13 both singly and in combination. Venus-expressing cells were selected by FACS, and, after DOX treatment for 5 days, Gα12 and Gα13 mRNA and protein levels were assessed (Fig. 2 c–e). Although we observed slightly reduced target knockdown in the cells expressing the tandem miR-shRNAs, we still achieved significant depletion of both Gα12 and Gα13 in the pSLIK-Venus-Gα12/13 cell line.
Fig. 2.
Multigene knockdown using pSLIK. (a) Specificity of miR-shRNAs against Gα12 and Gα13. MEFs transduced with pSLIK lentiviruses expressing Gα12 and Gα13 miR-shRNAs mediate specific knockdown of their target proteins in the presence of DOX (1 μg/ml). (b) Schematic showing creation of pSLIK lentivirus encoding tandem miR-shRNAs. miR-shRNAs are concatenated in the pEN_TmiRc2 entry vector by directional cloning (S, SpeI; X, XbaI; P, PstI), then subcloned to pSLIK-Venus by site-specific recombination. (c–e) Assessment of Gα12 (c) and Gα13 (d) mRNA and protein (e) levels in MEF cell lines transduced with control, Gα12, Gα13, and Gα12 plus Gα13 miR-shRNA-expressing pSLIK viruses. RNA and protein were harvested from cells cultured in the absence or presence of DOX (1 μg/ml) for 5 days. mRNA levels were assessed by quantitative RT-PCR and normalized to the expression level in untreated control cells. (f) LPA induced SRE-dependent transcription in MEF cell lines transduced with control, Gα12, Gα13, and Gα12 plus Gα13 miR-shRNA-expressing pSLIK viruses. Cells transfected with SRE-Luc and pCSK-lacZ were stimulated with 5 μM LPA for 7 h. Luciferase activity in cell lysates was assessed from cells cultured in the absence or presence of DOX (1 μg/ml) for 5 days and was normalized to the level of cotransfected β-galactosidase.
The serum response element (SRE) in the c-fos promoter binds to both the serum response factor and the ternary complex factor. Transcriptional activity of serum response factor induced by activation of the lysophosphatidic acid (LPA) receptor is mediated by the small G protein RhoA (24, 26), and LPA-induced RhoA activation has been proposed to be mediated by the Gα12 family (27, 28). Therefore, we investigated the effect of suppression of Gα12 and Gα13 on SRE-dependent transcription induced by LPA. The pSLIK-Venus transduced MEFs described above were transfected with an SRE-luciferase construct, and LPA-induced luciferase expression was compared in cells with and without DOX treatment. Luciferase induction was reduced ≈80% in the cells depleted in Gα13 alone, whereas the cells with depletion of both Gα12 and Gα13 showed a 45% decrease in luciferase induction (Fig. 2f). It seems likely that the lesser phenotype in the cells expressing miR-shRNAs against both Gα12 and Gα13 may be due to the lower level of knockdown of Gα13 in these cells, and the data support the conclusion derived from studies with a number of receptors (29) that Gα13 can be primarily responsible for transmitting specific receptor-mediated activation of serum response factor. The Tet-inducible knockdown of multiple genes after transduction of cells with a single pSLIK lentivirus provides an invaluable experimental platform for analysis of redundancy in signaling gene families.
pSLIK-Mediated Knockdown of Endogenous Signaling Genes in Hematopoietic Cells and Coupling of Conditional Knockdown with Transgene Expression.
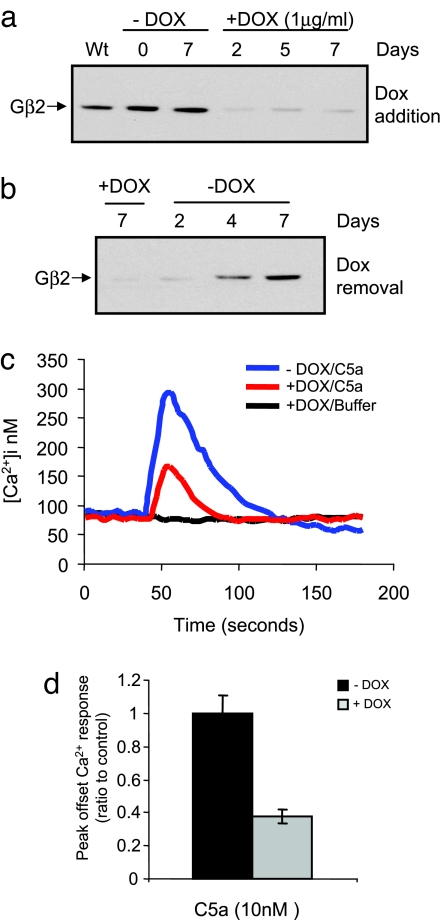
In less tractable cell types, such as those of hematopoietic origin, expression of shRNAs from lentivirus is a widely used approach to knockdown levels of endogenous genes (12, 13). Expression of shRNAs as pol III promoter-driven stem loops remains the most popular methodology in this respect, and we have used this strategy extensively in studies of signaling in the murine macrophage cell lines RAW264.7 (www.signaling-gateway.org/data/fxm/query?type=desc) and J774A.1 (14, 15). Although we have found this approach to be satisfactory in the majority of cases, there are certain drawbacks. We find that during long-term cell culture, and especially after freeze–thaw of cell lines, target gene knockdown diminishes (R.R., T.R., J.L., and I.D.C.F., unpublished observations). Similar effects have been reported by others in studies of colorectal carcinoma cells (6). Moreover, we have found that currently available platforms for Tet-regulated expression from pol III promoters provide unsatisfactory DOX dose–response and kinetics in RAW cells (L.A.S. and I.D.C.F., unpublished observations). To test the applicability of the pSLIK vector in RAW cells, we generated a potent miR-shRNA against the heterotrimeric G protein subunit Gβ2 and cloned it into pSLIK-Neo (Fig. 1a). RAW cells were transduced with lentivirus at MOI <1, and cells were selected for Neo resistance. Addition of DOX induced rapid and robust knockdown of Gβ2 after 2 days that remained stable for 1 week (Fig. 3a), and knockdown was quickly reversed after DOX removal (Fig. 3b). An important function of macrophages is their capacity to respond to chemokines such as complement factor C5a, and this response is mediated by the G protein-coupled C5a receptor (14, 30). We assessed the effect of depleting the Gβ2 subunit on the C5a-mediated Ca2+ response in RAW cells, and we found that 2 days after DOX treatment the response was diminished significantly relative to cells cultured in the absence of DOX (Fig. 3 c and d). These studies validate the applicability of the pSLIK system to introduce conditional RNAi in less tractable cells from a single viral infection and open up the attractive possibility of stable, robust, and reversible gene depletion in both primary cells and animal models.
Fig. 3.
pSLIK-based conditional knockdown of Gβ2 in RAW264.7 macrophages demonstrates Gβ2 selectivity in C5a-mediated Ca2+ response. (a) Western blot analysis of RAW cells transduced with a pSLIK-Neo-Gβ2 lentivirus. (b) Time course of target protein knockdown in response to 1 μg/ml DOX for 7 days and recovery kinetics through 7 days after DOX removal. (c) Ca2+ response profiles in pSLIK-Neo-Gβ2 transduced RAW cells treated with C5a (10 nM). Cells were cultured in the absence or presence of 1 μg/ml DOX for 2 days before assay. (d) Effect of DOX-dependent Gβ2 knockdown on the C5a-induced Ca2+ peak in pSLIK-Neo-Gβ2 transduced RAW cells. Data were averaged from cells induced with 1 μg/ml DOX for 2 and 4 days and normalized to the peak offset in cells cultured in the absence of DOX.
Recent reports suggest that expression of miR-shRNA and mRNA together in a monocistronic transcript can lead to both effective processing of the miR-shRNA and translation of protein from the mRNA (10, 11). To determine whether the pSLIK system would support conditional expression of a transgene together with a miR-shRNA, we constructed a pSLIK-Neo lentiviral vector expressing both GFP and the Gβ2 miR-shRNA from the TRE promoter (Fig. 4a). RAW cells were infected at MOI <1 and, after selection for a Neo-resistant population, treated with DOX for 9 days. As reported elsewhere (10), the presence of the intervening GFP sequence between the promoter and the miR-shRNA appeared to facilitate slightly more robust target gene knockdown, with almost complete depletion of Gβ2 throughout the 9 days of DOX induction, and this was accompanied by robust induction of GFP expression (Fig. 4b Left). Upon DOX removal, both Gβ2 knockdown and GFP expression were readily reversed (Fig. 4b Right). These results add significantly to the applicability of the pSLIK system by demonstrating the capacity for inducible knockdown of an endogenous target gene with simultaneous reexpression of a transgene.
Fig. 4.
Coupling of conditional miR-shRNA and transgene expression from pSLIK in RAW264.7 macrophages. (a) Schematic showing insertion of a GFP transgene in a pSLIK-Neo lentivirus encoding the Gβ2 miR-shRNA. (b) Western blot analysis demonstrates that conditional GFP expression is tightly correlated with conditional Gβ2 knockdown. Upon DOX removal, Gβ2 expression is restored. GFP expression is minimal after 2 days and is completely absent after 4 days.
Discussion
These data demonstrate a proof of principle for tightly regulated knockdown of multiple genes from a single infection of naïve mammalian cells. Previous efforts to combine all of the components of a regulated expression system into a single vector have met with mixed success, suffering from leaky expression (16) and poor in vivo transgene expression levels (17). Recently, an elegant study by Aebischer and colleagues (18) used the promiscuous transcriptional repression activity of the Kruppel-associated box domain to create a lentiviral vector that could promote conditional RNAi. Although versatile, their system does not support constitutive expression of a transduction marker while shRNA expression is in the repressed state, and the application of their system in multiple cell types is done at higher MOI, where multiple viral integration is likely. The high MOI also raises the possibility that the nonspecific Kruppel-associated box domain-mediated transcriptional repression could impact endogenous genes in the vicinity of the viral integration site(s). The pSLIK system supports constitutive expression of any transduction marker before induction of miR-shRNA expression, the Venus fluorescent protein, and Neo drug selection used in this study can be easily replaced with cell surface markers for cell sorting. We observe no evidence of leaky expression from the TRE promoter used in this study, but this does not discount the possibility that there is a low basal expression of miR-shRNA, which could be problematic with highly potent shRNA sequences. Replacement of the TRE promoter in the pEN_TmiRc2 entry vector with the TRE-tight promoter could address this concern. We believe that the rapid kinetics and potency of knockdown with pSLIK are supported by the nature of the vector topology, where TRE-driven rtTA3 expression in the presence of DOX creates a positive feedback loop driving higher rtTA3 (and presumably miR-shRNA) expression. Our use of an optimized rtTA3 variant may facilitate this effect (20, 21). Tissue-specific regulatable RNAi can also be accommodated in the pSLIK vector design by replacement of the Ubi-c promoter with a promoter that would restrict expression of the rtTA3 and marker gene to a particular cell lineage. Finally, the concomitant reexpression of a transgene, coupled to regulated miR-shRNA expression (Fig. 4), opens up an exciting possibility for gene therapy applications, whereby a mutant gene could be essentially replaced with an RNAi-resistant functional copy. This could have broad implications for both basic and applied research programs.
Methods
Plasmid Construction.
The U6 promoter and human mir30 sequence from pSM2 (generously provided by G. Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) was subcloned to the pENTR1A-Gent entry vector [American Type Culture Collection no. 10326362 and Alliance for Cell Signaling (AfCS) no. A84E0EN1AGXG]. A XhoI/EcoRI linker was inserted between the 5′ and 3′ mir30 sequences to restore the entire mir30 flanking sequence and to insert two BfuAI sites positioned to cut precisely at the mir30 junctions (Fig. 5b). The ccdB gene was cloned into a HindIII site between the BfuAI sites to create pEN_hUmiRc2. miR-shRNA sequences were designed by using the RNAi Codex algorithm (http://katahdin.cshl.org:9331/portal/scripts/main2.pl) and were cloned by linker annealing and ligation into BfuAI cut pEN_hUmiRc2 (Fig. 5c). The most potent miR-shRNAs identified for murine Gα12, Gα13, and Gβ2 targeted the sequences GCGACACCATCTTCGACAACAT, CTGGGTGAGTCTGTAAAGTATT, and TGCTCATGTATTCCCACGACAA, respectively. For pEN_hUmiRc2 vectors carrying effective miR-shRNAs, the hU6 promoter was excised by SalI digest and replaced with the TRE minimal promoter. For coupled conditional expression of a GFP transgene with a miR-shRNA (Fig. 4), the GFP cDNA sequence was cloned into a unique SacII site between the TRE promoter and the miR-shRNA sequence. For the lentiviral vectors, GFP was removed from the pL_UG vector (American Type Culture Collection no. 10326370 and AfCS no. L01GLUG001XA) and replaced with a linker containing several unique cloning sites. Cassettes were inserted containing rtTA3+IRES+Venus and rtTA3+IRES+Neo to create pSLIK-Venus and pSLIK-Neo, respectively. The optimized rtTA3 transactivator (rtTA S12G F86Y A209T) was generously provided by A. Das (University of Amsterdam, Amsterdam, The Netherlands). miR-shRNA-expressing pSLIK lentiviral vectors were generated by gateway recombination between the TRE-miR-shRNA entry vectors and the pSLIK destination vectors (Fig. 1a).
Consistent with the policy of the AfCS, all of the plasmid reagents described in this study have been made available to the research community through the American Type Culture Collection. Specific details are provided at the AfCS plasmid database (www.signaling-gateway.org/data/plasmid).
Cell Culture and Transfection.
HEK293 and HEK293T cells were maintained in DMEM, 10% FBS, and 2 mM glutamine. MEF cells were maintained in DMEM, 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. RAW264.7 cells were maintained in DMEM, 10% FBS, 20 mM Hepes, and 2 mM glutamine. For miR-shRNA testing, HEK293 cells (5 × 105) were seeded into six-well plates the day before transfection and transfected with 1.5 μg of YFP target gene expression plasmid and 0.5 μg of pEN_hU-miR-shRNA plasmid using Lipofectamine 2000 (Invitrogen, San Diego, CA). Two days after transfection, knockdown of the YFP-tagged target gene was assessed by YFP Western blot. For SRE-luciferase assay, MEF cells (7 × 104) were seeded into six-well plates the day before transfection. Cells were transfected with 1 μg of SRE-Luc (Stratagene, San Diego, CA) and 0.1 μg of pCsk lac Z plasmid using Lipofectamine 2000. After starvation for 18 h with DMEM containing 0.5% FBS, cells were treated with LPA and harvested for luciferase assays.
Lentivirus Production and Infection.
The pSLIK lentivector expression vector was transfected along with third-generation lentivirus packaging and pseudotyping plasmids (19) into HEK293T cells using Lipofectamine 2000 reagent (Invitrogen). HEK293T cells were cultured in DMEM (GIBCO Invitrogen), 10% FCS (Gemini), and 2 mM glutamine. Plasmids were cotransfected by using 10 μg of pSLIK plasmid, 7.5 μg of each of the two packaging plasmids pMDLg/pRRE and pRSVREV, and 5 μg of the vesicular stomatitis virus (VSV) G envelope plasmid pVSV diluted in Opti-MEM (Gibco Invitrogen). The medium was replaced after 12 h with Ultraculture medium (Cambrex, Baltimore, MD). The viral supernatant was collected 48 h after transfection and concentrated by using a Centricon Plus-70 filter unit (Millipore). Cells were infected at a low MOI to ensure <30% infection frequency such that the majority of transduced cells contained single viral integrants. Infection frequency was correlated with Venus expression assessed by FACS.
mRNA and Protein Expression Analysis.
Total RNA was isolated from cells by using RNeasy mini kits (Qiagen), and cDNA was prepared by using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was carried out as described (31). Sense and antisense amplification primers and probe primer sequences were as follows: Gα12, 5′-GAGGGTTCTTGTGGACGCTC-3′, 5′-AAACATCCCGTGCTTCTCGTT-3′, and 5′-FAM-CTCGGCATTCCCTGGCAGCACTCT-BHQ1–3′; Gα13, 5′-ACAAGTTGATGGCATTTGATACCC-3′, 5′-AGGCTCTGATAGCAGGAAGATACT-3′, and 5′-Texas red-TCGAGTCTCCACCATCCCCTGGGC-BHQ2–3′; Gβ2, 5′-ATGCGGGGATTCCACACTGA-3′, 5′-GGGTCCTCCGTGTTCTCATCT-3′, and 5′-FAM-TCGCCCCACTGGGTCCAGCCC-BHQ1–3′; β-actin reference, 5′-TCCATGAAATAAGTGGTTACAGGA-3′, 5′-CAGAAGCAATGCTGTCACCTT-3′, and 5′-HEX-TCCCTCACCCTCCCAAAAGCCACC-BHQ1–3′. rtTA3 and Neo mRNA was quantified by SYBR-Green incorporation using the following amplification primers: rtTA3, 5′-GCGAGTCATGGCAAGACTTTC-3′ and 5′-GAGCTGATTTTCCAGGGTTTCG-3′; Neo, 5′-TGGCTACCCGTGATATTGCTG-3′ and 5′-AAGGCGATAGAAGGCGATGC-3′. Isolation of cell protein lysates and Western blotting was carried out as described (31). The following antisera were used: anti-YFP (BD Clontech, catalog no. 8371-2), anti-Gα12 (Santa Cruz Biotechnology, catalog no. sc-409), anti-Gα13 (Santa Cruz Biotechnology, catalog no. sc-410), and anti-Gβ2 (Santa Cruz Biotechnology, catalog no. sc-380). Fluorescence intensity of Venus was assessed in WT and pSLIK-Venus-Gα12 transduced MEFs (10,000 cells) by FACS.
Luciferase Assay.
Luciferase assays were performed with a luciferase assay kit (Promega, Madison, WI), and the activity of cotransfected β-gal was measured with a β-gal assay kit (Roche, Palo Alto, CA). Transfection efficiency was corrected by the ratio of luciferase activity to β-gal activity in the same sample.
Measurement of Intracellular Ca2+ Mobilization.
RAW cells (6 × 104) were seeded into 96-well clear-bottom/black-wall plates (Corning, Frederick, CO) 24 h before assay. Cells were incubated with 4 μM fura-2/AM in Hanks’ balanced salt solution, 0.5% BSA, and 2.5 mM Probenecid (pH 7.45) (HBP) for 30 min at room temperature and then washed twice with HBP, and the volume was replaced after final wash. After 30 min of incubation at 37°C, Ca2+ mobilization in response to 10 nM C5a (Sigma Aldrich, catalog no. C5788) was assessed by using a FlexStation (Molecular Devices, Sunnyvale, CA).
Supplementary Material
Acknowledgments
We thank colleagues in the AfCS for criticism and insight during the course of this study. We thank Greg Hannon for the pSM2 plasmid, provision of manuscripts before publication, and helpful discussions. This work was supported by contributions from public and private sources, including the National Institute of General Medical Sciences Glue Grant Initiative (U54 GM062114). K.-J.S. and J.-I.H. were supported by National Institutes of Health Grant R37 GM034236 (to M.I.S.).
Abbreviations
- miR
microRNA
- shRNA
short hairpin RNA
- Tet
tetracycline
- rtTA
reverse Tet transactivator
- DOX
doxycycline
- MEF
mouse embryonic fibroblast
- Neo
neomycin
- TRE
Tet response element
- SRE
serum response element
- LPA
lysophosphatidic acid
- MOI
multiplicity of infection
- pol
RNA polymerase
- AfCS
Alliance for Cell Signaling
- pSLIK
single lentivector for inducible knockdown.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ, Rossi JJ. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 4.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul CP, Good PD, Winer I, Engelke DR. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 6.Fish RJ, Kruithof EK. BMC Mol Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng Y, Cullen BR. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 10.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 12.Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 13.Samakoglu S, Lisowski L, Budak-Alpdogan T, Usachenko Y, Acuto S, Di Marzo R, Maggio A, Zhu P, Tisdale JF, Riviere I, Sadelain M. Nat Biotechnol. 2006;24:89–94. doi: 10.1038/nbt1176. [DOI] [PubMed] [Google Scholar]
- 14.Hwang JI, Fraser ID, Choi S, Qin XF, Simon MI. Proc Natl Acad Sci USA. 2004;101:488–493. doi: 10.1073/pnas.0307549100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang JI, Choi S, Fraser ID, Chang MS, Simon MI. Proc Natl Acad Sci USA. 2005;102:9493–9498. doi: 10.1073/pnas.0503503102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafri T, van Praag H, Gage FH, Verma IM. Mol Ther. 2000;1:516–521. doi: 10.1006/mthe.2000.0083. [DOI] [PubMed] [Google Scholar]
- 17.Vogel R, Amar L, Thi AD, Saillour P, Mallet J. Hum Gene Ther. 2004;15:157–165. doi: 10.1089/104303404772679968. [DOI] [PubMed] [Google Scholar]
- 18.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 19.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das AT, Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Berkhout B. J Biol Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 21.Markusic D, Oude-Elferink R, Das AT, Berkhout B, Seppen J. Nucleic Acids Res. 2005;33:e63. doi: 10.1093/nar/gni062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riobo NA, Manning DR. Trends Pharmacol Sci. 2005;26:146–154. doi: 10.1016/j.tips.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Gohla A, Offermanns S, Wilkie TM, Schultz G. J Biol Chem. 1999;274:17901–17907. doi: 10.1074/jbc.274.25.17901. [DOI] [PubMed] [Google Scholar]
- 24.Mao J, Yuan H, Xie W, Simon MI, Wu D. J Biol Chem. 1998;273:27118–27123. doi: 10.1074/jbc.273.42.27118. [DOI] [PubMed] [Google Scholar]
- 25.Kim VN. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 26.Fromm C, Coso OA, Montaner S, Xu N, Gutkind JS. Proc Natl Acad Sci USA. 1997;94:10098–10103. doi: 10.1073/pnas.94.19.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 28.Gohla A, Harhammer R, Schultz G. J Biol Chem. 1998;273:4653–4659. doi: 10.1074/jbc.273.8.4653. [DOI] [PubMed] [Google Scholar]
- 29.Kabarowski JH, Feramisco JD, Le LQ, Gu JL, Luoh SW, Simon MI, Witte ON. Proc Natl Acad Sci USA. 2000;97:12109–12114. doi: 10.1073/pnas.97.22.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerard NP, Gerard C. Nature. 1991;349:614–617. doi: 10.1038/349614a0. [DOI] [PubMed] [Google Scholar]
- 31.Fraser ID, Liu W, Rebres R, Roach T, Zavzavadjian J, Santat L, Liu J, Wall E, Mumby M. Methods Mol Biol. 2006 doi: 10.1385/1-59745-267-X:261. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.