Abstract
In this article we report the in vivo and in vitro characterization of single chain tetracycline repressor (scTetR) variants in Escherichia coli. ScTetR is genetically and proteolytically stable and exhibits the same regulatory properties as dimeric TetR in E.coli. Urea-dependent denaturation of scTetR is independent of the protein concentration and follows the two-state model with a monophasic transition. Contrary to dimeric TetR, scTetR allows the construction of scTetR mutants, in which one subunit contains a defective inducer binding site while the other is functional. We have used this approach to establish that scTetR needs occupation of both inducer binding sites for in vivo and in vitro induction. Single mutations causing loss of induction in dimeric TetR lead to non-inducible scTetR when inserted into one half-side. The construction of scTetR H64K S135L S138I (scTetRi2) in which one half-side is specific for 4-dedimethylamino-anhydrotetracycline (4-ddma-atc) and the other for tetracycline (tc) leads to a protein which is only inducible by the mixture of tc and 4-ddma-atc. Fluorescence titration of scTetRi2 with both inducers revealed distinct occupancy with each of these inducers yielding roughly a 1:1 stoichiometry of each inducer per scTetRi2. The properties of this gain of function mutant clearly demonstrate that scTetR requires the binding of two inducers for induction of transcription.
INTRODUCTION
Gene regulation in bacteria occurs mostly at the level of transcription mediated by DNA binding proteins like activators or repressors (1). Most of them are homodimers, as was demonstrated for members of the LacI/GalR, TetR/CamR, FadR, MerR or LysR families (2–4). They exert their regulatory function by binding effectors upon which a conformational change is triggered which allosterically affects DNA binding.
The bacterial tetracycline (tc) responsive repressor, TetR, exhibits high affinity (5.6 ± 2 × 109 M−1) for the tet operator (tetO) and is very sensitive to induction by tc and its analogues (5–10). These features combined with the capacity of tc to penetrate cell membranes by passive diffusion have led to its wide use as a tool for gene regulation in prokaryotes and eukaryotes (11–14). Crystal structures of TetR revealed that two identical subunits each consisting of 10 α-helices form the active homodimer. Two DNA reading heads are connected to a core domain in which dimerization takes place and which harbours two inducer binding pockets. The [tc–Mg]+ complex binds into each pocket and triggers a movement of helix 6 which is transferred via helices 4 and 1 to the DNA binding heads (9,15) moving them apart by ∼3 Å. As a consequence, the affinity to the tetO is lowered by four orders of magnitude (16) and TetR leaves the DNA.
When bound to TetR, each [tc–Mg]+ complex is contacted by 13 residues from 1 monomer and 4 from the other. Since one inducer contacts residues from both subunits it is conceivable that the allosteric change may be triggered by binding of a single tc molecule. There is only very little information available regarding the correlation between the number of occupied inducer binding sites necessary for induction in regulatory proteins. This has been discussed for MerR, a member of the MerR/SoxR family (17) involved in the regulation of bacterial mercury resistance. Preliminary data would be in agreement with induction by binding of one Hg+ to the MerR dimer (18,19). The multi-drug resistance repressor QacR is, like TetR, a member of the TetR/CamR family of regulators (20). In vitro binding studies revealed a 1:2 stoichiometry of inducer per QacR dimer, and it has been suggested that this is sufficient for induction in vivo (21). Addressing this question in vivo is generally hampered by the exchange of subunits in a dimer as has been clearly demonstrated for TetR (7). Thus, a strain containing a wild type and an induction deficient allele of tetR would contain a mixture of all three possible dimers.
We describe here single chain TetR (scTetR) variants, in which the two subunits of TetR are linked by a 25 amino acids (SG4)5 sequence and cannot form heterodimers (22), and their use to construct and analyze TetR variants with mutations in just one half-side in Escherichia coli. We establish clearly, that TetR needs to bind two inducers to accomplish induction.
MATERIALS AND METHODS
General materials and methods
Anhydrotetracycline (atc) was purchased from Acros (Geel, Belgium). All other chemicals were from Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany) or Sigma (Munich, Germany) at the highest purity available. Enzymes for DNA restriction and modification were from New England Biolabs (Frankfurt/Main, Germany) or Roche (Mannheim, Germany). Isolation and manipulation of DNA as well as strain transformation was performed as described previously (23). All plasmids constructed were sequenced between the restriction sites employed for cloning. Oligonucleotides for PCR and sequencing were obtained from MWG Biotech (Ebersberg, Germany) unless stated otherwise. Sequencing was carried out according to the protocol provided by Applied Biosystems for cycle sequencing and analyzed with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany). For β-galactosidase (β-gal) assays, E.coli WH207λtet50 bearing a Tn10 tetA–lacZ trancriptional fusion was used. Cells were grown in Luria–Bertani (LB) medium supplemented with the required antibiotics to an OD600 of 0.4. β-gal activity was determined as published previously (24).
Culture and growth conditions
E.coli was generally grown in LB medium at 37°C. Antibiotics for selection were added to the following final concentrations: Ampicillin (Ap) 100 mg/l, Kanamycin (Kan) 30 mg/l. When necessary, the cultures were adjusted to final concentrations of atc, tc or 4-dedimethylamino-anhydrotetracycline (4-ddma-atc) as indicated. All atc and 4-ddma-atc solutions were protected from light. Tc and atc were dissolved in 70% ethanol, 4-ddma-atc was solved in dimethyl sulfoxide.
Design and construction of genes and plasmids
Components for construction of scTetR
The following DNA fragments were already available: wt-tetR(BD) and a (SG4)5 encoding linker sequence together with the first 50 codons of the synthetic tetR(B) sequence (termed tetR(sB) originationg from sctetR(BsB) (22). TetR(BD) is a chimera consisting of the first 50 codons of tetR(B) fused to the last 158 codons of tetR(D).
We designed the DNA sequence of the synthetic class (D) portion de novo by in silico back-translation of the primary protein sequence online at http://www.entelechon.com/eng/backtranslation.html. Settings were attuned to human codon usage and putative eukaryotic splicing-sites were eliminated. For use in E.coli rare arginine codons were silently exchanged (as found at www.kazusa.or.jp/codon). Care was taken that the length of identical DNA stretches to tetR(BD) did not exceed 20 bp to minimize recombination events (25). The final sequence of sctetR(BDsBsD) was deposited in the NCBI GenBank (accession no. DQ 392985). A total of 20 oligonucleotides, 39–48 bp in length (MWG Biotech, Ebersberg, Germany), entirely covering both strands of the desired ‘tetR(D) gene fragment (471 bp in size), with an overlap of 12 bp to complementary oligos were used. Assembling was done in a PCR-like method, as described previously (26). A DNA-sequence alignment of the gene portions ‘tetR(D)’ and ‘tetR(sD)’ displayed an identity of ∼76%. The assembled product was amplified with primers syn(D)c_fw1 (5′-ctagcagtcgagatactggcccggcaccatgactacagt-3′) and syn(D)c_rev10 (5′-catggcctcacacaatctgaaggagggctgtaagctgga-3′) and temporarily subcloned into pCR™2.1-Topo™ (Invitrogen, Karlsruhe, Germany) as recommended by the manufacturer.
Construction of scTetR(BDsBsD)
Construction of sctetR(BD) was performed in pWH1926 (like pWH1925 (6), however with tetR(BD) running in opposite direction). The 3′ end of tetR(BD) was modified by silently inserting a ClaI restriction site upstream of the stop-codon (PCR primers were sc0_fw and sc0_rev, restriction was done with MluI and NcoI).
After amplification and modification by PCR using primers sc1_fw (5′-gtgaaagtatcgattcaggaggcggtgg-3′) and sc1_rev (5′-gtatgatccatggccagcatctcgattgctagcgcatcgag-3′), the (SG4)5 linker sequence together with the first 50 codons of the synthetic tetR(sB) sequence was fused to the 3′ end of tetR(BD) via ClaI and NcoI, simultaneously inserting a new NheI site directly in front of NcoI. The tetR(sD) portion of the pCRO™2.1-TopoO™ derivative described above was finally inserted as an NheI/NcoI fragment into the likewise cut pWH1926 derivative, resulting in a full-length sctetR(BDsBsD) gene, which gave rise to pWH1926(BDsBsD). The gene was entirely sequenced on both directions. Finally sctetR(BDsBsD) was cloned into a pWH1925 derivative to enable exact comparison with other variants, most of which are encoded on this vector. All tetR and sctetR variants were cloned and analyzed as pWH1925 derivatives. For overexpression of wild-type scTetR, the synthetic part was set in front of the gene. For the sake of convenience all tetR(BD) genes and proteins are termed tetR or TetR, while sctetR(BDsBsD) and the corresponding protein are denoted sctetR or scTetR, respectively.
Lysozyme tratment of cells and western blot analysis
Cells from a 20 ml culture were harvested at an OD600 of 0.4 by centrifugation at 8000 r.p.m. at 4°C and the pellet was resuspended in 500 μl buffer containing 10 mM Tris–HCl (pH 8.0), 200 mM NaCl, 0.4 mM EDTA and 20 g/l lysozyme. The lysate was incubated at ambient temperatures for 60 min followed by centrifugation at 13 000 r.p.m., 4°C for 60 min. The protein concentration of the soluble fraction was determined using the Biorad Assay (Bio-Rad Laboratories, Munich, Germany). TetR was detected by SDS–PAGE of 35 μg E.coli cell extract on a 10% polyacrylamide gel. Proteins were transferred by electroblotting onto a PhotoGeneO™ nylon membrane (GibcoBRL, Karlsruhe, Germany). Further steps were performed as described previously (27).
Overexpression and purification of proteins
For overexpression we used the plasmid pWH610 and the strain RB791. Proteins were purified as described previously (28) except scTetRi2, which was overproduced at 22°C. After harvesting, cell pellets were stored on ice instead of freezing. The temperature of all subsequent steps was kept below 8°C.
Electrophoretic mobility shift assays and fluorescence measurements
For electrophoretic mobility shift assays (EMSA), the complementary synthetic 40 bases tetO containing oligonucleotides designated tetO1 and tetO2 were hybridized. Equimolar amounts of each oligonucleotide were mixed in water, heated at 96°C for 5 min and allowed to cool to room temperature within 2 h. A double stranded oligonucleotide of the same size without palindromic sequence was used as a control. TetR proteins were added at indicated amounts. All samples were incubated in a complex buffer containing 0.02 M Tris–HCl (pH 8.0) and 5 mM MgCl2. Atc and 4-ddma-atc were added to final concentrations of 0.1 mM to the sample. After incubation for 10 min at ambient temperatures, the DNA was electrophoresed on an 8% polyacrylamide gel at 50 V in TBM buffer (0.09 M Tris, 0.09 M boric acid and 5 mM MgCl2), and the DNA was detected by ethidium bromide staining. Fluorescence titrations were performed as described previously (16) with excitation/emission wavelengths of 370/515 nm to observe tc- and 420/540 nm to observe 4-ddma-atc-binding.
Denaturation of scTetR
All measurements were performed in F-buffer [100 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA and 1 mM dithiothreitol]. Equilibrium denaturation was carried out by incubation of protein samples overnight at given urea concentrations. Protein samples which had been incubated for 3 h at 20°C in F-buffer containing 8 M urea, were renaturated by dilution them 200-fold in F-buffer without urea. All measurements were performed at 25°C. Fluorescence intensities were measured at an excitation wavelength of 280 and 295 nm, respectively, emission intensity was recorded at 324 nm and corrected by the substraction of the buffer fluorescence.
Thermodynamic calculations
For scTetR, a two-state model of denaturation was applied in which a native monomeric protein N exists in equilibrium with a denatured monomer, U:
| 1 |
The equilibrium constant Ku for every measured point within the linear transition was calculated according to two step denaturation of a monomeric protein, in which fu is the fraction of unfolded protein.
| 2 |
The linear extrapolation method (33) was used to derive protein stability at zero urea concentration.
| 3 |
where stands for Gibbs free energy of unfolding at a 1 M concentration of all reactants, (H2O) is the extrapolated Gibbs free energy in buffer without urea and m represents the slope of the straight line in the plot of ΔGu versus denaturant concentration. At the midpoint of the unfolding transition at urea1/2, ΔGo = 0 and Ku = 1. To determine urea1/2, we rearranged Equation 3 to Equation 4.
| 4 |
RESULTS
Construction and in vivo stability of scTetR
The scTetR variant was constructed as described for the mammalian applications (22) except that a sequence variant with optimal regulatory properties in bacteria was used (see Figure 1). Since the in vivo properties of scTetR with mutations in one inducer binding pocket may be masked by small amounts of dimeric TetR which could result from recombination of the repeat tetR sequence or from proteolysis within the peptide linker, it was important to exclude such events. To minimize recombination, we constructed a synthetic tetR allele (see Materials and Methods for details) so that the DNA sequence identity is only ∼76%. Identical sequences longer than 20 bp were avoided to further minimize recombination. The genetic stability of the resulting sctetR in E.coli DH5α harbouring pWH1925sctetR was determined by growing at 37°C over night on LB/Ap plates. Colonies were streaked out for three successive times and then used to inoculate a liquid culture. Plasmids isolated from these cultures gave restriction patterns indicating the presence of only the full-length sctetR which was also confirmed by sequencing (data not shown).
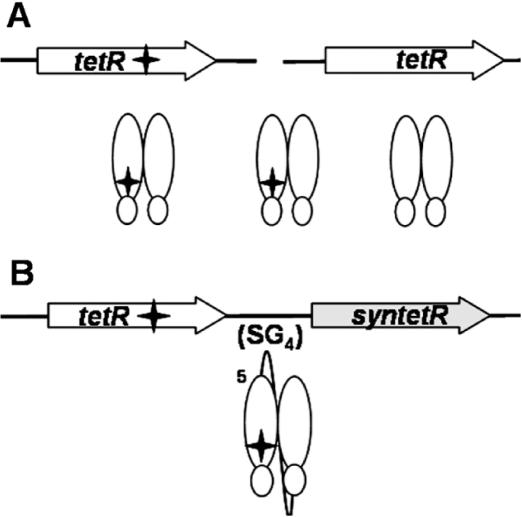
Figure 1.
Schematic presentation of dimeric and scTetR mutants. The protein arrangements possible with different genetic situations are shown. (A) The TetR dimers arising from two different tetR alleles expressed in the same cell. The filled star indicates a mutation. The three possible dimers are two homodimers and one heterodimer. (B) The scTetR gene (sctetR) in which one half-side carries the mutation (filled star). The first tetR sequence (white arrow) contains the desired mutation (filled star), followed by the linker encoding sequence designated (SG4)5 and the second synthetic tetR sequence (grey arrow). The resulting monomeric protein contains the mutation (filled star) in one half-side.
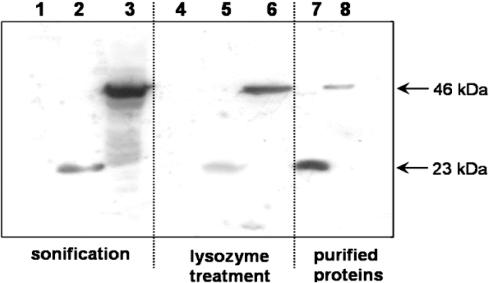
The proteolytic stability of scTetR was examined by western blots. As presented in Figure 2, a single band of TetR at ∼23 kDa is observed in extracts from cells containing tetR after sonification or lysozyme treatment (lanes 2 and 5). The scTetR band appears at ∼46 kDa in the extract from the respective strain containing sctetR (lanes 3 and 6). A smear of additional bands indicating fragmentation is visible below the scTetR signal when cells are broken by sonification (lane 3), while these are absent when cells are treated with lysozyme (lane 6). Taken together, these results indicate that scTetR exhibits sufficient genetic and proteolytic stability for functional analysis in vivo.
Figure 2.
Western blots of TetR and scTetR. The influence of different cell disruption methods on the integrity of TetR proteins is shown. Soluble proteins from crude cell extracts (35 μg) were loaded in lanes 1–6. Proteins from E.coli WH207λtet50 cells carrying pWH1925ΔtetR were loaded in lanes 1 and 4; proteins from cells carrying pWH1925 (tetR) were loaded in lanes 2 and 5, and cells carrying pWH1925sc (sctetR) were loaded in lanes 3 and 6. Lane 7 contains 60 ng of purified TetR, and lane 8 contains 20 ng of purified scTetR. The lysis methods employed are indicated below the respective lanes.
ScTetR with a single inducer binding site is induction deficient
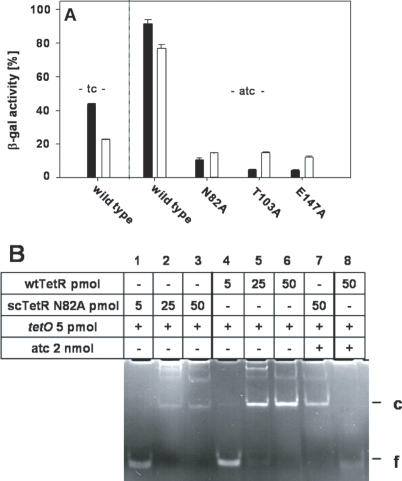
We constructed three scTetR variants, each with the single mutation N82A or T103A or E147A in only one half-side. The residues N82, T103 and E147 in TetR are essential for tc and atc binding, and each mutation to A leads to induction deficiency of the respective TetR mutant (29,30). The encoded scTetR proteins will thus contain one deficient and one functional inducer binding pocket (see Figure 1). The inducibility of these half-side scTetR mutants was determined by β-gal measurements with and without tc or atc (Figure 3A). β-gal expression in the absence of tetR defines the 100% level (data not shown). TetR and all single chain variants repress lacZ in the absence of inducer to <1% (data not shown). Tc leads to 45 and 22%, while atc leads to 92 and 78% induction of TetR and scTetR, respectively. The three TetR variants carrying mutations in both subunits (black bars) are only weakly inducible with the more efficient inducer atc, resulting in β-gal activities between 9 and 4%, while the corresponding single chain half-side mutants are inducible to ∼12%. All three TetR and scTetR half-side mutants are not inducible with tc (values <1% β-gal activity; not shown). Thus, the inactivation of one inducer binding site in scTetR reduces induction to about the same level as the elimination of both inducer binding sites.
Figure 3.
Induction efficiencies of sctetR with mutations in one inducer binding pockets. (A) β−gal activities of E.coli WH207λtet50 transformed with plasmids expressing tetR (black columns) or sctetR (white columns) with mutations causing induction deficiency in tetR (designated N82A, T103A or E147A) in one half-side are shown. The β−gal activity in the absence of tetR was ∼8000 Miller Units and was set to 100% (data not shown). β−gal activities in the absence of inducer were <1% for all variants (data not shown). β−gal expression in the presence of inducers is shown for 0.4 μM tc and 0.4 μM atc as indicated in the figure. (B) EMSA with purified TetR and scTetR N82A carrying a mutation in one inducer binding pocket are shown in the insert. The compounds and the amounts present in the respective reaction mixtures are listed in the table above the lanes. The positions of bands corresponding to free (f) and complexed (c) DNA are indicated on the right side.
To analyse TetR- and scTetR-tetO interactions in vitro, we purified scTetR N82A and performed EMSA (Figure 3B). This mutant binds tetO similar to dimeric TetR (compare lanes 1–3 with lanes 4–6) in the absence of effector, where a 5-fold molar excess of protein is needed for complete binding of tetO. ScTetR N82A is not released from tetO upon addition of atc, since no band of free tetO DNA is visible in lane 7, while dimeric wild-type TetR is efficiently removed from tetO by atc, and the free operator DNA appears in lane 8. Neither dimer nor scTetR bind a control fragment which contains no tetO sequence (data not shown). We conclude that the in vitro results are consistent with the non-inducible phenotype of scTetR with mutations in one inducer binding pocket.
ScTetR variants with different half-side inducer specificities
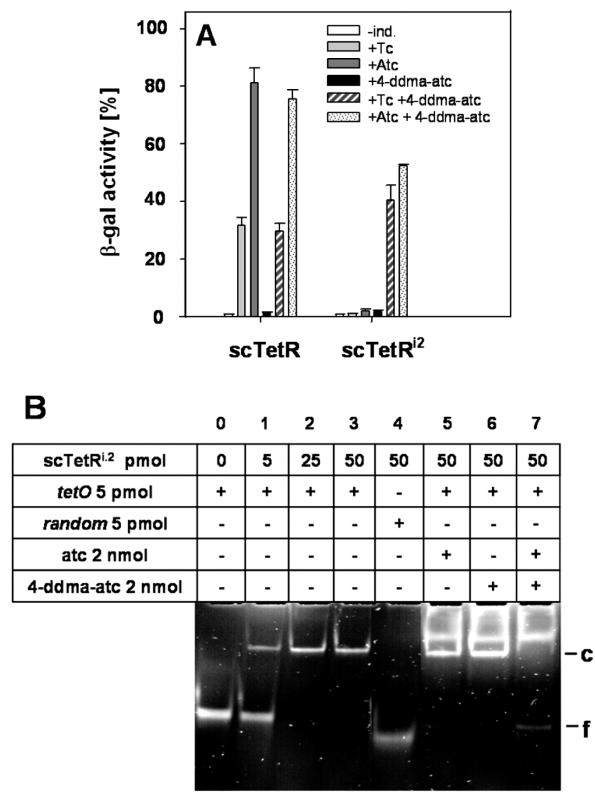
In order to investigate whether the lack of inducibility can be restored, we constructed a scTetR variant with different inducer specificities in each half-side. The TetRi2 mutant H64K S135L S138I is specifically induced by 4-ddma-atc and not by tc or atc (31). We introduced these mutations into one half-side of scTetR and expected the resulting variant to harbour two distinct inducer specificities combined in one protein called scTetRi2. The regulatory properties of this mutant were determined by β-gal measurements with tc, atc, 4-ddma-atc and combinations of them (Figure 4A). β-gal expression in the absence of tetR defines the 100% level (data not shown). ScTetR is not inducible with 4-ddma-atc, while β-gal activities in the presence of tc and atc are ∼35 and 80%, respectively, regardless of the presence of 4-ddma-atc. ScTetRi2 is not inducible with tc, atc or 4-ddma-atc alone. Induction of 40% β-gal expression is observed in the presence of tc and 4-ddma-atc, and induction of 55% in the presence of atc and 4-ddma-atc. Although the β-gal activities are not as high as those of scTetR in the presence of atc, scTetRi2 is apparently induced in the presence of both effectors. This is in agreement with in vitro results obtained by EMSA (Figure 4B). ScTetRi2 binds to tetO in the absence of inducer (lanes 1–3), whereas the control DNA is not bound (lane 4). The bulk of tetO remains bound in the presence of atc or 4-ddma-atc, as indicated by the strong complex band, and only a small amount of free DNA is visible in the presence of atc (lane 5). No band representing the complexed DNA is visible in the presence of atc and 4-ddma-atc, and the band corresponding to free tetO is more intense (lane 7). The free operator DNA appears in lane 0. Bands migrating above the complex appear only in lanes loaded with samples containing inducer and scTetR. Their fluorescence exhibits a different colour than the DNA stained with ethidium bromide and they are visible in control lanes where only scTetR and inducer without DNA have been loaded (data not shown). We conclude that they originate from the scTetR-inducer complex and visualize the well-known tetracycline fluorescence which is enhanced in complex with TetR. These results clearly indicate that two occupied binding pockets are required for induction.
Figure 4.
Induction efficiencies of scTetR harbouring different inducer specificities. (A) β−gal activities of E.coli WH207λtet50 transformed with plasmids expressing sctetR or sctetRi2 (H64K, S135L, S138I in one half-side) are depicted. The β−gal activity in the absence of tetR is ∼8000 Miller Units and was set to 100% (not shown). Measurements were performed in the absence and presence of 0.4 μM of each inducer as indicated in the figure. (B) EMSA were performed with purified scTetRi2 and the compounds and amounts indicated in the table above the lanes. ‘Random’ refers to a non-tetO containing DNA. The positions of bands corresponding to free (f) and complexed (c) DNA are indicated on the right side.
Binding pockets with different specificities are bound only by their cognate inducers
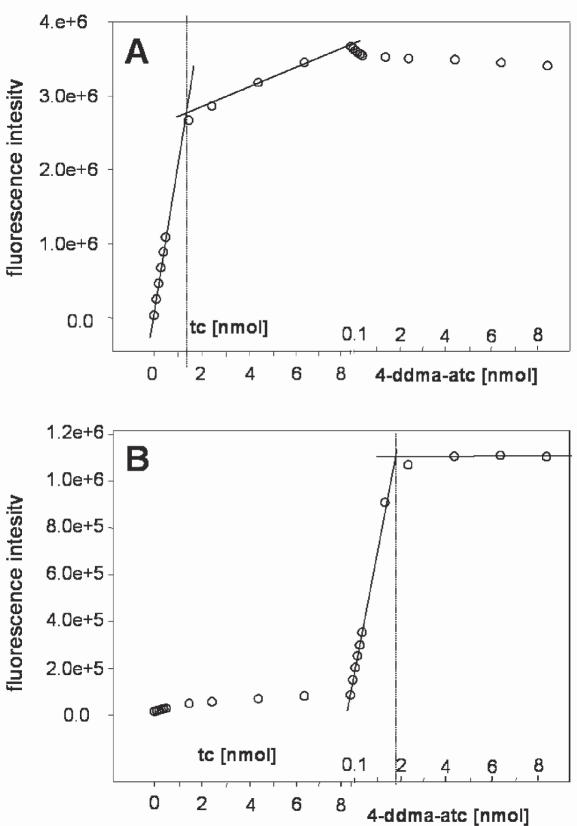
Distinct occupancy of the effector-specific binding pockets with the respective inducers was assayed by fluorescence titration of purified scTetRi2 first with tc and subsequently with 4-ddma-atc in the same cuvette (Figure 5A and B). As shown in Figure 5A, tc binding to TetR can clearly be detected based upon the excitation/emission wavelengths specific for tc. The subsequent addition of 4-ddma-atc leads to only a small further change of fluorescence intensity. An increase in fluorescence cannot be detected upon addition of tc when observed at excitation/emission wavelengths specific for 4-ddma-atc (Figure 5B), although tc binds to the first wild-type binding pocket (cf. Figure 5A). Thus, the increase in fluorescence during the following addition of 4-ddma-atc must result from binding of 4-ddma-atc in its respective binding pocket. We analyzed a concentration of 2 nmol of protein harbouring 4 nmol of binding pockets. As shown in Figure 5, the equivalence point occurs at 1.3 nmol of tc and 1.8 nmol of 4-ddma-atc. Thus, roughly half of the available inducer binding sites are occupied by each of the two effectors.
Figure 5.
Titration of scTetRi2 with tc and 4-ddma-atc. (A) The fluorescence intensity of 2 nmol of purified scTetRi2 titrated with up to 8.5 nmol of tc (corresponding to a concentration of 8.5 μM) followed by titration with up to 8.5 nmol of 4-ddma-atc (corresponding to a concentration of 8.5 μM) is shown. Since fluorescence was excited at 370 nm and emission was measured at 515 nm only the binding of tc to the protein is detected. (B) The same experiment except that the excitation and emission wavelengths were 420 and 540 nm, repectively, where only 4-ddma-atc binding is observed. The equivalence points are indicated by the dotted lines.
ScTetR stability
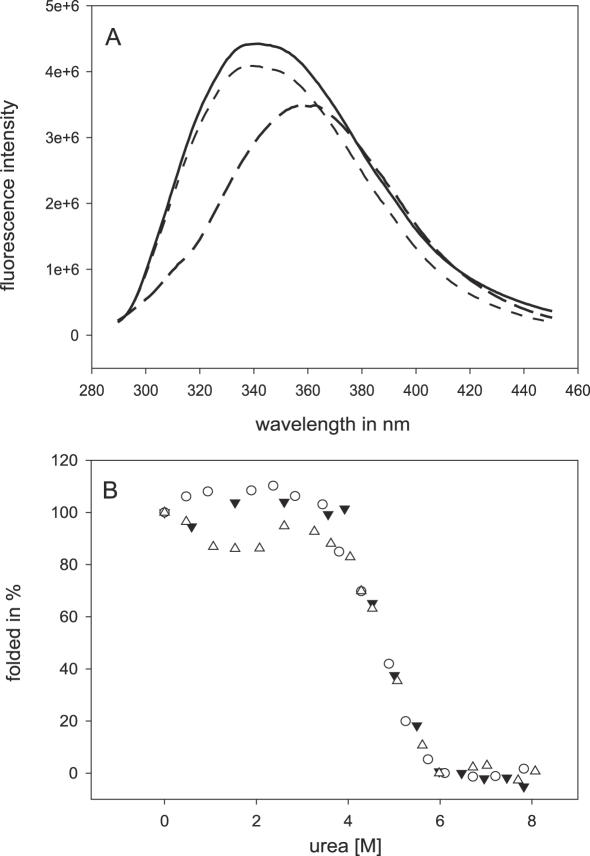
A bimolecular single transition denaturation reaction has been shown for dimeric TetR (32,33), which was the basis for the two-state model of unfolding. We investigated urea-dependent denaturation of scTetR. Conformational changes during denaturation were monitored by the fluorescence of W43 and W75, located in the helix–turn–helix motif and in the core domain, respectively. The fluorescence quantum yield decreases in dependence of the urea concentration, and the emission maximum is shifted from 342 to 363 nm upon denaturation. Fluorescence emission was determined at 324 nm where the highest change in fluorescence occurs (Figure 6A). The urea-dependent unfolding of scTetR shows a sigmoidal, monophasic decrease in fluorescence which is independent of the scTetR concentration (Figure 6B). Thus, there is no indication for any stable intermediate, indicating that scTetR also denatures in an all or none process.
Figure 6.
Urea-dependent denaturation of scTetR. (A) Fluorescence spectra of native (solid line), denatured (long-dashed line) and renatured (dashed–dotted line) scTetR are shown. The difference spectrum between native and denatured forms is shown by the dotted line. (B) The figure shows the change of fluorescence in dependence of the urea concentration. The fluorescence in the absence of urea was set to 100% of folded protein. The denaturation curves were determined at different concentrations of scTetR (filled triangle, 5 μM; circle, 1 μM; open triangle, 0.4 μM, scTetR).
The midpoint of transition is not dependent on the protein concentrations between 0.4 and 5 μM of scTetR (Table 1), and the determination of urea1/2 (Equation 4) concentrations yields values of 4.7, 4.7 and 4.8 M urea for 0.4, 1 and 5 μM scTetR, respectively (Table 1). Since the transition is not dependent on protein concentration, Gibbs free energy was calculated employing Equation 2. The (H2O) values determined by Equation 3 for each scTetR concentration are identical within the standard deviations and range from 26 to 27 kJ/mol (Table 1).
Table 1.
Thermodynamic stability of scTetR and TetR(D) in urea-dependent denaturation determined by fluorescencea
| 0.4 μM | 1 μM | 5 μM | ||||
|---|---|---|---|---|---|---|
| (kJ/mol) | Urea1/2 (M) | (kJ/mol) | Urea1/2 (M) | (kJ/mol) | Urea1/2 (M) | |
| scTetR | 26 ± 1.1 | 4.7 | 27 ± 3.8 | 4.7 | 26 ± 0.8 | 4.8 |
| TetR(D)b | 60 ± 3 | 3.8 | n.d. | n.d. | 61 | 4.2 |
n.d., not determined.
aUnfolding was followed by the change of the fluorescence signal at 330 nm using protein concentrations of 0.1, 1 (excitation at 280 nm) and 5 μM (excitation at 295 nm) in 1 cm cells.
bValues taken from Schubert et al.(33).
DISCUSSION
We have constructed a bacterial scTetR and characterized the modified protein in vivo and in vitro. Since TetR mutations always occur in each subunit of the native dimer, we have not been able up to now to determine whether induction requires occupation of one or both inducer binding pockets. We have overcome this limitation by ‘monomerizing’ TetR to scTetR (Figure 1). The sctetR gene is genetically as stable and the encoded protein is as functional in E.coli as dimeric TetR. Apparently, the human codon usage implemented into the synthetic part of sctetR did not notably reduce the translational efficiency in E.coli. It is surprising that this is possible with the (SG4)5 linker without any loss of activity. In fact, we consistently observed a slightly increased repression exerted by scTetR compared with the dimer (data not shown), and induction is slightly impaired, which is probably due to an increased amount of intracellular scTetR compared with dimeric TetR (Figure 2). Another reason could be the de facto duplication of the tetR gene dosis in sctetR. Although the linker length was chosen to be long and flexible enough to allow assembly of both domains properly (22), it could influence the entrance of inducer into the binding pocket. However, increased regulatory effects have also been observed when similar sctetR constructs were used in eukaryotes (22). Other dimeric repressors, like lambda Cro, P22 Arc or the N-terminal domains of the bacteriophage 434 repressor cI have been successfully modified such that they are also expressed as a single chain protein to investigate their stability and DNA binding (34–36). However, the allosterical conformational change occurring upon effector binding has to our knowledge not yet been analyzed in a transcriptional regulator.
To clarify if one or two occupied binding pockets are necessary for induction, we introduced three different mutations conferring a non-inducible phenotype owing to lack of inducer binding into one half-side of scTetR. The respective dimeric TetR mutants exhibit a massive drop of inducer affinity (29,30). We assume that they exert a similar influence on the respective scTetR half-side containing the alteration. Indeed, half-side induction deficient mutants are efficient non-inducible repressors as demonstrated in vivo and in vitro (Figure 3). While the tetO binding clearly underscores that the half-side mutant proteins are functional, the lack of induction is nevertheless a negative result, and could thus be attributed to local folding problems around the inducer binding pocket.
To obtain a gain of function mutation we have examined the inducibility of scTetR with two different effector specificities harbored in both half-sides. The TetRi2 protein shows specificity for 4-ddma-atc (5) in one half-side and for tc or atc in the other. As a result, scTetRi2 is only inducible by mixtures of tc/4-ddma-atc or atc/4-ddma-atc. This clearly points out that (i) both binding pockets need to be occupied with suitable inducers to lead to the allosteric conformational change necessary for induction and (ii) both half-sides are folded properly and are active. EMSA and tc- and 4-ddma-atc-dependent titrations verified the distinct occupancy of the two binding pockets by the suitable inducer as well as a 1:2 stoichiometry for each of them. Furthermore, the titrations demonstrate that the respective specificities of the binding pockets, although each binding pocket is formed by residues of both monomers, are totally independent on its counterpart's sequence and specificity (Figure 5).
Despite the participation of residues of both monomers in binding of one inducer, it is not likely in the light of the crystal structure (9) that the second monomer undergoes major conformational changes when the [tc–Mg]+ complex binds to the pocket mainly formed by the other monomer. Hence, a suitable movement of both binding heads requires the allosteric process to occur in both half-sides. It is shown here that this requires occupation of both binding pockets. It is not clear whether induction by different effectors in both half-sides differs from that of wild-type TetR. As shown for another homodimeric repressor, LacI (37), an asymmetric mechanism of induction could be possible.
The TetR family of bacterial regulators is mainly defined by sequence similarities in the DNA reading heads (20). Hence it is conceivable that members of this diverse family may follow different induction modes. This seems to be the case for TetR requiring two inducers bound per dimer and QacR, for which one inducer bound per dimer seems to be sufficient for induction. One possibility would be that TetR contacts tetO more strongly than QacR binds its respective cognate DNA. In addition, there is a very remarkable difference in DNA binding between the two repressors, as QacR uses a tetramer to bind its operator (21). Hence, there might also be a very different allosteric process underlying induction. Given the lack of sequence similarity within the inducer binding regions of the TetR family proteins, there might be a number of more different induction pathways present in this family.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Christian Berens, Oliver Scholz and Martin Köstner for helpful discussions and Irina Pimenta for titration experiments. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 473 and the Fonds der Chemischen Industrie. Funding to pay the Open Access publication charges for this article was provided by SFB 473.
Conflict of interest statement. None declared.
REFERENCES
- 1.Browning D.F., Busby S.J. The regulation of bacterial transcription initiation. Nature Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 2.Huffman J.L., Brennan R.G. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 3.Zaim J., Kierzek A.M. The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 2003;31:1444–1454. doi: 10.1093/nar/gkg234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busby S., Ebright R.H. Transcription activation by catabolite activator protein (CAP) J Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 5.Henssler E.M., Bertram R., Wisshak S., Hillen W. Tet repressor mutants with altered effector binding and allostery. FEBS J. 2005;272:4487–4496. doi: 10.1111/j.1742-4658.2005.04868.x. [DOI] [PubMed] [Google Scholar]
- 6.Scholz O., Henssler E.M., Bail J., Schubert P., Bogdanska-Urbaniak J., Sopp S., Reich M., Wisshak S., Köstner M., Bertram R., et al. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol. Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 7.Schnappinger D., Schubert P., Pfleiderer K., Hillen W. Determinants of protein–protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J. 1998;17:535–543. doi: 10.1093/emboj/17.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubert P., Pfleiderer K., Hillen W. Tet repressor residues indirectly recognizing anhydrotetracycline. Eur. J Biochem. 2004;271:2144–2152. doi: 10.1111/j.1432-1033.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 9.Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system [see comments] Nature Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 10.Kedracka-Krok S., Gorecki A., Bonarek P., Wasylewski Z. Kinetic and thermodynamic studies of tet repressor-tetracycline interaction. Biochemistry. 2005;44:1037–1046. doi: 10.1021/bi048548w. [DOI] [PubMed] [Google Scholar]
- 11.Ehrt S., Guo X.V., Hickey C.M., Ryou M., Monteleone M., Riley L.W., Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen M., Bujard H. Tetracyclines in the control of gene expression in eukaryotes. In: Nelson M., Hillen W., Greenwald R.A., editors. Tetracyclines in Biology, Chemistry and Medicine. Switzerland: Birkhäuser Verlag; 2001. pp. 139–158. [Google Scholar]
- 13.Weinmann P., Gossen M., Hillen W., Bujard H., Gatz C. A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J. 1994;5:559–569. doi: 10.1046/j.1365-313x.1994.5040559.x. [DOI] [PubMed] [Google Scholar]
- 14.Berens C., Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 15.Kisker C., Hinrichs W., Tovar K., Hillen W., Saenger W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J Mol. Biol. 1995;247:260–280. doi: 10.1006/jmbi.1994.0138. [DOI] [PubMed] [Google Scholar]
- 16.Kamionka A., Bogdanska-Urbaniak J., Scholz O., Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 18.Dieckmann G.R., McRorie D.K., Lear J.D., Sharp K.A., DeGrado W.F., Pecoraro V.L. The role of protonation and metal chelation preferences in defining the properties of mercury-binding coiled coils. J. Mol. Biol. 1998;280:897–912. doi: 10.1006/jmbi.1998.1891. [DOI] [PubMed] [Google Scholar]
- 19.Shewchuk L.M., Verdine G.L., Nash H., Walsh C.T. Mutagenesis of the cysteines in the metalloregulatory protein MerR indicates that a metal-bridged dimer activates transcription. Biochemistry. 1989;28:6140–6145. doi: 10.1021/bi00441a002. [DOI] [PubMed] [Google Scholar]
- 20.Ramos J.L., Martinez-Bueno M., Molina-Henares A.J., Teran W., Watanabe K., Zhang X., Gallegos M.T., Brennan R., Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher M.A., Miller M.C., Grkovic S., Brown M.H., Skurray R.A., Brennan R.G. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 22.Krueger C., Berens C., Schmidt A., Schnappinger D., Hillen W. Single-chain Tet transregulators. Nucleic Acids Res. 2003;31:3050–3056. doi: 10.1093/nar/gkg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Watt V.M., Ingles C.J., Urdea M.S., Rutter W.J. Homology requirements for recombination in Escherichia coli. Proc. Natl Acad. Sci. USA. 1985;82:4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz C., Karosi S., Niederweis M. High-level expression of the mycobacterial porin MspA in Escherichia coli and purification of the recombinant protein. J Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2003;790:337–348. doi: 10.1016/s1570-0232(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 27.Kamionka A., Bertram R., Hillen W. Tetracycline-dependent conditional gene knockout in Bacillus subtilis. Appl. Environ. Microbiol. 2005;71:728–733. doi: 10.1128/AEM.71.2.728-733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ettner N., Müller G., Berens C., Backes H., Schnappinger D., Schreppel T., Pfleiderer K., Hillen W. Fast large-scale purification of tetracycline repressor variants from overproducing Escherichia coli strains. J. Chromatogr. A. 1996;742:95–105. doi: 10.1016/0021-9673(96)00232-4. [DOI] [PubMed] [Google Scholar]
- 29.Müller G., Hecht B., Helbl V., Hinrichs W., Saenger W., Hillen W. Characterization of non-inducible Tet repressor mutants suggests conformational changes necessary for induction. Nature Struct. Biol. 1995;2:693–703. doi: 10.1038/nsb0895-693. [DOI] [PubMed] [Google Scholar]
- 30.Scholz O., Schubert P., Kintrup M., Hillen W. Tet repressor induction without Mg2+ Biochemistry. 2000;39:10914–10920. doi: 10.1021/bi001018p. [DOI] [PubMed] [Google Scholar]
- 31.Henssler E.M., Scholz O., Lochner S., Gmeiner P., Hillen W. Structure-based design of Tet repressor to optimize a new inducer specificity. Biochemistry. 2004;43:9512–9518. doi: 10.1021/bi049682j. [DOI] [PubMed] [Google Scholar]
- 32.Backes H., Berens C., Helbl V., Walter S., Schmid F.X., Hillen W. Combinations of the alpha-helix–turn–alpha-helix motif of TetR with respective residues from LacI or 434Cro: DNA recognition, inducer binding, and urea-dependent denaturation. Biochemistry. 1997;36:5311–5322. doi: 10.1021/bi961527k. [DOI] [PubMed] [Google Scholar]
- 33.Schubert P., Schnappinger D., Pfleiderer K., Hillen W. Identification of a stability determinant on the edge of the tet repressor four-helix bundle dimerization motif. Biochemistry. 2001;40:3257–3263. doi: 10.1021/bi001927e. [DOI] [PubMed] [Google Scholar]
- 34.Simoncsits A., Chen J., Percipalle P., Wang S., Toro I., Pongor S. Single-chain repressors containing engineered DNA-binding domains of the phage 434 repressor recognize symmetric or asymmetric DNA operators. J. Mol. Biol. 1997;267:118–131. doi: 10.1006/jmbi.1996.0832. [DOI] [PubMed] [Google Scholar]
- 35.Robinson C.R., Sauer R.T. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl Acad. Sci. USA. 1998;95:5929–5934. doi: 10.1073/pnas.95.11.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jana R., Hazbun T.R., Fields J.D., Mossing M.C. Single-chain lambda Cro repressors confirm high intrinsic dimer-DNA affinity. Biochemistry. 1998;37:6446–6455. doi: 10.1021/bi980152v. [DOI] [PubMed] [Google Scholar]
- 37.Flynn T.C., Swint-Kruse L., Kong Y., Booth C., Matthews K.S., Ma J. Allosteric transition pathways in the lactose repressor protein core domains: asymmetric motions in a homodimer. Protein Sci. 2003;12:2523–2541. doi: 10.1110/ps.03188303. [DOI] [PMC free article] [PubMed] [Google Scholar]