Abstract
The genome-wide mapping of gene-regulatory motifs remains a major goal that will facilitate the modelling of gene-regulatory networks and their evolution. The repressor element 1 is a long, conserved transcription factor-binding site which recruits the transcriptional repressor REST to numerous neuron-specific target genes. REST plays important roles in multiple biological processes and disease states. To map RE1 sites and target genes, we created a position specific scoring matrix representing the RE1 and used it to search the human and mouse genomes. We identified 1301 and 997 RE1s inhuman and mouse genomes, respectively, of which >40% are novel. By employing an ontological analysis we show that REST target genes are significantly enriched in a number of functional classes. Taking the novel REST target gene CACNA1A as an experimental model, we show that it can be regulated by multiple RE1s of different binding affinities, which are only partially conserved between human and mouse. A novel BLAST methodology indicated that many RE1s belong to closely related families. Most of these sequences are associated with transposable elements, leading us to propose that transposon-mediated duplication and insertion of RE1s has led to the acquisition of novel target genes by REST during evolution.
INTRODUCTION
It is clear that much of what was once termed ‘junk’ DNA represents highly evolved, functional sequence containing amongst other things, numerous transcriptional regulatory motifs. A major challenge of post-genome biology is to identify these motifs and the genes they regulate, and to incorporate these results into accurate models of the transcriptional regulatory networks underlying processes such as development and disease. In addition to helping us understand gene regulation within a species, identification of transcription factor-binding sites (TFBSs) is an important tool in understanding phenotypic differences between species. Although the majority of regulatory sequences are phylogenetically conserved (1), a significant fraction of TFBSs are not conserved among closely related species, indicating that regulatory DNA can experience high rates of evolutionary change (2–6). This finding provides important support for the notion that changes in gene-regulatory networks, brought about by mutations to TFBSs, are an important contributor to phenotypic evolution (7). Little is known about the mechanisms responsible for such TFBS ‘turnover’. Computational and theoretical studies have found that short binding sites can appear rapidly through random DNA mutation (8,9); however, this rate of generation falls exponentially with motif length, suggesting that additional mechanisms may be responsible for the generation of longer binding sites. Therefore, in addition to providing insights into the role of transcription factors in gene regulation, whole-genome maps of TFBSs in multiple species may yield important insight into how gene-regulatory networks evolve.
The past decade has seen rapid advances in the complexity of motif-identification techniques: consensus sequences have given way to position-specific scoring matrices (PSSMs) (10), Bayesian-based interdependency models (11) and non-parametric approaches (12) amongst others. Such techniques must confront a serious hurdle to TFBS discovery, namely the short, degenerate nature of most binding motifs. Although this property no doubt confers evolutionary advantages (13), in terms of the high rates of site generation by random sequence mutation, it presents grave challenges to the confident identification of bona fide binding sites against a background of similar, non-functional motifs. A number of approaches have been developed to improve motif identification, most recently and successfully phylogenetic footprinting (14,15) which has been made possible by the sequencing of increasing numbers of genomes. This approach weights putative regulatory DNA sequences by their degree of conservation amongst related species, with the assumption that evolutionary conservation is indicative of functional importance. However, it is likely that those regulatory motifs which are not strongly conserved between species are those that are responsible for their phenotypic differences. Inlight of these considerations, a contemporary project to make a comparative map of TFBSs and the genes they regulate across multiple species might avoid such challenges by selecting a regulatory motif which is long and highly conserved enough to be unambiguously identified using current, non-phylogenetic methodologies.
REST (also known as neuron-restrictive silencing factor) is an essential vertebrate transcriptional repressor (16) involved in multiple biological processes and disease states (17–20). REST is the sole transcription factor to bind the highly conserved repressor element 1 (RE1, also known as neuron-restrictive silencing element, NRSE), to which it recruits various histone-modifying and chromatin-remodelling complexes (21–24). At 21 bp long, the RE1 represents an ideal model for bioinformatic regulatory element prediction, and in the past consensus sequence models have successfully predicted novel REST target genes de novo in a number of vertebrate species (25,26), most recently when Bruce et al. (26) published the first exhaustive TFBS search of the newly published human genome. In comparison to well-constructed probabilistic models of TFBSs, consensus sequences suffer from relatively high false negative and false positive rates (27). For example, the RE1 consensus falsely identifies large numbers of human endogenous retrovirus (hERV) Class I sequences as RE1s [(26) and R. Johnson, unpublished data]. Consensus sequences give no indication of a sequence's degree of similarity to the RE1 motif; consequently, the user has no control of the stringency with which searches are carried out, and no indication of a test sequence's likely affinity for REST. For similar reasons, the inability to identify less well-conserved sites rules out more exhaustive searches for weak affinity sites.
REST's target genes include many necessary for terminally differentiated neuronal function, such as synapse formation (SYN1) (28), neurotransmitter secretion (SNAP25) (26) and signalling (CHRM4) (29). Calcium signalling is also an essential process in neurons where it mediates transduction of electrical signals into cellular responses (30), and regulation of the voltage-gated calcium channel subunit gene CACNA1H by REST is necessary for normal heart function in mouse (19). Voltage-gated calcium channels are composed of multiple subunits. The α1 subunits, encoded by the CACNA1 family of genes, are responsible for pore formation and this subunit defines the pharmacological properties of the channel (31). The Cav2.1 subunit confers a P/Q type calcium current, initiating rapid synaptic transmission and the secretion of neurotransmitters and neuropeptides. The CACNA1A gene encoding Cav2.1 is highly expressed in the Purkinje cells of the cerebellum (32) and mutations in CACNA1A are responsible for a number of cerebellar disorders including migraine (33), epilepsy (34) and ataxias (35) but little is known about the transcriptional regulation of this gene.
The aim of this study was to accurately map the RE1s of human and mouse, as well as their target genes. To this effect, we develop and characterize an RE1 PSSM which is capable of predicting functional RE1s in genomic sequence with high sensitivity and selectivity. Using the RE1s identified in this way, we present a thorough analysis of the genomic distribution of this regulatory motif and its target genes. We also apply the RE1 PSSM to the exhaustive analysis of the REST-regulatory apparatus of a model gene, the novel target CACNA1A. We find that CACNA1A is regulated by a combination of binding sites of various affinities and degrees of phylogenetic conservation. Finally, we identify widespread duplication of functional RE1s, principally located within or beside transposable elements (TEs), which leads us to propose that transposon-mediated duplication has been an important mechanism of evolutionary expansion in the REST regulon.
MATERIALS AND METHODS
RE1 identification and database construction
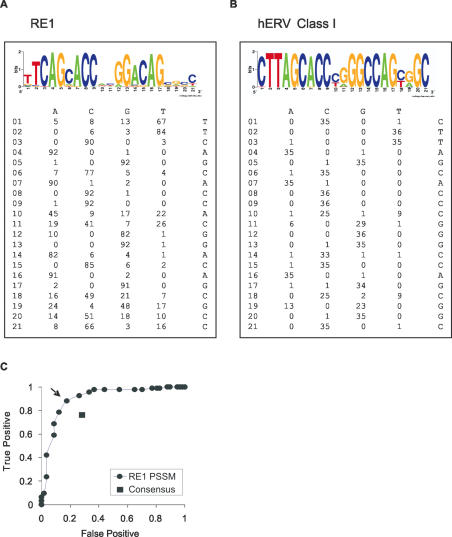
A PSSM was created by combining 93 experimentally verified REST-binding sequences from the primary literature and personal communications—the ‘positive training set’ (Figure 1A). Using the C program SeqScan (R. J. Gamblin and R. M. Jackson, manuscript in preparation), based on the scoring function of Stormo and Hartzell III (36,37), this PSSM was used to score both the positive training set, and 57 RE1-like sequences known not to bind REST in electrophoretic mobility shift assay (EMSA)—the ‘negative training set’. Query scores fall between 0 (no similarity to PSSM) and 1 (full identity). True and false positive rates at incremental cutoff scores were plotted in a receiver-operator curve (ROC) curve, which indicated that a cutoff of 0.91 produced an optimal sensitivity and selectivity for this set (Figure 1C). Unmasked NCBI builds 35 and 34 of the human and mouse genomes, respectively, were downloaded from Ensembl and searched for RE1s using SeqScan. All 21mers scoring >0.83 were saved to an updated version of the relational database described in Ref. (26) along with Swiss-Prot and TrEMBL annotations of nearest genes (accessible at http://bioinformatics.leeds.ac.uk/RE1db_mkII/).
Figure 1.
(A) The RE1 PSSM. Ninety-three sequences known to bind REST were combined to form a PSSM, here shown below a Weblogo recording the sequence conservation at each nucleotide position of this set (43). The height of each letter is proportional to its information content. (B) The hERV Class I PSSM. Thirty-six hERV Class I sequences containing conserved RE1-like sequences were used to create a PSSM. (C) Measuring the performance of the RE1 PSSM using a ROC curve. A ROC curve was generated by scoring the positive and negative training sets with the RE1 PSSM. The true positive (number of true sites to score above cutoff/total number of true sites) and false positive (number of non-binding sites to score above cutoff/total number of non-binding sites) rates were plotted at incremental cutoff scores (circles). The optimal combination of 88.2% true positive and 17.5% false positive rates were achieved at a cutoff of 0.91 (arrow). A similar analysis was carried out using the RE1 consensus sequence (square).
hERV Class I motifs that fell within the RE1 consensus were collected by submitting a representative set of sequences from the consensus RE1 database (http://www.bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm) to the motif-finding tool MEME (http://bioweb.pasteur.fr/seqanal/motif/meme/). Approximately 40% were found to have highly similar flanking regions, identified as hERV Class 1 elements by RepeatMasker (http://www.repeatmasker.org/). Inspection of their core motif showed that they corresponded to the common non-binding RE1 motif identified in Ref. (26). A PSSM was constructed using the RE1-like motif of 36 of these examples (Figure 1B). By scoring the contents of the RE1 database it was clear that all hERV sequences had hERV PSSM scores >0.9, while genuine RE1s lay below this score. All RE1s identified in the whole-genome PSSM scan were also scored using the hERV PSSM.
To assess the number of RE1 motifs expected by chance alone, six control PSSMs were constructed by shuffling the RE1 PSSM, and used to re-search the genome. The randomization process was constrained such that shuffled PSSM motifs had a similar propensity for CG dinucleotides as the RE1, in light of the under-representation of this pair in genomic DNA.
Adenoviral Infection
Recombinant adenoviruses expressing transgenes for REST DNA-binding domain (Ad DN:REST), or full-length REST (Ad REST), or no transgene (Ad), were amplified in HEK293 cells and purified by centrifugation in a CsCl gradient as described in Ref. (38). Viruses also contained a GFP transgene. Virus was added to cell media such that after 48 h >90% displayed green fluorescence, at which point cells were harvested.
Electrophoretic mobility shift assay
Nuclear protein extract from rat fibroblast JTC-19 cells was prepared as described previously (39). Protein was mixed with unlabelled competitor 28 bp oligonucleotides and 32P-labelled rat SCN2A2 promoter double-stranded DNA (dsDNA) sequences containing a strong RE1 sequence (40), then run on a 4% non-denaturing polyacrylamide gel. Unlabelled rat CHRM4 RE1 was used as positive control, while a non-binding, RE1-like sequence was used as non-specific dsDNA control. Competitor DNA was added in molar ratios of 100:1, 10:1 and 1:1 to labelled probe. Anti-REST (P18; Santa Cruz) antibodies were used to super-shift REST. A complete list of oligonucleotides used can be found in Supplementary Data.
RT–PCR
RT–PCR was carried out on RNA harvested from human HeLa cells using the protocol described in Ref. (26). cDNA samples were interrogated by quantitative PCR using the real-time iQ system (Bio-Rad), with primers to the coding regions of CACNA1A and cyclophilin genes. The specificity of PCR was verified by melt curve analysis of products obtained from cDNA, as well as controls in which the reverse transcriptase was omitted. Expression changes were inferred using the ΔΔCt method (41). The sequences of all primer sets used in this study are available in Supplementary Data.
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out essentially as described in Ref. (42) on chromatin from HeLa cells. IPs were performed using 10 μg of anti-REST (P18; Santa Cruz) or the same amount of non-specific goat IgG. ChIP DNA was interrogated by real-time quantitative PCR, using primers designed adjacent to RE1 sites. Starting concentrations of ChIP DNA were calculated with reference to a standard curve.
Multispecies alignment
Whole-genome multispecies alignments were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). Pairwise alignments were carried out using ClustalW provided by the EBI (www.ebi.ac.uk) at default settings. For each case where aligned sequence could not be found in the other species, the result was verified by BLAST search.
RESULTS
Identification of RE1s and target genes in human and mouse genomes
The RE1 is unusually long (21 bp) and highly conserved compared to a typical TFBS of 5–8 bp. We sought to improve on previous bioinformatic studies of the REST regulon by employing a more sensitive probabilistic PSSM model to identify RE1 sites, and apply it to whole-genome mapping of REST's binding sites. PSSMs are representations of sequence motifs, constructed using a set of known sequences, the ‘training set’, which can be used in conjunction with an appropriate scoring algorithm to identify other similar sequences (10). Query sequences are scored by the product of the similarity of each individual nucleotide to that in the corresponding position of the PSSM. The contribution of each nucleotide to the final score is weighted by its degree of conservation in the training set. For these reasons, PSSMs are both more sensitive and selective at identifying DNA motifs over a similar consensus sequence. In order to construct an RE1 PSSM, we compiled 93 RE1 sequences which have been shown to bind REST either in vitro (by EMSA) or in vivo (by ChIP). These sequences were combined to yield a probability value for each nucleotide at each position in the RE1 motif, resulting in a 21 bp motif with 63% GC content [represented as a Weblogo (43) in Figure 1A]. The composition of this RE1 differs in several important respects from that used in previous consensus sequences, having stronger constraint in positions 1, 3 and 7, as well as including additional conserved positions at the 3′ end (25,26). PSSMs assign all query sequences a score, regardless of whether they are bona fide binding sites or not; therefore it is necessary to empirically determine an optimal cutoff score for each PSSM, to include the maximum number of true binding sites while excluding spurious sequences. Using the program SeqScan (R. J. Gamblin and R. M. Jackson, manuscript in preparation), based on the method of Stormo et al. (36,37), the RE1 PSSM was used to score each sequence of the positive training set. A negative training set, composed of sequences that were shown to be incapable of binding REST in EMSA was scored in a similar way. We created a ROC by counting the numbers of true positives (TP) and false positives (FP) identified in both training sets at incremental cutoff scores; optimal sensitivity and selectivity occurred at a score of 0.91, which we subsequently defined as the RE1 PSSM cutoff score (Figure 1C). Any sequence having a score >0.91 is henceforth defined as an RE1, while sequences scoring below this value will be designated ‘below-cutoff’ RE1. Nevertheless, a population of bona fide RE1s exists in the training set below the cutoff (including functional RE1s in the human NPPA and rat SYN1 genes). The sequences of both training sets were similarly examined for identity with the RE1 consensus sequence, and by plotting the resulting TP/FP values on a ROC curve we confirmed that the RE1 PSSM is both more sensitive and selective at identifying RE1s from this training set (Figure 1C).
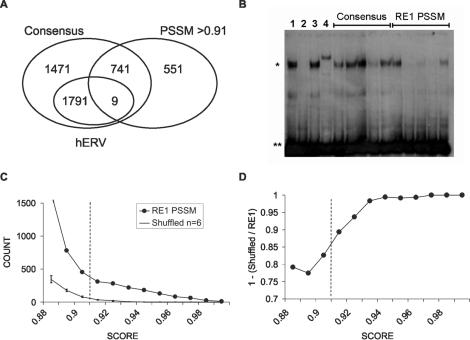
We next used the RE1 PSSM to identify potential REST-binding sites in genomic sequence of human and mouse.The full, unmasked genome sequence of both species was scanned using the RE1 PSSM with SeqScan. This search identified 1301 RE1s above cutoff in the human genome, including 551 which do not conform to the RE1 consensus and therefore principally constitute novel REST-binding sites (Figure 2A). Overall, we identified 995 REST target genes (defined as the closest Ensembl-annotated gene within 100 kb of an RE1), including 418 which were not identified in previous searches and hence represent novel REST target genes (a full list of novel target genes and disease-associated target genes identified in this study can be found in Supplementary Data). A similar search of the mouse genome identified 1039 RE1s and 822 target genes. The majority of consensus RE1 sequences failed to meet the RE1 PSSM cutoff, including 99.5% of contaminating hERV Class I RE1-like sequences (Figure 1B), which do not bind REST. The results of both searches, including the score, location and sequence of all RE1s and their target genes, are available in a searchable online database RE1db Mk II (http://bioinformatics.leeds.ac.uk/RE1db_mkII).
Figure 2.
(A) Identification of many novel RE1s by RE1 PSSM. The Venn diagram shows the number of RE1s identified in the human genome (Ensembl Build 35) by the RE1 PSSM, using the empirically determined cutoff score of 0.91, as well as those found by the RE1 consensus sequence. Non-functional, RE1-like hERV Class I sequences identified by each technique are also shown. (B) RE1s identified by PSSM in genomic DNA can bind REST. Randomly selected consensus RE1s with PSSM scores below cutoff, and non-consensus RE1s with PSSM scores above cutoff, were tested for the ability to compete REST off a radiolabelled RE1 in EMSA. Unlabelled test sequences were tested at 100:1 excess over probe. Single asterisk indicates the REST-bound probe and double asterisks indicate unbound probe. Controls: 1, no competitor; 2, CHRM4 RE1 site; 3, non-specific oligonucleotide; 4, Anti-REST antibody (P18; Santa Cruz). (C) The RE1 PSSM identifies more sequences in genomic DNA than expected by chance. The number of RE1s in the human genome were plotted at 0.01 score increments (circles). The same sequence was also scanned with six shuffled matrices, for which the mean count in each score bin is also shown (squares). Error bars represent standard deviation, and the PSSM cutoff score is indicated by a dotted line. (D) Excess of RE1s over shuffled sequences in the human genome. The data from (C) were re-plotted to emphasize the excess number of sequences discovered by the RE1 PSSM over shuffled PSSMs.
We next confirmed that the RE1 PSSM could identify functional RE1s in genomic sequence, and that such prediction was more effective than the RE1 consensus sequence. From human Chromosome 1 we randomly selected five non-consensus, PSSM-predicted RE1s, and five consensus RE1s with PSSM scores below cutoff, and tested their ability to interact with REST in vitro by EMSA (Figure 2B). Nuclear protein extracts containing REST protein were incubated with a radiolabelled promoter fragment containing the rat SCN2A2 RE1 (40), as well as unlabelled competitor oligonucleotides representing test sequences. We found that no oligonucleotides representing consensus RE1s with below-cutoff PSSM scores were capable of fully competing REST from the radiolabelled probe at a ratio of 100:1, while one competed partially. Conversely, sequences representing PSSM-predicted, non-consensus RE1s completely competed in three cases, with one competing partially. We concluded that the RE1 PSSM is capable of correctly identifying functional REST-binding sites in genomic DNA in the majority of cases, and that it has greater predictive power than previous techniques.
In order to identify the maximum number of potential RE1s, the whole-genome RE1 scans were carried out with minimum score cutoffs of 0.88, yielding ∼4000 hits in each species. The number of sequences identified for each PSSM score range in the human genome is shown in Figure 2C. To gauge the background probability of finding sequences with similar conservation and length as the RE1, the RE1 PSSM was repeatedly shuffled and used to scan the same genomic sequence. The shuffled matrices identified on average 58 sequences with scores >0.91 in the human genome, indicating that the 1301 sequences with scores >0.91 RE1s identified by the RE1 PSSM are highly significant. Interestingly, the RE1 PSSM also identifies significantly more sequences than the shuffled PSSMs at the lower score range 0.88–0.91. We re-plotted these data to emphasize the ratio of discovered RE1s to random sequences (Figure 2D); this clearly shows that there is an excess of RE1s over shuffled sequences at all measured score ranges, and that this excess increases steeply with score until 0.93, above which the background count is negligible. We performed similar searches on the genome of Caenorhabditis elegans, which has no known REST homologue: for this genome, the number of sequences identified by the RE1 PSSM and shuffled PSSMs were similar and low (data not shown). Therefore, in addition to a well-defined population of above-cutoff RE1s, the human genome contains an excess of below-cutoff RE1-like sequences, the majority of which might be expected to be unable to recruit REST.
REST is known to participate in diverse biological processes, including neuronal development (44,45), axonal pathfinding (46), heart development and function (19) and smooth muscle cell proliferation (42). This is reflected in the functions of REST's known target genes which predominantly encode ion channels, neurotransmitter transporters and receptors, SNARE proteins and transcription factors. We hypothesized that biological processes in which REST plays a role might be overrepresented in the ontology classifications of its target genes. To test this, we compared the prevalence of gene ontology (GO) terms associated with RE1-containing genes to that of all Ensembl genes. This analysis identified a large number of GO terms that were significantly enriched in REST target genes (P < 0.01, Fisher's exact test), of which a selection is shown in Table 1. For most cases, multiple related terms corresponding to the same underlying process were identified; e.g. the terms ‘synaptosome’, ‘synapse’, ‘synaptic vesicle’ and ‘synaptic transmission’ are all enriched in RE1-bearing genes, suggesting that REST regulates synaptic transmission at multiple levels. Similarly, enrichment was found for terms corresponding to various classes of ion channel (potassium, sodium, calcium channels) and neurotransmitter receptor (glutamate, GABA A, GABA B, glycine) involved in cell excitability and neurotransmission. In such cases, a single representative term is shown in Table 1. Overall, ontology terms correspond to most specialized functions of differentiated neurons, such as synaptic transmission, ion conductance and transport, neurotransmitter secretion and reception, axonal guidance, cell structure and cell adhesion. Interestingly, the term ‘sugar metabolism’ is significantly enriched, which may reflect specialized programmes of metabolic gene expression in neurons, as has been observed in previous studies (47). A number of terms including ‘cell differentiation’, ‘central nervous system development’ and ‘neurogenesis’ support REST's documented role in neuronal development (44,45). Target genes were also enriched for multiple terms related to calcium signalling (‘calcium ion binding’, ‘calcium ion transport’, ‘calmodulin binding’, ‘voltage-gated calcium channel complex’, ‘voltage-gated calcium channel activity’), a process in which REST has been implicated previously in relation to regulation of normal heart function through the repression of the α1H voltage-gated calcium channel subunit gene, CACNA1H (19). REST's role in regulating gene expression in the vasculature is reflected in enrichment of the terms ‘regulation of blood vessel size’ and ‘regulation of blood pressure’. Not all ontology terms were enriched in the REST target set; however, a number of GO terms such as ‘protein biosynthesis’ and ‘ribosome’ were underrepresented amongst RE1-containing genes, indicating that enrichment of particular GO terms described above is genuine.
Table 1.
Enrichment of GO terms in REST target genes
| GO code | GO term | Na | Obs.b | Exp.c | Foldd | Pe |
|---|---|---|---|---|---|---|
| GO:0007268 | Synaptic transmission | 187 | 32 | 5.4 | 5.9 | 6.1E−16 |
| GO:0005509 | Calcium ion binding | 1008 | 72 | 29.2 | 2.5 | 2.9E−12 |
| GO:0048699 | Neurogenesis | 275 | 29 | 8.0 | 3.6 | 2.7E−09 |
| GO:0006811 | Ion transport | 347 | 33 | 10.1 | 3.3 | 3.0E−09 |
| GO:0007155 | Cell adhesion | 455 | 28 | 13.2 | 2.1 | 1.8E−04 |
| GO:0005529 | Sugar binding | 194 | 15 | 5.6 | 2.7 | 5.8E−04 |
| GO:0050880 | Regulation of blood vessel size | 4 | 2 | 0.1 | 17.2 | 4.9E−03 |
| GO:0007611 | Learning and/or memory | 14 | 3 | 0.4 | 7.4 | 7.0E−03 |
| GO:0005200 | Structural constituent of cytoskeleton | 101 | 8 | 2.9 | 2.7 | 9.2E−03 |
aNumber of Ensembl genes associated with this ontology term.
bNumber of REST target genes associated with this ontology term.
cExpected number of REST target genes.
dFold enrichment of REST target genes over expectation.
eProbability, by Fisher's exact test.
Genic and genomic distributions of RE1s
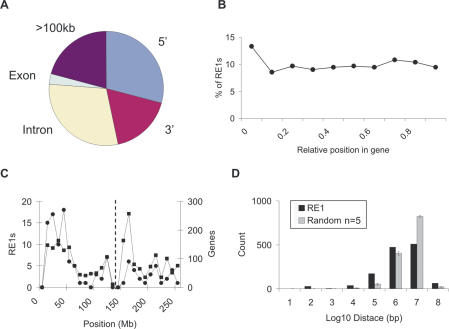
Using the positional information from the RE1 PSSM search, we next measured how RE1s are distributed on a number of scales. Conventional models have transcription factors regulating target genes from proximal upstream promoter regions or distal enhancer modules. Recent in vivo measurements of transcription factor occupancy have demonstrated recruitment to a variety of positions relative to target genes, including introns (48,49). To investigate whether this is the case for REST, we next used the human whole-genome RE1 data to define the most prevalent locations of an RE1 relative to its target gene. We found the greatest number of RE1s are located in the introns of genes (29.4%) (Figure 3A); indeed, this figure is almost certainly an underestimate given the large number of transcripts that remain to be properly annotated, and the uncertainty of the transcription start site (TSS) of many genes. RE1s showed a weak preference for locations upstream over downstream of target genes (29.3 and 17.5%, respectively), while one-fifth of RE1s are located >100 kb from the nearest annotated gene (20.7%), suggesting that either REST can have long-range (>100 kb) interactions with target genes, or that many target genes remain to be annotated. Surprisingly, intronic RE1s are rather uniformly distributed within their target genes, with a small excess situated within 0.1 gene lengths of the TSS (Figure 3B). Together these data suggest that RE1s are only weakly constrained in their position relative to target genes.
Figure 3.
(A) RE1s are most frequently located in the introns of target genes. The fraction of strong RE1s at each position relative to human target genes was calculated. 5′, <100 kb upstream of annotated TSS; 3′, <100 kb downstream of gene end. (B) RE1s have little preference for location within genes. The proportion of internal (i.e. intronic and exonic) RE1s was plotted according to its relative position along the target gene's length (circles), defining the TSS to be 0 and the gene end to be 1. (C) Peaks of RE1 and gene density do not correlate on Chromosome 1. The number of RE1s (circles, left scale), and the number of Ensembl genes (squares, right scale) are plotted for 1 Mb windows along Chromosome 1. The approximate position of the centromere is indicated by a dashed line. (D) RE1s are clustered over distinct distance scales in the human genome. The distance from each RE1 to the next, moving in a positive direction along each chromosome, is plotted as a histogram for human RE1s. As a reference, equivalent data were generated for five sets of equivalent random genomic coordinates. Error bars represent standard deviation.
Genes have a heterogeneous distribution on chromosomal scales, tending to be highly clustered in G-C rich regions near the ends of chromosomes (50); we next tested whether REST target genes show a similar tendency. By plotting the density of PSSM RE1s and Ensembl genes along each chromosome, we found that while the RE1 density generally mirrors that of annotated genes, there are regions where the RE1 density is markedly lower or higher than corresponding gene density. This phenomenon is well illustrated on Chromosome 1, where one gene-rich region towards the tip of p-arm is highly enriched for RE1s, while another at the centromeric end of the q-arm is markedly under-enriched (Figure 3C). We further sought to quantify the degree of clustering of RE1s by calculating the distance from each RE1 to its nearest neighbour (Figure 3D). Comparison of the RE1 distance distribution to that of randomly generated sets clearly demonstrates that the distribution of RE1s in the human genome is significantly non-random over all distance scales (at most P < 0.001, Student's t-test). In particular, RE1s are clustered on scales of both 10–100 kb and 100–1000 kb; this distance is similar to, or smaller than the length of a typical gene, suggesting that some genes might be regulated by multiple RE1s. Subsequent inspection of the RE1 PSSM data revealed that 101 human genes are closest to, and within 100 kb of two or more RE1s. Nevertheless the majority of RE1s are separated by 1–10 Mb, indicative of typical gene–gene distance or greater. A number of REST target genes have been found to contain pairs of RE1s arranged in tandem: human SNAP25 and L1CAM recruit REST more effectively than single sites in the same cells (26), while KCNN4 contains a strong, well-conserved RE1 together with a second, more weakly conserved site that is capable of interacting with REST in mouse but not human (42). The nearest-neighbour analysis identified 27 examples of pairs of RE1s within 100 bp of each other, compared to none in the random sets. A number of these tandem RE1s were in fact members of groups of up to five motifs. All of these sequences are aligned in a ‘head to tail’ configuration (P = 7 × 10−9, Binomial distribution). Subsequent inspection of the flanking regions of human RE1s revealed an additional 32 RE1-like sequences with scores in the score range 0.88–0.91, compared to zero found by six shuffled matrices, indicating that RE1s tend to colocalize. A list of all human tandem RE1s can be found in Supplementary Data.
Dissecting the REST-regulatory sequences of a model gene, CACNA1A
We next wished to test the predictive power of the RE1 PSSM by making an in-depth study of the REST-regulatory sequences within a single model gene. We intended for this approach to shed light on the functional significance of below-cutoff RE1s, and by performing the study in both human and mouse we hoped to measure the degree of evolutionary conservation of RE1s. We selected as a model the CACNA1A gene, which was found to contain multiple high-scoring RE1s by the PSSM search in human and mouse. The gene is classified in the RE1-enriched ‘voltage-gated calcium channel complex’ ontology classification, and is a paralogue of the REST target CACNA1H. CACNA1A is a large, multi-exon gene encoding the Cav2.1 neuron-specific voltage-gated calcium channel subunit. Little is known about the regulatory mechanisms governing CACNA1A expression, although the mouse homologue does have two functional Sp1 sites (51), while the human gene contains numerous clusters of potential TFBSs, including Sp1, Pax4 and Myc (R. Johnson, unpublished data).
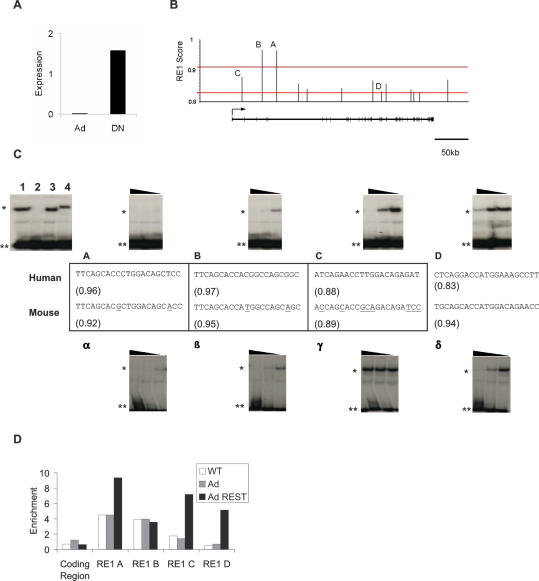
We first confirmed that expression of CACNA1A can be regulated by REST in human cells. HeLa cells are non-neuronal and express relatively high levels of REST [L. Ooi, unpublished data]. Virally mediated overexpression of a dominant-negative REST construct resulted in 75-fold de-repression of CACNA1A mRNA levels in HeLa cells (Figure 4A), indicating that REST is capable of strongly repressing this gene. In addition to the two strong RE1s located in introns 3 and 5 of the gene, the RE1 PSSM identified 13 more RE1-like sequences with scores in the range 0.83–0.91 in this gene (Figure 4B). We tested all potential RE1s in EMSA, and identified four which were capable of interacting with REST (data not shown). We further investigated the relative in vitro binding affinity of these sequences by testing their ability to compete in EMSA at decreasing molar ratios to labelled probe (100:1, 10:1, 1:1) (Figure 4C). Both RE1s with scores above cutoff strongly competed: Site A (PSSM score 0.96) at 1:1, and Site B (0.97) at 10:1. In addition, two other below-cutoff sites displayed detectable affinity for REST: Site C (0.88) competed at 100:1, and Site D (0.83) competed partially at 100:1. These results underlined the accuracy of using a 0.91 cutoff in predicting high-affinity RE1s, while demonstrating that a minority of below-cutoff RE1s can interact with REST, albeit more weakly.
Figure 4.
(A) REST regulates transcription of CACNA1A in HeLa. mRNA from HeLa cells infected with either empty adenovirus (Ad), or adenovirus expressing the REST DNA-binding domain (DN), was harvested and reverse transcribed. These cDNAs were interrogated in real-time quantitative PCR using primers specific to the coding sequence of CACNA1A and the housekeeping gene, cyclophilin. Axis indicates expression level relative to cyclophilin X107. (B) Identification of RE1s in the human CACNA1A gene. The RE1 PSSM was used to scan the human CACNA1A gene, and the scores of putative RE1s were plotted as a function of position. Upper and lower lines represent scores of 0.91 and 0.83, respectively. A cartoon of the gene is shown below, with exons represented by wide lines. (C) Human and mouse CACNA1A orthologues contain multiple functional RE1s that are only partially conserved. Sequences identified by the RE1 PSSM in the human (upper panel) and mouse (lower panel) CACNA1A genes were tested in EMSA competition assay. Only those sequences that displayed detectable affinity for REST are shown. Decreasing molar ratios (100:1, 10:1, 1:1) of unlabelled oligonucleotides were used in EMSA competition assay against a radiolabelled RE1. Single asterisk indicates the REST-bound probe and double asterisks indicate unbound probe. Controls: 1, no competitor; 2, CHRM4 RE1 site; 3, non-specific oligonucleotide; 4, anti-REST antibody (P18; Santa Cruz). The sequence of each RE1 is shown with its RE1 PSSM score in brackets. Boxes denote homologous pairs of RE1s, as determined by a combination of whole-genome alignment from the UCSC Genome Browser and BLAST. Underlined bases indicate those mouse positions which are not conserved in human. (D) REST is recruited to CACNA1A RE1s in vivo. DNA was immunoprecipitated with anti-REST (P18; Santa Cruz) and control non-specific IgG antibodies from wild-type HeLa (WT), as well as cells infected with empty adenovirus (Ad) and adenovirus carrying the full-length REST gene (Ad REST). ChIP DNA was interrogated in quantitative real-time PCR using primers flanking the RE1s of CACNA1A, as well as the coding region of the CHRM4 gene, which is distal to any RE1. Values represent the fold enrichment of anti-REST immunoprecipitate over IgG.
We next wished to ask whether the CACNA1A RE1 sites recruit REST in vivo, and whether the degree of that recruitment reflects their in vitro binding affinity. We assayed REST occupancy at the four functional CACNA1A RE1s in HeLa by ChIP assay. In wild-type HeLa both above-cutoff RE1s, Sites A and B, were strongly immunoprecipitated by an anti-REST antibody compared to a non-specific IgG, indicating their occupancy by REST (Figure 4D). Neither Site C nor D were enriched >2-fold. In order to investigate whether these weaker sites can recruit REST at higher cellular concentrations, similar ChIP assays were carried out on HeLa cells overexpressing virally delivered, full-length REST protein. In contrast to wild-type cells, all four CACNA1A RE1s were found to be occupied in cells overexpressing REST. This effect was not observed in cells infected with control adenovirus, nor was overexpressed REST recruited non-specifically to DNA lacking an RE1. Therefore REST is only recruited to the high-affinity RE1s of the CACNA1A gene in HeLa cells under normal conditions, but weaker RE1s retain the ability to specifically recruit REST at elevated concentrations.
With the publication of the genomes of multiple species, interest has focussed on phylogenetic comparison as ameans of identifying conserved gene-regulatory elements (52), and for understanding variations in gene expression characteristics between species (53). A number of studies have demonstrated gain and loss (turnover) of TFBSs regulating orthologous genes in closely related species (2–6). Having mapped the regulatory sites through which REST can regulate the CACNA1A gene in human cells, we wished to determine the degree to which these elements are conserved in mouse, both in terms of their DNA sequence and affinity for REST. The RE1 PSSM identified three RE1s in the mouse CACNA1A orthologue. Examination of the predicted RE1s by global alignment in UCSC genome browser (54,55) and by local alignment in ClustalW (http://www.ebi.ac.uk/clustalw/index.html) showed that three mouse sequences are homologous to human Sites A, B and C (designated α, β, γ for mouse) (Figure 4C) (Note: Human Site B cannot be aligned to mouse in UCSC; however, the similarity of both RE1 sequences, as well as ClustalW alignment of their respective flanking regions suggests they are indeed homologous, and will be considered as such). This was not the case for human site D and mouse site δ, which appear to bear no evolutionary relationship to each other. The two homologous pairs of high-affinity sites, A/α and B/β, are highly conserved both in PSSM score and sequence, a fact reflected in their identical capacity to bind REST in EMSA (Figure 4C). On the other hand, although Site C in human has detectable affinity for REST in EMSA, its mouse homologue of similar PSSM score does not. Separate experiments showed that the homologous sequence in dog is similarly incapable of interacting with REST (data not shown). Finally, each species has one RE1 (Site D/Site δ) which has no homologue in the other, and appears to reside in inserted/deleted sequence blocks. The mouse sequence, Site δ, has a high RE1 PSSM score and in vitro affinity for REST. ChIP assays carried out on the mouse neural stem cell line, NS5 (56) found Site δ to be specifically occupied by REST (R. Johnson, unpublished data). Together, these data indicate that conservation of functional REST-binding sites between human and mouse is not total. Non-conserved functional sites come in two distinct types, suggestive of alternative mechanisms of generation: human Site C can be aligned with non-functional mouse sequence, whereas mouse Site δ cannot be aligned with any human sequence and is only shared by one other mammal, rat. The former example is suggestive of RE1 creation or loss through random DNA mutation, while the latter appears to be the result of an insertion or deletion event. These hypotheses are supported by the degree of conservation of these sequences across multiple vertebrate species (Supplementary Data): Sites A/α and B/β have high-conservation scores by PhastCons (52) (notwithstanding alignment issues for Site B discussed above), while Sites C/γ, D and δ do not.
Non-conservation of RE1s between human and mouse is not confined to the CACNA1A gene. We investigated the degree of conservation of PSSM-predicted RE1s in other voltage-gated calcium channel genes of the α1, β, γ, α2δ subunit families. We identified a distinct RE1 sequence to that identified by Kuwahara et al. (19) proximal to the CACNA1G gene. Altogether we identified 10 human voltage-gated calcium channel subunit genes associated with 14 strong RE1s. By inspection of the alignment of these RE1s to genomic sequence of mouse, we found that just seven(50%) were well conserved in mouse in terms of RE1 PSSM score, while four had strongly reduced score (mouse sequence below cutoff) and three could not be aligned (Table 2). The three non-conserved human RE1s we tested by ChIP (CACNB2, CACNA1G, CACNG7) recruit REST in vivo, suggesting that these are functional sites (Figure 5D). We infer that non-conservation of RE1 sequence and affinity is a more general feature of REST target genes.
Table 2.
Conservation of RE1s between human and mouse CACNA1 family genes
| Gene | Human Ensembl ID | Human RE1 score | RE1 ID | TE? | Mouse Ensembl ID | Mouse RE1 score | RE1 ID | TE? |
|---|---|---|---|---|---|---|---|---|
| CACNA1A | ENSG00000141837 | 0.96 | hum39611 | No | ENSMUSG00000034656 | 0.92 | mus16973 | No |
| 0.97 | hum39604 | No | 0.95 | mus16983 | No | |||
| — | — | — | 0.94 | mus16981 | No | |||
| CACNA1B | ENSG00000148408 | 0.92 | hum23208 | No | ENSMUSG00000004113 | — | — | — |
| 0.97 | hum23209 | No | <0.83 | — | — | |||
| 0.98 | hum23210 | No | 0.96 | mus2523 | No | |||
| CACNA1D | ENSG00000157388 | 0.92 | hum8374 | No | ENSMUSG00000015968 | <0.83 | — | — |
| CACNA1H | ENSG00000196557 | 0.97 | hum33973 | No | ENSMUSG00000024112 | 0.98 | mus31150 | No |
| CACNA1G | ENSG00000006283 | 0.93 | hum37140 | LINE2 | ENSMUSG00000020866 | <0.83 | — | — |
| CACNA2D2 | ENSG00000007402 | 0.95 | hum8255 | No | ENSMUSG00000010066 | 0.88 | mus19450 | No |
| 0.91 | hum8267 | No | 0.93 | mus19435 | No | |||
| CACNA2D3 | ENSG00000157445 | 0.97 | hum8379 | No | ENSMUSG00000021991 | 0.96 | mus27091 | No |
| CACNB2 | ENSG00000165995 | 0.93 | hum23514 | Alu | ENSMUSG00000057914 | — | — | — |
| CACNG2 | ENSG00000166862 | 0.93 | hum43594 | No | ENSMUSG00000019146 | 0.94 | mus29100 | No |
| — | — | — | 0.93 | mus29059 | No | |||
| CACNG7 | ENSG00000105605 | 0.93 | hum40713 | Alu | ENSMUSG00000069806 | — | — | — |
Dash (—) indicates no aligned sequence exists.
TE?: The identity of transposable elements overlapping or in the immediate flanking region of RE1s is indicated.
Figure 5.
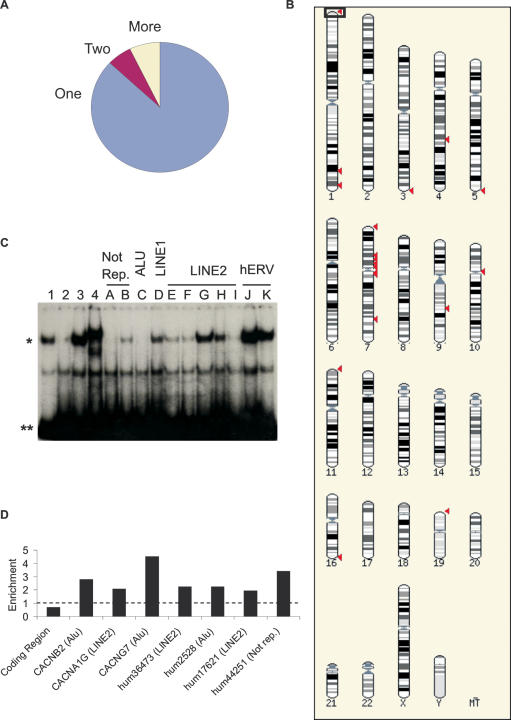
(A) The human genome contains families of related RE1 sequences. All above-cutoff human RE1s, including 100 bp of flanking sequence, were used in a BLAST search of the remaining RE1s. Sequences identified with similarity to at least one other with P < 10−30, are considered ‘related’. The proportions of RE1s which are unique (One), in pairs of high similarity (Two) or in groups of three or more highly related sequences (More), are shown. (B) Dispersal of RE1s from a single parent site. The largest family of related RE1s contains 28 members, all sharing significant homology with the RE1 hum3 located on Chromosome 1 (boxed). The locations of these sites in the human genome were plotted (red arrowheads) using the Ensembl tool Karyoview. The presumptive parent site, hum3, is boxed. A number of duplicated RE1s cannot be distinguished in this view due to their close proximity. (C) RE1s associated with transposon sequences can bind REST (Table 3). A selection of duplicated RE1 sequences were tested for their ability to interact with REST by EMSA competition assay. ‘Not Rep.’ indicates sequences that have no detectable repetitive characteristics, as judged by the tool RepeatMasker (www.repeatmasker.org). Single asterisk indicates the REST-bound probe and double asterisks indicate unbound probe. Controls: 1, no competitor; 2, CHRM4 RE1 site; 3, non-specific oligonucleotide; 4, anti-REST antibody (P18; Santa Cruz). (D) Transposon-associated and duplicated RE1s recruit REST in vivo (Tables 2 and 3). ChIP assay was carried out on endogenous HeLa cells using an anti-REST antibody (P18; Santa Cruz) and non-specific IgG. Enrichments were measured as in Figure 4D, using primers flanking the indicated RE1s. TEs associated with each RE1 are indicated in brackets.
Duplication of RE1s
The identification of RE1s which are not conserved between human and mouse, as well as the greater number of RE1s in the genome of the former, suggested to us that novel RE1s had arisen since both species diverged. We hypothesized that the duplication and insertion of sequence blocks containing RE1s might be a mechanism of novel TFBS creation, perhaps leading to the acquisition of novel target genes by REST over time. Pairs of RE1s which had been duplicated recently would be characterized not only by the similarity of their core RE1 sequences, but also by that of their flanking regions. To test whether this was the case, each RE1 identified by the RE1 PSSM in the human genome was sequentially used to search for similar sequences in the RE1 database using the BLAST algorithm. Searches were carried out using the RE1 itself and 100 bp of flanking sequence, with a stringent P-value cutoff of 1 × 10−30 to identify only those sequences with highly significant homology. In this way we found that at least 10% (126/1301) of RE1s in the human genome belong to evolutionarily related groups (Figure 5A). (A full list of the identifiers and locations of duplicated RE1s is available from the authors on request.) A similar effect was observed in mouse (data not shown). This analysis excluded all instances where the RE1 in a duplicated sequence had an RE1 PSSM score below cutoff, as well as the tandem RE1s mentioned earlier.
We reasoned that duplication and insertion by TEs might be a potential mechanism of RE1 duplication. We therefore tested the duplicated RE1s for repetitive or transposon characteristics. We submitted the flanking sequences of duplicated RE1s to the online tool RepeatMasker (www.repeatmasker.org), which indicated that the majority of duplicated RE1s are located in TEs of most major classes, including long interspersed repeats (LINEs, principally LINE2s), short interspersed repeats (SINEs, principally Alus) and hERV sequences. In addition, a number of duplicated RE1s are located in sequence with no characteristics of TEs. The largest single family of RE1s, located in the coding region of a LINE2 element, had 28 members with significant similarity to the hum3 RE1 sequence located in the subtelomeric region of the Chromosome 1 p-arm (Figure 6). These sites had an apparently non-random distribution, with 29% (8/28) located within 1 Mb of a telomere, 25% (7/28) within 1 Mb of a centromere and 43% (12/28) located on Chromosome 7 (Figure 5B). To confirm that duplicated RE1s are functional binding sites, we tested a selection of these RE1s’ ability to interact with REST in vitro by EMSA competition assay (Figure 5C). Most of those sequences tested, including those associated with Alu, LINE1 and LINE2 sequences, as well as two pairs residing in non-repetitive DNA, were capable of interacting with REST. The most common LINE2-derived hum3 sequence was amongst this group. In agreement with previous findings (26), neither hERV Class I RE1 showed detectable affinity for REST. In addition to binding REST in vitro, we found that all four of the duplicated RE1s we tested (including two associated with LINE2s and one with an Alu) could be enriched by an anti-REST antibody in ChIP (Figure 5D). We conclude that functional RE1s have been duplicated and inserted at new positions in the human genome by both transposon-dependent and independent processes, and that a high proportion recruit REST in vivo.
Figure 6.
Location of hum3 RE1 within the LINE2 element. The RE1 sequence is boxed. ‘-’ indicates an insertion/deletion, ‘i’ a transition (G↔A, C↔T) and ‘v’ a transversion (all other substitutions).
We next investigated the target genes of duplicated RE1s; in particular, we used the Ensembl database to check whether such genes had a mouse homologue, and if so, whether the homologue is also a REST target (defined as being the closest gene within 100 kb of an RE1 with PSSM score >0.88). A list of human targets of duplicated RE1s is shown in Table 3. We identified at least six human REST target genes whose mouse orthologue contain no identifiable RE1, as well as seven for which no mouse orthologue has been identified. TEs have gone through bursts of active transposition during distinct periods of evolutionary history: although LINE2 elements were thought to be active ∼200 million years ago and before human–mouse divergence, LINE1 and Alu elements continue to retrotranspose in humans (57). This is reflected in the phylogenetic conservation of human TE-associated RE1s: those associated with Alu and LINE1 elements have no aligned sequences other than in chimp, while a number of ancient LINE2 elements are conserved amongst multiple species (Table 3). Interestingly, multispecies alignment of LINE2 RE1s is only possible in a minority of cases, again suggesting that significant gain and loss of RE1s has taken place since human–mouse divergence.
Table 3.
Target genes of duplicated RE1s
| Ensembl ID | Description | RE1 ID | Element | EMSA lanea | Conserved? |
|---|---|---|---|---|---|
| No mouse homologue exists | |||||
| ENSG00000196164 | Q96HX1_HUMAN | hum21648 | LINE2 | H | m |
| ENSG00000101825 | Adlican | hum44251 | Not rep. | B | m |
| ENSG00000185164 | Nodal modulator 2 precursor (pM5 protein 2) | hum34541 | LINE2 | N/A | m |
| ENSG00000154608 | KIAA0470L protein | hum11432 | LINE2 | E | c |
| ENSG00000189281 | PREDICTED: hypothetical protein XP_375668 | hum11438 | LINE2 | E | c |
| ENSG00000182053 | PREDICTED: similar to tripartite motif-containing 51 | hum26460 | Not rep. | N/A | m |
| ENSG00000182111 | PREDICTED: similar to Zinc finger protein 479 | hum17621 | LINE2 | F | c, d |
| ENSG00000197123 | Zinc finger protein 679 | hum17654 | Not rep. | N/A | m |
| ENSG00000163040 | NP_620125.1 | hum6034 | LINE1 | D | c |
| Mouse homologue has no RE1 | |||||
| ENSG00000106078 | Cordon-bleu homologue | hum17527 | LINE2 | N/A | c |
| ENSG00000105198 | Galactoside-binding soluble lectin 13 (PP13) | hum40246 | Not rep. | N/A | c |
| ENSG00000006659 | Placental protein 13-like (CLC2) | hum40243 | Not rep. | N/A | c |
| ENSG00000184330 | S100 calcium-binding protein A7-like 1 | hum2528 | Alu | C | c |
| ENSG00000143556 | S100 calcium-binding protein A7 (Psoriasin) | hum2515 | Alu | N/A | c |
| ENSG00000196396 | Tyrosine-protein phosphatase, non-receptor type 1 (PTP-1B) | hum41830 | LINE2 | N/A | c, d |
| Mouse homologue has RE1 | |||||
| ENSG00000152953 | STK32B | hum10317 | Not rep. | A | c, d |
| ENSG00000007171 | NOS2 | hum36473 | LINE2 | I | m |
Not rep., flanking sequence has no repetitive characteristics; m, sequence can be aligned to multiple species; c, sequence can be aligned to chimp; d, sequence can be aligned to dog.
aSee Figure 5C.
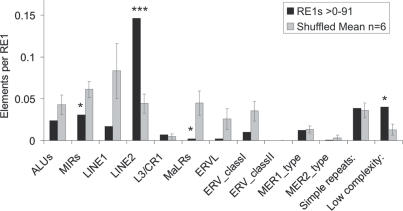
If duplication and insertion of RE1s by TEs has contributed to the current population of human RE1s, one might expect to observe a statistically significant association of the two sequence features. The proximal flanking regions of all above-cutoff human RE1s was searched for TEs using RepeatMasker. The same operation was performed on the control sets of sequences identified by shuffled RE1 matrices using the same cutoff score (Figure 2C). These data, presented in Figure 7, clearly show that the association of RE1s with TEs is non-random. RE1s are under-associated with most classes of TE; in the case of MaLRs and MIRs this effect is statistically significant (P < 0.05, Student's t-test). In contrast, LINE2 are highly significantly associated with RE1s elements (P < 0.001), with approximately one in seven RE1s overlapping or flanking a LINE2. This finding suggests that LINE2 retrotransposition in particular has been an important driver of RE1 generation and insertion in the human lineage. We tested this idea by compiling the putative target genes of the 190 LINE2-associated RE1s in the human genome, and testing their GO classifications for significantly overrepresented terms. We found that this set of genes is significantly enriched for a number of important terms identified for the set of all human REST target genes (Supplementary Data). We consider this to be a strong evidence for the functional importance of gene regulation by LINE2-associated RE1s.
Figure 7.
Association of RE1s with TE classes. The numbers of TEs identified in the proximal flanking region (<100 bp) of all human RE1s were measured using the tool RepeatMasker. As a reference, the sequences identified by the six shuffled RE1 PSSMs (Figure 2C) were analysed in the same way. Statistical significance was calculated using Student's t-test (*P < 0.05, ***P < 0.001).
DISCUSSION
The genomic population of RE1s is open-ended
We have mapped the REST-regulatory sequences and target genes in the human and mouse genomes with greater accuracy than possible previously. Although the approach we have used is not applicable to most TFBSs, owing to their low information content, nevertheless conclusions from this study on REST will be applicable to transcription factors in general. By searching for RE1s over a wide range of PSSM scores, we showed that instead of a well-defined population of unambiguous, high-affinity binding sites, the human genome may be more accurately considered to have an open-ended continuum of RE1 sites of varying conservation, similarity to the RE1 motif, and affinity. This resonates with the emerging view of dynamically evolving gene-regulatory networks based on a constantly changing set of genomic-binding sites. Therefore statements of absolute numbers of binding sites discovered might better be replaced by ratios of discovered sites to those expected by chance for each PSSM score range. By this regime, there is a 23-fold (1301/57) excess of RE1s above cutoff in the human genome, and a 5-fold (2890/600) excess of RE1s in the below-cutoff score range 0.88–0.91. Furthermore, a distinct change in this ‘RE1 excess’ occurs at scores >0.93, suggesting that 706 sequences in the human genome above this score are highly significant. One might expect that as score constraints are progressively relaxed, the ratio of sequences identified by RE1 and shuffled PSSMs should approach 1; the fact that there is still a 5:1 excess of RE1s between 0.88 and 0.91, strongly suggests that the population of below-cutoff RE1s has genuine biological significance, despite evidence that the majority cannot bind REST. There may be several reasons for this excess. First, the RE1 PSSM no doubt falsely rejected a number of high affinity by scoring them below cutoff; the fact that a PSSM is incapable of perfectly predicting REST-RE1 affinity may indicate that the training set is incomplete or skewed by too many RE1s discovered by consensus search, or more fundamentally, that significant interdependency occurs between nucleotides of the RE1, which cannot be represented by a PSSM. As we have shown, some below-cutoff sites represent low-affinity RE1s which are only capable of recruiting REST at elevated concentrations and/or permissive chromatin states, such as occurs in ischemic neurons (17) or neural stem cells (44,45), respectively. Furthermore, a significant fraction of below-cutoff RE1s represent a sequence motif of hERV Class I elements which we know to be unable to interact with REST. Finally, and perhaps most intriguingly, a number of below-cutoff RE1 sequences might represent evolutionarily turned-over RE1s, i.e. sites which were functional at some point during history, but which are no longer under selection pressure. Over time, random mutation of such RE1s would give rise to RE1-like sequences which are degenerate and non-functional, but nevertheless bear vestigial similarity to the RE1 motif and are detected below the score cutoff by the RE1 PSSM. Future studies of genome-wide phylogenetic conservation of RE1s should identify turned-over RE1s if they exist. The number and state of mutation of such an evolutionary ‘footprint’ of turned-over TFBSs in vertebrate genomes would be an important basis for attempting to estimate the rate of evolution of gene-regulatory DNA.
REST regulation of CACNA1A through multiple binding sites
The RE1 PSSM was also an effective tool in understanding how REST regulates individual genes. Our thorough analysis of the CACNA1A REST-regulatory DNA showed that the gene contains a number of functional REST-binding sites, with a range of affinities and degrees of evolutionary conservation in mouse. This suggests that, in some cases, regulation of REST target genes may be more complex than previous models suggest. It is conceivable that the number of occupied RE1s in CACNA1A, and hence the degree of transcriptional repression of the gene, can assume a number of well-defined states, determined by the concentration of REST relative to the kd of each RE1. This resonates with the model of Ballas et al. (45) regarding the progressive loss of REST from target genes during neuronal differentiation, where REST recruitment is lost from those genes with weaker RE1s first, as REST levels in the nucleus drop during development. Alternatively, variation in REST protein levels through organs such as the brain might lead to finely patterned levels of CACNA1A transcription (58). In a similar way, the existence of closely spaced tandem RE1 clusters strongly suggests that this arrangement of sites has biological relevance, perhaps through their ability to recruit REST at low concentrations, or by simultaneous recruitment of multiple REST complexes at once to a target gene. Tandem RE1s were originally identified in the SNAP25 gene (26), which could recruit REST at low cellular concentrations such as that found in the U373 astrocytoma cell line. Although the head-to-tail orientation of tandem RE1s seems to be ubiquitous, the distance separating the tandem sites is not, suggesting that REST–REST interactions are not an important element of binding. Rather, we propose that the tandem configuration might instead reflect the process by which the site was generated through a tandem duplication event (59,60).
Evolutionary turnover of RE1s through mutation and sequence duplication
Comparison of the RE1s of orthologous human and mouse genes of the voltage-gated calcium channel subunit family provided strong evidence of evolutionary turnover in RE1s. We showed that a large proportion of human RE1s from this set are not conserved in aligned genomic sequence of mouse. Such non-conserved RE1s fall into two categories; first, there are sites which have aligned sequence in mouse that cannot bind REST (e.g. human/mouse Site C of CACNA1A). Second, there are RE1s which are aligned to gaps in the other species’ genome sequence (e.g. human Site D of CACNA1A). What mechanisms might be responsible for the observed differences in the RE1s of human and mouse? The generation of novel regulatory sequences by random DNA mutation is thought to be important in instances where the motif is short enough that it occurs at high frequency in a given stretch of DNA (13). However, the exponential relationship between ‘waiting time’ (i.e. the average time it takes for a particular motif to appear through random DNA mutation) for a sequence in a promoter-sized DNA sequence and the motif length (8,9) suggests that other mechanisms might be necessary to explain the generation of motifs as long as the RE1. Nevertheless, random DNA mutation would appear to be responsible for the appearance of Site C in the human orthologue of CACNA1A.
We also addressed possible mechanisms of creation of the second type of non-conserved RE1, which appear to be due to insertion. We identified a significant population of duplicated RE1s, principally but not exclusively associated with TEs, which are capable of recruiting REST and have been inserted throughout the human genome. Indeed, a number of such inserted RE1s are to be found proximal to annotated genes. The insertion of functional gene-regulatory motifs by Alu elements has been observed previously (61), and it is likely that insertion of novel gene-regulatory sequences by TEs has been important in the evolution of gene regulation (62). Although we identified RE1s associated with all the major classes of TE, including Alus, LINE1 and LINE2 elements, high-scoring RE1s are more strongly associated with ancient LINE2 elements than expected by chance and occur in its coding sequence. We infer that LINE2-mediated RE1 duplication was an important agent of RE1 creation in the human lineage. In support of this, the genome of the fish Fugu rubripes, from which the human lineage diverged before the historical period of LINE2 activity 100–200 million years ago, contains ∼3-fold fewer consensus RE1s than either human or mouse (554 versus 1892 or 1894, respectively) (26). Such results point to episodic transposon-mediated duplication as an important mechanism by which the REST regulon has acquired new targets over evolutionary history.
Through the identification of RE1s in two mammalian species, we have produced evidence for the evolutionary change in the regulon of an essential transcription factor. Since the majority of REST target genes are neuronal-specific, this provides an insight into how genome evolution may have given rise to the differential gene expression regimes observed in human brain compared to closely related species (63,64). Intriguingly, many neuronal genes with rapidly evolving coding sequences have been identified as REST targets by this study (65), while evolutionary changes in brain developmental processes, in which REST plays a central role, have been important drivers of brain evolution (66). Therefore it is conceivable that the mutation and insertion of RE1s we have observed have played important roles in vertebrate brain evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank Dr Gos Micklem (University of Cambridge) for reviewing the manuscript, Dr Michael I. Sadowski (University College, London) for help with the RE1 database, as well as Professor Haig H. Kazazian (University of Pennsylvania), Megan L. Cooper (University of Leeds) and Dr Nikolai D. Belyaev (University of Leeds) for advice and discussions. This work was supported by the Wellcome Trust. R.J. and L.O. are Wellcome Trust PhD students. R.G. is a BBSRC PhD student. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interst statement. None declared.
REFERENCES
- 1.Cooper G.M., Stone E.A., Asimenos G., NISC Comparative Sequencing Program, Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dermitzakis E.T., Clark A.G. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 3.Dermitzakis E.T., Bergman C.M., Clark A.G. Tracing the evolutionary history of Drosophila regulatory regions with models that identify transcription factor binding sites. Mol. Biol. Evol. 2003;20:703–714. doi: 10.1093/molbev/msg077. [DOI] [PubMed] [Google Scholar]
- 4.Costas J., Casares F., Vieira J. Turnover of binding sites for transcription factors involved in early Drosophila development. Gene. 2003;310:215–220. doi: 10.1016/s0378-1119(03)00556-0. [DOI] [PubMed] [Google Scholar]
- 5.Smith N.G.C., Brandstrom M., Ellegren H. Evidence for turnover of functional noncoding DNA in mammalian genome evolution. Genomics. 2004;84:806–813. doi: 10.1016/j.ygeno.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Wittkopp P.J., Haerum B.K., Clark A.G. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 7.Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 8.Berg J., Willmann S., Lassig M. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone J.R., Wray G.A. Rapid evolution of cis-regulatory sequences via local point mutations. Mol. Biol. Evol. 2001;18:1764–1770. doi: 10.1093/oxfordjournals.molbev.a003964. [DOI] [PubMed] [Google Scholar]
- 10.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 11.Barash Y., Elidan G., Friedman N., Kaplan T. Modeling dependencies in protein–DNA binding sites. Proceedings of the Seventh Annual International Conference on Computational Biology; 10–13 April; Berlin, Germany: ACM press; 2003. pp. 28–37. [Google Scholar]
- 12.King O.D., Roth F.P. A non-parametric model for transcription factor binding sites. Nucleic Acids Res. 2003;31:e116. doi: 10.1093/nar/gng117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll S.B., Grenier J.K., Weatherbee S.D. From DNA To Diversity: Molecular Genetics And The Evolution Of Animal Design. 2nd edn. Blackwell, Oxford; 2004. [Google Scholar]
- 14.Berezikov E., Guryev V., Plasterk R.H.A., Cuppen E. CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting. Genome Res. 2004;14:170–178. doi: 10.1101/gr.1642804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z.-F., Paquette A.J., Anderson D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nature Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 17.Calderone A., Jover T., Noh K.-m., Tanaka H., Yokota H., Lin Y., Grooms S.Y., Regis R., Bennett M.V.L., Zukin R.S. Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T., et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nature Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 19.Kuwahara K., Saito Y., Takano M., Arai Y., Yasuno S., Nakagawa Y., Takahashi N., Adachi Y., Takemura G., Horie M., et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong A,B.A., Li J., Kumar B., Sukumar P., Munsch C., Buckley N.J., Neylon C.B., Porter K.E., Beech D.J., Wood I.C. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell. 2005;20:45–52. doi: 10.1016/j.molcel.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 21.Roopra A., Sharling L., Wood I.C., Briggs T., Bachfischer U., Paquette A.J., Buckley N.J. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3–histone deacetylase complex. Mol. Cell. Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andres M.E., Burger C., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battaglioli E., Andres M.E., Rose D.W., Chenoweth J.G., Rosenfeld M.G., Anderson M.E., Mandel G. REST repression of neuronal genes requires components of the hSWI SNF complex. J. Biol. Chem. 2002;277:41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- 24.Roopra A., Qazi R., Schoenike B., Daley T.J., Morrison J.F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Schoenherr C.J., Paquette A.J., Anderson D.J. Identification of potential target genes for the neuron-restrictive silencer factor. PNAS. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce A.W., Donaldson I.J., Wood I.C., Yerbury S.A., Sadowski M.I., Chapman M., Gottgens B., Buckley N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl Acad. Sci. USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osada R., Zaslavsky E., Singh M. Comparative analysis of methods for representing and searching for transcription factor binding sites. Bioinformatics. 2004;20:3516–3525. doi: 10.1093/bioinformatics/bth438. [DOI] [PubMed] [Google Scholar]
- 28.Schoenherr C.J., Anderson D.J. The neuron-restrictive silencing factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 29.Wood I.C., Roopra A., Buckley N.J. Neural specific expression of the m4 muscarinic acetylcholine receptor gene is mediated by a RE1/NRSE-type silencing element. J. Biol. Chem. 1996;271:14221–14225. doi: 10.1074/jbc.271.24.14221. [DOI] [PubMed] [Google Scholar]
- 30.Catterall W.A., Striessnig J., Snutch T.P., Perez-Reyes E. International Union of Pharmacology. XL. Compendium of voltage-gated ion channels: calcium channels. Pharmacol. Rev. 2003;55:579–581. doi: 10.1124/pr.55.4.8. [DOI] [PubMed] [Google Scholar]
- 31.Diriong S., Lory P., Williams M.E., Ellis S.B., Harpold M.M., Taviaux S. Chromosomal localization of the human genes for [alpha]1A, [alpha]1B, and [alpha]1E voltage-dependent Ca2+ channel subunits. Genomics. 1995;30:605–609. doi: 10.1006/geno.1995.1284. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa K., Fujigasaki H., Saegusa H., Ohwada K., Fujita T., Iwamoto H., Komatsuzaki Y., Toru S., Toriyama H., Watanabe M., et al. Abundant expression and cytoplasmic aggregations of α1A voltage-dependent calcium channel protein associated with neurodegeneration in spinocerebellar ataxia type 6. Hum. Mol. Genet. 1999;8:1185–1193. doi: 10.1093/hmg/8.7.1185. [DOI] [PubMed] [Google Scholar]
- 33.Terwindt G.M., Ophoff R.A., van Eijk R., Vergouwe M.N., Haan J., Frants R.R., Sandkuijl L.A., Ferrari M.D. Involvement of the CACNA1A gene containing region on 19p13 in migraine with and without aura. Neurology. 2001;56:1028–1032. doi: 10.1212/wnl.56.8.1028. [DOI] [PubMed] [Google Scholar]
- 34.Chioza B., Wilkie H., Nashef L., Blower J., McCormick D., Sham P., Asherson P., Makoff A.J. Association between the {α};1a calcium channel gene CACNA1A and idiopathic generalized epilepsy. Neurology. 2001;56:1245–1246. doi: 10.1212/wnl.56.9.1245. [DOI] [PubMed] [Google Scholar]
- 35.Jodice C., Mantuano E., Veneziano L., Trettel F., Sabbadini G., Calandriello L., Francia A., Spadaro M., Pierelli F., Salvi F., et al. Episodic ataxia type 2 (EA2) and spinocerebellar ataxia type 6 (SCA6) due to CAG repeat expansion in the CACNA1A gene on chromosome 19p. Hum. Mol. Genet. 1997;6:1973–1978. doi: 10.1093/hmg/6.11.1973. [DOI] [PubMed] [Google Scholar]
- 36.Stormo G.D., Hartzell G.W., III Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl Acad. Sci. USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hertz G.Z., Hartzell G.W., III, Stormo G.D. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- 38.Wood I.C., Belyaev N.D., Bruce A.W., Jones C., Mistry M., Roopra A., Buckley N.J. Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J. Mol. Biol. 2003;334:863–874. doi: 10.1016/j.jmb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Andrews N.C., Faller D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraner S.D., Chong J.A., Tsay H.-J., Mandel G. Silencing the Type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Cheong A., Bingham A.J., Li J., Kumar B., Sukumar P., Munsch C., Buckley N.J., Neylon C.B., Porter K.E., Beech D.J., et al. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell. 2005;20:45–52. doi: 10.1016/j.molcel.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 43.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y.-M., Greenway D.J., Johnson R., Street M., Belyaev N.D., Deuchars J., Bee T., Wilde S., Buckley N.J. Distinct Profiles of REST Interactions with Its Target Genes at Different Stages of Neuronal Development. Mol. Biol. Cell. 2005;16:5630–5638. doi: 10.1091/mbc.E05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout Neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 46.Paquette A.J., Perez S.E., Anderson D.J. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl Acad. Sci. USA. 2000;97:12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugino K., Hempel C.M., Miller M.N., Hattox A.M., Shapiro P., Wu C., Huang Z.J., Nelson S.B. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nature Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X., Odom D.T., Koo S.-H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl Acad. Sci. USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martone R., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl Acad. Sci. USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lander E., Linton M., Birren B., Nusbaum C., Zody M., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi E., Murata Y., Oki T., Miyamoto N., Mori Y., Takada N., Wanifuchi H., Wanifuchi N., Yagami K., Niidome T. Isolation and functional characterization of the 5′-upstream region of mouse P/Q-type Ca2+ channel alpha1A subunit gene. Biochem. Biophys. Res. Commun. 1999;260:54–59. doi: 10.1006/bbrc.1999.0857. [DOI] [PubMed] [Google Scholar]
- 52.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rockman M.V., Hahn M.W., Soranzo N., Zimprich F., Goldstein D.B., Wray G.A. Ancient and recent positive selection transformed Opioid cis-regulation in humans. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapitonov V.V., Pavlicek A., Jurka J. Anthology of Human Repetitive DNA. Wiley, NY: 2004. [Google Scholar]
- 58.Palm K., Belluardo N., Metsis M., Timmusk T.o. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas E.E., Srebro N., Sebat J., Navin N., Healy J., Mishra B., Wigler M. Distribution of short paired duplications in mammalian genomes. Proc. Natl Acad. Sci. USA. 2004;101:10349–10354. doi: 10.1073/pnas.0403727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 61.Shankar R., Grover D., Brahmachari S., Mukerji M. Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol. Biol. 2004;4:37. doi: 10.1186/1471-2148-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hedges D.J., Batzer M.A. From the margins of the genome: mobile elements shape primate evolution. BioEssays. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 63.Enard W., Khaitovich P., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 64.Caceres M., Lachuer J., Zapala M.A., Redmond J.C., Kudo L., Geschwind D.H., Lockhart D.J., Preuss T.M., Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dorus S., Vallender E.J., Evans P.D., Anderson J.R., Gilbert S.L., Mahowald M., Wyckoff G.J., Malcom C.M., Lahn B.T. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 66.Gilbert S.L., Dobyns W.B., Lahn B.T. Genetic links between brain development and brain evolution. Nature Rev. Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]