Abstract
Proteins containing the 21st amino acid selenocysteine (Sec) are present in the three domains of life. However, within lower eukaryotes, particularly parasitic protists, the dependence on the trace element selenium is variable as many organisms lost the ability to utilize Sec. Herein, we analyzed the genomes of Trypanosoma and Leishmania for the presence of genes coding for Sec-containing proteins. The selenoproteomes of these flagellated protozoa have three selenoproteins, including distant homologs of mammalian SelK and SelT, and a novel multidomain selenoprotein designated SelTryp. In SelK and SelTryp, Sec is near the C-terminus, and in all three selenoproteins, it is within predicted redox motifs. SelTryp has neither Sec- nor cysteine-containing homologs in the human host and appears to be a Kinetoplastida-specific protein. The use of selenium for protein synthesis was verified by metabolically labeling Trypanosoma cells with 75Se. In addition, genes coding for components of the Sec insertion machinery were identified in the Kinetoplastida genomes. Finally, we found that Trypanosoma brucei brucei cells were highly sensitive to auranofin, a compound that specifically targets selenoproteins. Overall, these data establish that Trypanosoma, Leishmania and likely other Kinetoplastida utilize and depend on the trace element selenium, and this dependence is due to occurrence of selenium in at least three selenoproteins.
INTRODUCTION
Trypanosomatids are a group of exclusively parasitic kinetoplastid protozoa, which are responsible for several major human diseases. The most notable of these are sleeping sickness and South American Chagas' disease, caused by Trypanosoma spp., and the different forms of leishmaniasis, caused by Leishmania spp. Sleeping sickness is endemic in certain regions of Sub-Saharan Africa that encompass 36 countries and 60 million people. It is estimated that 300–500 thousand people are infected and 40 000 die every year of this disease. According to the World Health Organization, Chagas' disease currently affects 16–18 million people, particularly in the South America. Leishmaniasis adds another 12 million people living in 88 different countries (1–3).
Recently, significant efforts have been placed on genome sequencing and annotation of both Trypanosoma and Leishmania, and several completely sequenced genomes of these organisms are currently available (4–6). Correct genome annotation and understanding of protein functions in these organisms are considered crucial for drug development and disease prevention (7,8). However, the use of existing annotation tools did not result in identification of genes coding for selenocysteine (Sec)-containing proteins because Sec, the 21st naturally occurring amino acid in the genetic code, is encoded by UGA, one of three signals that terminate protein synthesis (9,10). Leishmania major (accession no. AAG35734), Trypanosoma cruzi (XM_805940) and Trypanosoma brucei (XP_823164) were reported to contain a gene coding for a homolog of selenophosphate synthetase, an enzyme that generates selenophosphate, a selenium donor compound used for biosynthesis of Sec (11). However, whether this protein is functional in L.major is not known, and in addition, selenophosphate synthetase is also involved in pathways other than Sec biosynthesis (12). Thus, whether Leishmania or other Kinetoplastida utilize Sec remains unknown.
Sec is inserted into nascent polypeptides with the help of an RNA structure, designated Sec insertion sequence (SECIS) element (9). We previously reported that SECIS elements in closely related species show a significant level of homology and that evolutionary criteria could be applied to carry out computational searches with increased specificity for these structures (13). Since multiple completely sequenced Kinetoplastida genomes are available, simultaneous analysis of these genomes could help identify selenoprotein genes encoded in these organisms.
Identification of selenoprotein genes in Kinetoplastida is also interesting from an evolutionary point of view: although selenoproteomes have been previously identified in the three domains of life, lower eukaryotes show variable dependence on selenium, with yeast and higher plants lacking selenoproteins (14). Few attempts have been made to explore a larger protozoan community (15,16). Thus, information about similarities and differences among protozoan selenoproteomes may provide valuable insights into evolution of selenium utilization.
In this study, we carried out bioinformatics analyses and identified three selenoprotein genes in Kinetoplastida. Two of them correspond to already known SelK and SelT families, while the third selenoprotein showed no homology to known proteins and thus represents a new selenoprotein family. Metabolic labeling of cells with 75Se and auranofin inhibition studies supports these in silico findings. These data are discussed with respect to the dependence of Kinetoplastida on selenium.
MATERIALS AND METHODS
Databases and programs
Nucleotide sequences of Trypanosoma congolense, T.cruzi, Trypanosoma vivax, Trypanosoma gambiense, Trypanosoma brucei brucei, L.major, Leishmania infantum and Leishmania braziliensis genomes, as well as predicted proteins sequences, were downloaded from The Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). SECISearch 2.19 (14) was used for identification of SECIS elements. FASTA package (17) and BLAST were used for similarity search. An online version of MFOLD version 3.2 (18) was used for RNA secondary structure prediction and preliminary analysis of SECIS-like structures. ClustalX was used for calculation of distances between each pair of sequences used to construct the phylogenetic tree.
Identification of distant homologs of known selenoprotein genes
In the search for homologs of known selenoprotein, query sequences were represented by Chlamydomonas MsrA (19,20), four Plasmodium falciparum selenoproteins (16), Gallus gallus SelU (21), protein disulfide isomerase from Emiliania huxleyi (22) and the full set of human selenoproteins (14). A stand-alone version of TBLASTN was used to detect nucleotide sequences corresponding to known selenoprotein families. Downstream regions of detected sequences were analyzed for presence of SECIS elements with SECISearch. In addition, nucleotide sequences were analyzed with MFOLD (18) to identify SECIS-like structures. All SECIS-like structures were screened for compliance with elements of the current SECIS consensus model (e.g. a non-Watson–Crick quartet in the SECIS core and unpaired AA, AG or CC nucleotides in the apical loop).
Searches for SECIS elements
The default pattern of SECISearch was modified to accommodate Trypanosoma SECIS elements identified with the loose pattern of SECISearch and those not detectable by SECISearch but identified by manual searches with MFOLD. The modifications were as follows: (i) the threshold of the free energy of the overall structure was −11.5 kcal/mol, (ii) the minimum length of the stem was 10 bp and (iii) the apical loop was 3–17 nt. Genomic sequences of Kinetoplastida were searched using this modified version of SECISearch. Details of the procedures have been previously described (14,16). Briefly, nucleotide sequences were identified in the genome that meet primary and secondary sequence/structure requirements, satisfy free energy criteria and pass additional structural filters.
All SECIS candidates were analyzed for the presence of at least one homolog in other Kinetoplastida species by searches against a database containing SECIS candidates from other Kinetoplastida species. To align SECIS sequences, we used FASTA with an E-value of 1 × 10−8. Regions upstream of SECIS elements were further analyzed for occurrence of open reading frames (ORFs). An additional requirement was the presence of at least two homologous ORFs in Kinetoplastida. Candidates, in which SECIS elements and ORFs were on different DNA strands, were filtered out.
Cultivation of bloodstream and procyclic T.brucei brucei
For the auranofin experiments, culture-adapted bloodstream and procyclic T.brucei of the cell line 449 [descendants of strain Lister 427 (23) and stably transfected with pHD449 encoding the tetracycline repressor (24)] were used. Bloodstream T.brucei brucei was grown at 37°C in a humidified atmosphere with 5% CO2 in HMI-9 medium supplemented with 1.5 mM cysteine, 0.0014% (v/v) β-mercaptoethanol, 10% heat-inactivated fetal calf serum (FCS) (v/v), 50 U/ml penicillin, 50 μg/ml streptomycin and 0.2 μg/ml phleomycin. Procyclic T.brucei were grown in MEM-Pros medium (Biochrom) supplemented with 7.5 μg/ml hemin, 10% heat-inactivated FCS (v/v), 50 U/ml penicillin, 50 μg/ml streptomycin and 0.5 μg/ml phleomycin at 27°C.
Auranofin inhibition studies
Auranofin, a highly specific inhibitor of several selenoenzymes (25,26) was tested as an inhibitor of trypanosomal growth. A total of 1 × 105 bloodstream trypanosomes per ml or 2 × 105 procyclic parasites per ml were cultured as described above in the presence of 1 nM to 10 μM of auranofin dissolved in dimethyl sulfoxide (DMSO). To rule out that DMSO by itself might influence growth of cells, an additional control experiment without DMSO was carried out in parallel. A sample without auranofin but with an identical volume of DMSO served as an additional control. After 18 h, the cells were counted in a Neubauer chamber. No growth difference could be detected between these two negative controls. All experiments were carried out in duplicate.
Metabolic labeling with 75Se
T.cruzi (Tulahuen-2 strain) epimastigotes (proliferative and extracellular stage) were cultured at 28°C in BHI medium [33 mg/ml brain-heart infusion (Difco)] supplemented with 3 mg/ml tryptose, 20 μg/ml hemin, 5 mM KCl and 25 mM sodium phosphate, complement-inactivated 10% fetal bovine serum (v/v), 1.7 mM glucose, 200 μg/ml streptomycin sulfate and 200 U/ml penicillin at pH 7.3. A total of 100 ml exponential-phase parasites (2 × 107 cells per ml) were harvested by centrifugation at 800 g (Sorvall RC5Cplus, rotor F21S FiberLite) and washed twice with DMEM (Dulbecco's Modified Eagle Medium, SIGMA), without fetal bovine serum but supplemented with 1.5 mM l-glutamine, 5.6 mM glucose, 45 mM sodium bicarbonate, 200 μg/ml streptomycin sulfate and 200 U/ml penicillin. The collected parasites were resuspended in 5 ml of DMEM and cultured for 40 h at 28°C in the presence of 400 μCi of 75Se provided as 5 μM [75Se]selenite (University of Missouri Research Reactor). After 40 h, cells were harvested by centrifugation, washed twice, resuspended in 500 μl of phosphate-buffered saline (PBS) containing 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM (2S,3S)-3-(N-{(S)-1-[N-(4-guanidinobutyl) carbamoyl] 3-methylbutyl}carbamoyl)oxirane-2-carboxylic acid (E-64), and sonicated. A total of 10–20 μg of protein from parasite cell homogenates were subjected to 10% SDS–PAGE under reducing conditions and transferred onto a polyvinylidine difluoride (PVDF) membrane. Proteins were stained with Coomassie blue and radioactivity was visualized by autoradiography using a PhosphorImager (Fuji).
RESULTS AND DISCUSSION
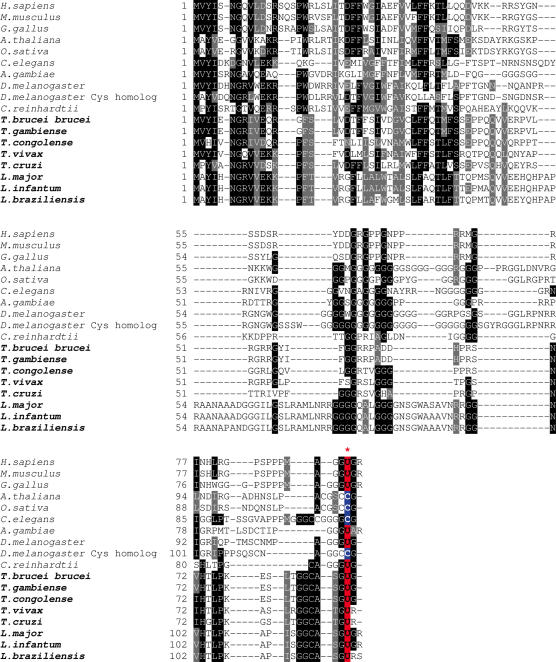
To identify selenoprotein genes in Trypanosoma and Leishmania, we initially searched available sequenced genomes of these organisms for occurrence of homologs of known selenoprotein genes with TBLASTN. Two selenoprotein families were found in several Kinetoplastida genomes, including homologs of human SelK (accession no. Q9Y6D0) and SelT (AAH26350).
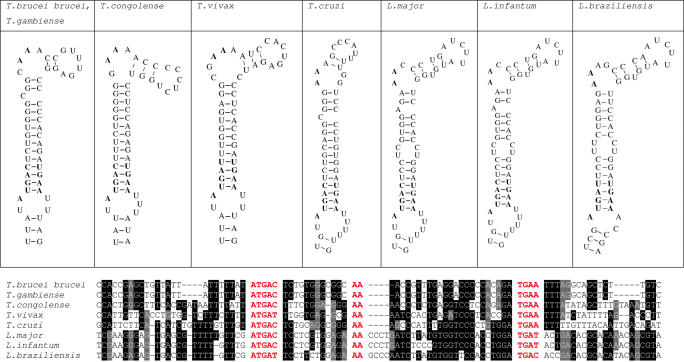
Alignments of SelK SECIS elements and protein sequences are shown in Figures 1 and 2. All SECIS elements in the SelK family could be found using the default pattern of SECISearch. In addition, both SECIS elements (Figure 1) and protein sequences (Figure 2) were highly conserved among Trypanosomas and Leishmania. Thus, the SelK SECIS elements found in Kinetoplastida sequences fit very well the eukaryotic SECIS consensus model.
Figure 1.
Structures and nucleotide sequence alignment of SelK SECIS elements. Functionally important nucleotides in the apical loop and the Quartet (SECIS core) are shown in bold (in the structure) or in red (in the alignment). Conserved nucleotides are highlighted.
Figure 2.
Amino acid sequence alignment of SelK sequences. Sec (indicated by U) is shown in red, and the corresponding Cys in blue. The position of these residues in the sequences is indicated by a red star above the sequences. Conserved residues are highlighted. ORFs were predicted in the following sequences: 4 265 315.c000313905.Contig1 (T.congolense), Tb927_10_v4 (T.brucei brucei), 1 585 712.c000312726.Contig1 (T.gambiense), Contig8734 (T.vivax), Tcruzi.chrunknown.4757 (T.cruzi), LmjF36_01_20040630_V4.0 (L.major), LI0706f02.p1k (L.infantum), brazil1129f08.q1k (L.braziliensis).
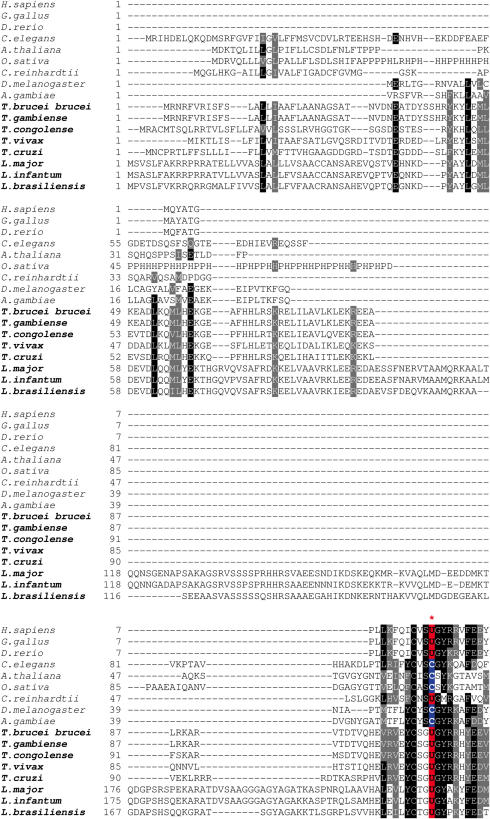
In contrast, SECIS elements in SelT genes (Figure 3) differed from the typical SECIS structure. While SelT protein sequences showed high conservation rate among Kinetoplastida (Figure 4), SECIS elements were much less conserved, and identification of several SelT SECIS elements posed a challenge even with the loose pattern of SECISearch.
Figure 3.
Structures and nucleotide sequence alignment of SelT SECIS elements. Functionally important nucleotides in the apical loop and the Quartet (SECIS core) are shown in bold (in the structure) or in red (in the alignment). Conserved nucleotides are highlighted. Separate alignments for Trypanosoma and Leishmania are shown due to lack of homology between SECIS elements from these two groups of organisms.
Figure 4.
Amino acid sequence alignment of SelT sequences. Conserved residues are highlighted. Sec (indicated by U) is shown in red, and the corresponding Cys in blue. The following sequences were used to construct the alignment: tviv326d03.p1k_1 and tviv326d03.p1k_2 (T.vivax), gamb21f07.q1k_2 (T.gambiense), Tb927.5.860 and Tb927.5.870 (T.brucei brucei), congo270d08.q1k_5 and congo270d08.q1k_6 (T.congolense), Tc00.1047053505163.60 and Tc00.1047053505163.70 (T.cruzi), LmjF35.1110 (L.major), LI0881h10.q1k (L.infantum), brazil1006d02.p1k and brazil74d01.q1k (L.braziliensis).
To adjust SECISearch for identification of Trypanosoma SECIS elements, we developed a modified version of the program (as described in Materials and Methods), which significantly improved specificity of the searches. Application of this program resulted in the detection of all SelT SECIS elements except that in the T.cruzi SelT gene. Interestingly, predicted SECIS elements in L.major and L.infantum SelT genes drastically differed from their T.gambiense, T.congolense and T.vivax counterparts.
We further applied the modified version of SECISearch to analyze entire genomes and genome survey sequences of T.congolense, T.cruzi, T.vivax, T.gambiense, T.brucei brucei, L.major, L.infantum and L.braziliensis. An additional requirement was the presence of SECIS homologs in other Kinetoplastida (cut-off value of 1 × 10−8 as determined from FASTA alignments of Trypanosoma and Leishmania SECIS elements in SelK and SelT genes). However, L.major, L.infantum and L.brasiliensis genomes exhibited very high sequence similarity; therefore, only L.major was included in the searches. Upstream regions of SECIS element candidates were analyzed for the presence of at least one homolog in other Kinetoplastida species. Predicted ORFs were then searched against NCBI non-redundant protein database, as well as against predicted Kinetoplastida proteins.
This analysis identified six homologous groups of candidates, including homologs of SelK and SelT, as well as a new selenoprotein family designated SelTryp (Table 1). Two other candidates were filtered out because SECIS elements and ORFs were on different DNA strands, and the last SECIS candidate corresponded to a predicted ORF with no suitable in-frame TGA triplet.
Table 1.
Computational analysis of Kinetoplastida genomes for SECIS elements and selenoprotein genes
| Organism | Database size, bp | Number of SECIS candidates that satisfy primary and secondary structure criteria | Number of SECIS candidates that satisfy free energy criteria | Selenoproteome |
|---|---|---|---|---|
| T.congolense | 36 631 425 | 5482 | 1646 | SelK, SelT, SelTryp |
| T.vivax | 52 907 595 | 5851 | 1848 | SelK, SelT, SelTryp |
| T.brucei brucei | 29 866 766 | 4601 | 1109 | SelK, SelT, SelTryp |
| T.gambiense | 29 310 016 | 4504 | 1106 | SelK, SelT, SelTryp |
| L.major | 38 793 468 | 4111 | 2334 | SelK, SelT, SelTryp |
| L.infantum | 194 921 525 | 18 267 | 9992 | SelK, SelT, SelTryp |
| L.braziliensis | 129 419 413 | 11 982 | 5861 | SelK, SelT |
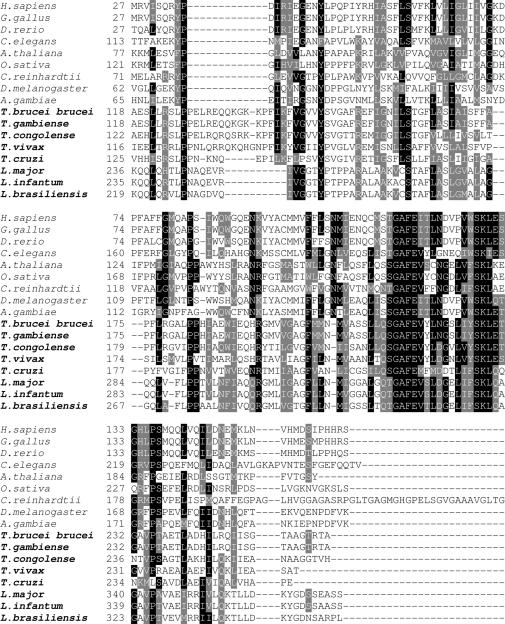
SelTryp SECIS elements, their alignments, and alignments of the corresponding selenoproteins are shown in Figures 5 and 6. Like SelT structures, SelTryp SECIS elements were conserved within Trypanosoma and within Leishmania, but little conservation was detected between Trypanosoma and Leishmania SelTryp SECIS elements (Figure 5).
Figure 5.
Structures and nucleotide sequence alignments of SelTryp SECIS elements. Functionally important nucleotides in the apical loop and the Quartet (SECIS core) are shown in bold (in the structure) or in red (in the alignment). Conserved nucleotides are highlighted. Separate alignments for Trypanosoma and Leishmania are shown due to lack of homology between SECIS elements from these two groups of organisms.
Figure 6.
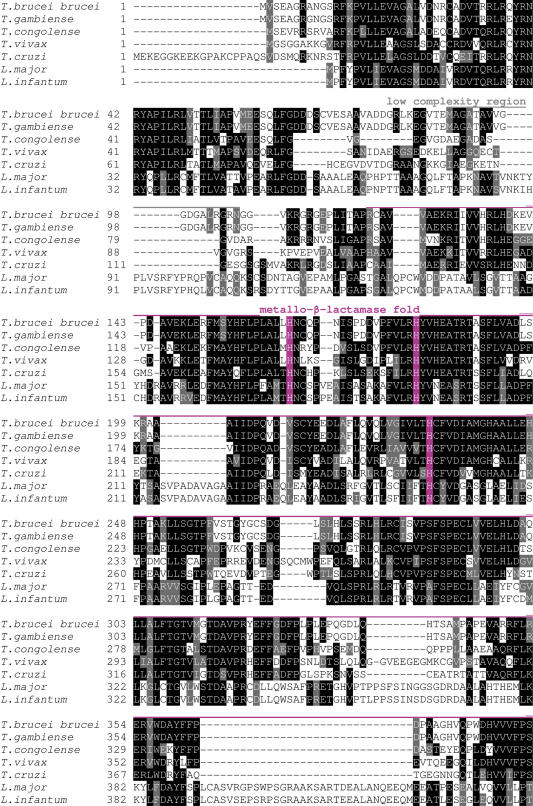
Amino acid sequence alignment of SelTryp sequences. Conserved residues are highlighted. Sec (indicated by U) is shown in red. Sequences used in the alignment were as follows: Tc00.1047053507485.100 (T.cruzi), tviv195d03.q1k_2 (T.vivax), Tb927.4.3410 (T.brucei brucei), gamb564d12.p1k_15 (T.gambiense), congo936h09.q1k_1 (T.congolense), LmjF34.0950 (L.major), LinJ34.0860 (L.infantum). Location of rhodanese domains, metallo-β-lactamase domain and the CxxU motif is indicated above the sequences. Active site cysteines in rhodanese domains are highlighted in blue and additional conserved cysteines in SelTryps in green. Conserved histidines that may be involved in metal coordination are shown in pink.
In the SelTryp ORF, Sec was present in the C-terminal region, within a conserved C-terminal peptide, SI(V)I(V)CI(V)SUPR (U is Sec). Although in known selenoproteins Sec is most often found in loops located between secondary structures, the C-terminal location is also common to eukaryotic selenoproteins (e.g. thioredoxin reductase, SelK, SelS and SelO). In SelTryp, Sec is present within a CxxU motif, which is often found in selenoproteins that carry our redox function through reversible formation of a selenenylsulfide bond. This observation suggests a redox function for the CxxU motif in SelTryp.
Analysis of SelTryp sequences revealed distant homology to a rhodanese-like protein from Thermobifida fusca YX (accession no. YP_291141). Two rhodanese homology domains (RHOD) could be seen in SelTryp by CD-Search (27) in the region spanning 500–585 and 621–774 amino acids. An additional analysis of amino acid sequences using SMART (28), AnDom (29) and PROSITE (30) predicted the occurrence of a metallo-β-lactamase fold (Figure 6). The presence of conserved cysteines in the rhodanese domains within a 6 amino acid active site loop (CXGGXR) suggested that this protein belonged to a YceA subfamily (31). In addition, the use of DisEMBL (32) revealed the lack of secondary structures in the C-terminal region of SelTryp, suggesting a flexible C-terminal Sec-containing tail.
The N-terminal sequences of SelTryp belong to a metallo-β-lactamase superfamily of proteins and are followed with rhodanese domains. Proteins with the metallo-β-lactamase fold catalyze a wide variety of reactions, partly because this fold allows selectivity for different metals. For example, hydrolytic metallo-β-lactamase proteins mostly bind zinc, redox-active rubredoxin:oxygen-oxidoreductases contain a di-iron cluster, and glyoxalases II (thiolesterases) contain iron, manganese or zinc (33).
Subcellular localization predictions using iPSORT [(34), http://hc.ims.u-tokyo.ac.jp/iPSORT/], TargetP 1.1 [(35), http://www.cbs.dtu.dk/services/TargetP/] and PredictProtein [(36), http://www.predictprotein.org/) suggested a mitochondrial localization of SelTryp, whereas the SelT sequence contains a potential export signal. In good agreement with its human homolog, the Kinetoplastida SelK has a predicted transmembrane motif.
All selenoprotein-coding genes that were found in Leishmania and Trypanosoma lacked introns. In Leishmania selenoproteins, the distance from Sec-encoding UGA codons to SECIS elements was between 800 bp (SelTryp) and 1100 bp (SelK). In Trypanosoma selenoprotein genes, this distance was more variable: ∼200 bp for SelK and SelTryp, and 850 bp for SelT.
Analysis of selenoprotein ORFs against annotated Leishmania and Trypanosoma genomes revealed that SelK genes were not annotated at all, except for the T.cruzi SelK gene, for which a wrong ORF was predicted. SelTryp genes were misannotated because the in-frame UGA codons were interpreted as stop signals. SelT genes were split into two parts. One part corresponded to the N-terminal regions of the proteins and was predicted to terminate at the Sec-encoding UGA codons, and the second was predicted to initiate from an AUG codon, which corresponded to the internal methionine downstream of the Sec UGA codon, continuing until the true stop signal.
In addition to protein components of the Sec insertion machinery (selenophosphate synthetase, Sec tRNA-specific elongation factor), Sec tRNA genes were identified. Although tRNAscan-SE used with default settings (37) failed to recognize Sec tRNA genes in L.major, the use of ARAGORN (38) identified Sec tRNAs in Trypanosoma and Leishmania genomes. The predicted Sec tRNAs were then successfully verified using COVE with a Sec tRNA profile. All Sec tRNAs in Kinetoplastida could also be found with a tool adapted for unusual tRNAs (39). All Sec insertion machinery genes and selenoprotein genes were located either on different chromosomes or, when on the same chromosome, they were distant from each other. Thus, although Kinetoplastida form operon-like structures, the Sec insertion trait genes are spread throughout the parasite genomes.
As a preliminary test to verify that these sequences are indeed expressed as predicted we exploited two peculiarities of Trypanosomes. First, mature T.brucei brucei mRNA molecules share a common 5′ sequence (the so called spliced leader, SL). Thus, a common forward primer can be used for PCR on a T.brucei brucei cDNA preparation and subsequent sequencing allows verification that a predicted start codon is correct and to rule out that our sequence is a part of a larger one. Second, as trypanosomal protein expression is primarily regulated at the transcriptional level, the detection of the respective mRNA (via cDNA) is essentially a proof that the respective protein is expressed. Using this approach, we were able to identify unequivocally the respective mRNA of SelT and SelK in T.brucei brucei (data not shown).
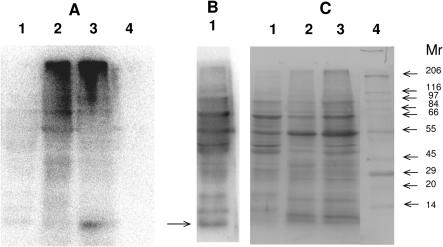
To directly test if selenium is inserted into Trypanosoma proteins, we metabolically labeled Trypanosoma cells with 75Se. Following SDS–PAGE, the 75Se profile was visualized with a PhosphorImager. A major 75Se-containing high-molecular weight band was detected at the top of the gel (Figure 7). This 75Se species was insoluble (Figure 7A), but selenium could be partially released by treatment with urea and high concentrations of reducing agents (data not shown). This band did not correspond to the three selenoproteins, and in fact this form of selenium has not been previously observed in other species. Determination of the nature of the high-molecular weight selenium species should await further studies.
Figure 7.
Selenoprotein expression in T.cruzi. T.cruzi epimastigote cells were radiolabeled with 75Se and collected after 40 h of DMEM culture. Proteins were electrophoresed under reducing conditions on a 10% SDS–PAGE gel, transferred onto a PVDF membrane, analyzed by PhosphorImager (A and B) exposure time 30 min and 72 h, respectively and stained with Coomassie blue (C). Predicted molecular masses of T.cruzi selenoproteins were 9.5 kDa (SelK), 28.8 kDa (SelT) and 89 kDa (SelTryp). Lane 1: supernatant of epimastigote cells after sonication; lane 2: resuspended pellet after epimastigote sonication; lane 3 total epimastigote extract; lane 4: molecular weight markers (Sigmawide). An arrow on (B) shows a specific selenoprotein band, probably corresponding to SelK.
The soluble fraction of Trypanosoma cell extracts had little 75Se (Figure 7A). However, longer exposure to a PhosphorImager screen revealed a 10 kDa band that migrated in accord with the predicted molecular mass of SelK (Figure 7B). In addition, minor bands that were labeled with 75Se could be detected, but they corresponded to a protein profile detected by Coomassie blue staining (Figure 7C). These additional bands probably derived from non-specific labeling of proteins with selenium wherein this trace element entered sulfur pathways and was inserted in place of sulfur in methionine and cysteine residues. However, the candidate radioactive SelK band had no corresponding protein band, further suggesting that this was a specific selenoprotein band. These data show that selenoproteins are expressed in Trypanosoma cells in the life-cycle stage at low level, and that only some selenoproteins could be visualized by metabolic 75Se labeling. Whereas SelT and SelTryp could not be detected with 75Se, specific 75Se insertion into 10 kDa and high-molecular weight bands verified our prediction of the use of selenium by Trypanosoma cells.
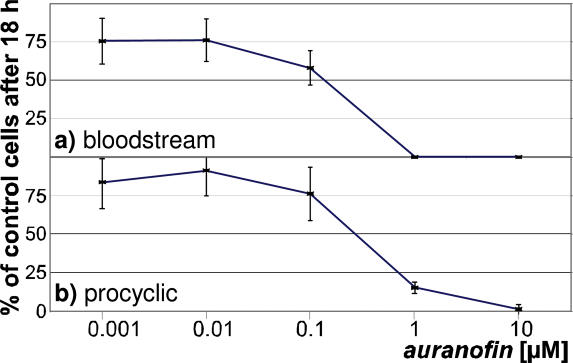
Gold(I) compounds, such as auranofin, are highly specific inhibitors of several eukaryotic selenoenzymes. We therefore studied the impact of auranofin on the growth of T.brucei brucei. As shown in Figure 8, this compound was highly toxic for bloodstream and procyclic stages of the parasite with IC50 values in the lower nanomolar range. This renders auranofin a highly interesting drug candidate per se. It should be noted that the growth medium contains significant amounts of plasma proteins. As auranofin is preferentially bound to plasma proteins (∼60%), it can be assumed that the active (i.e. free) drug concentration in our experiment was even lower (40). In this context, it is important to stress that the three selenoproteins contain putative redox centers (CxxU in the case of SelT and SelTryp, and CxxxU in SelK), and that impaired redox balance may influence Kinetoplastid infections (41). Yet, further studies are required to verify the selenium dependence and determine the concentration of this trace element that is required for viability of the parasites in different developmental stages.
Figure 8.
Effect of auranofin on parasite growth. (a) Bloodstream and (b) procyclic T.brucei brucei (strain 449) cell cultures were incubated with different concentrations (shown on a logarithmic scale) of the gold-compound auranofin. Indicated are the percentages of viable cells after 18 h of incubation compared to a culture in the absence of auranofin. All experiments were done in duplicate. The error bars indicate the standard deviation.
To examine the evolutionary history of the Sec trait in Kinetoplastida, we constructed a phylogenic tree for Sec tRNA (Figure 9). Trypanosoma sequences clustered with other eukaryotes, suggesting a common origin of the Sec insertion system. In the tree, Trypanosoma Sec tRNAs formed a cluster with animal Sec tRNAs and Chlamydomonas Sec tRNA. This cluster was separated from the plasmodial cluster. Together with the finding of a eukaryotic Sec-specific elongation factor, these data suggest that the Sec insertion system is Kinetoplastida is similar to the previously characterized eukaryotic Sec insertion systems. Nevertheless, identification of a protein specific for this group of organisms (SelTryp) highlights the fact that low eukaryotes may possess novel selenoproteins. One recent study revealed four such proteins in Plasmodia (16). With an ever increasing pace at which new genome sequences become completed, further computational analyses should reveal yet additional selenoproteins, and with them new pathways of selenium utilization in biology.
Figure 9.
Phylogenetic tree of Sec tRNAs. Phylogenetic tree of Sec tRNAs was constructed using ClustalX program for alignments of tRNA sequences and calculating the distances between them. TreeView was used for tree visualization.
In conclusion, we carried out an in silico analysis of all available sequenced Kinetoplastida genomes for the presence of selenoprotein genes. By computationally predicting SECIS elements, we characterized the Trypanosoma and Leishmania selenoproteomes, which consist of three selenoproteins. Among them, SelT and SelK were distant homologs of previously identified mammalian selenoproteins. A new selenoprotein, SelTryp, was also discovered. This selenoprotein has two rhodanese and one rubredoxin:oxygen oxidoreductase domains and appears to be a Kinetoplastida-specific multidomain redox protein of unknown function. All selenoprotein genes were previously misannotated in sequence databases. Metabolic labeling of Trypanosoma cells with 75Se revealed specific insertion of this radioisotope into a defined set of proteins, and in addition, Trypanosoma cells were found to be sensitive to a gold(I) compound, auranofin, which specifically targets selenoproteins. These findings, together with the presence of the Sec-decoding trait in Kinetoplastida genomes suggest that these organisms utilize selenium and depend on this trace element, and that this dependence is likely due to the occurrence of at least three selenoproteins in these organisms. The absence of SelTryp homologs (either Sec or Cys forms) in the human host may also be relevant to drug development: selective inhibition of this selenoprotein might lead to new drugs to treat typanosomatid infections. Finally, these findings highlight the fact that lower eukaryotes evolved unique selenoproteomes, whose analysis should suggest new uses of the trace element selenium in biology.
Acknowledgments
The authors would like to thank L. Krauth-Siegel and her group for providing T.brucei brucei strain 449, F. Irigoín for providing T.cruzi epimastigotes and helpful support, T. Irsch for PCR analyses, and U. Göbel for excellent experimental assistance. This work is supported by NIH grant GM061603 and Deutsche Forschungsgemeinschaft grant GR 2028/1-2. Funding to pay the Open Access publication charges for this article was provided by GM061603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rassi A., Jr, Rassi A., Little W.C. Chagas’ heart disease. Clin. Cardiol. 2000;23:883–889. doi: 10.1002/clc.4960231205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sternberg J.M. Human African trypanosomiasis: clinical presentation and immune response. Parasite Immunol. 2004;26:469–476. doi: 10.1111/j.0141-9838.2004.00731.x. [DOI] [PubMed] [Google Scholar]
- 3.Gelb M.H., Hol W.G.J. Drugs to combat tropical protozoan parasites. Science. 2002;297:343–344. doi: 10.1126/science.1073126. [DOI] [PubMed] [Google Scholar]
- 4.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 5.El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 6.Ivens A.C. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krauth-Siegel R.L., Bauer H., Schirmer R.H. Dithiol proteins as guardians of the intracellular redox milieu in parasites: old and new drug targets in Trypanosomes and Malaria-causing Plasmodia. Angew. Chem. Int. Ed. 2005;44:690–715. doi: 10.1002/anie.200300639. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia V., Sinha M., Luxon B., Garg N. Utility of the Trypanosoma cruzi. Sequence database for identification of potential vaccine candidates by in silico and in vitro screening. Infect. Immun. 2004;72:6245–6254. doi: 10.1128/IAI.72.11.6245-6254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low S.C., Berry M.J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends. Biochem. Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 10.Tujebajeva R.M., Copeland P.R., Xu X.M., Carlson B.A., Harney J.W., Driscoll D.M., Hatfield D.L., Berry M.J. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayakumar P.C., Musande V.V., Shouche Y.S., Patole M.S. The selenophosphate synthetase gene from Leishmania major. DNA Seq. 2004;15:66–70. doi: 10.1080/10425170310001623653. [DOI] [PubMed] [Google Scholar]
- 12.Romero H., Zhang Y., Gladyshev V.N., Salinas G. Evolution of selenium utilization traits. Genome Biol. 2005;6:R66. doi: 10.1186/gb-2005-6-8-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kryukov G.V., Gladyshev V.N. Mammalian selenoprotein gene signature: identification and functional analysis of selenoprotein genes using bioinformatics methods. Meth. Enzymol. 2002;347:84–100. doi: 10.1016/s0076-6879(02)47010-3. [DOI] [PubMed] [Google Scholar]
- 14.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 15.Mourier T., Pain A., Barrell B., Griffiths-Jones S. A selenocysteine tRNA and SECIS element in Plasmodium falciparum. RNA. 2005;11:119–122. doi: 10.1261/rna.7185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobanov A.V., Delgado C., Rahlfs S., Novoselov S.V., Kryukov G.V., Gromer S., Hatfield D.L., Becker K., Gladyshev V.N. The Plasmodium selenoproteome. Nucleic Acids Res. 2006;34:496–505. doi: 10.1093/nar/gkj450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novoselov S.V., Rao M., Onoshko N.V., Zhi H., Kryukov G.V., Xiang Y., Weeks D.P., Hatfield D.L., Gladyshev V.N. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu L.H., Wang X.F., Eyal Y., She Y.M., Donald L.J., Standing K.G., Ben-Hayyim G. A selenoprotein in the plant kingdom: mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J. Biol. Chem. 2002;277:25983–25991. doi: 10.1074/jbc.M202912200. [DOI] [PubMed] [Google Scholar]
- 21.Castellano S., Novoselov S.V., Kryukov G.V., Lescure A., Blanco E., Krol A., Gladyshev V.N., Guigo R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obata T., Shiraiwa Y. A novel eukaryotic selenoprotein in the haptophyte alga Emiliania huxleyi. J. Biol. Chem. 2005;280:18462–18468. doi: 10.1074/jbc.M501517200. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham M.P., Vickerman K. Antigenic analysis in the Trypanosoma brucei group, using the agglutination reaction. Trans. R. Soc. Trop. Med. Hyg. 1962;56:48–59. doi: 10.1016/0035-9203(62)90088-3. [DOI] [PubMed] [Google Scholar]
- 24.Biebinger S., Wirtz L.E., Lorenz P., Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 25.Gromer S., Arscott L.D., Williams C.H., Jr, Schirmer R.H., Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme steady state kinetics and inhibition by therapeutic gold compounds. J. Biol. Chem. 1998;273:20096–200101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 26.Alger H.M., Williams D.L. The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase. Mol. Biochem. Parasitol. 2002;121:129–139. doi: 10.1016/s0166-6851(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt S., Bork P., Dandekar T. A versatile structural domain analysis server using profile weight matrices. J. Chem. Inf. Comput. Sci. 2002;42:405–407. doi: 10.1021/ci010374r. [DOI] [PubMed] [Google Scholar]
- 30.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C.J., Hofmann K., Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordo D., Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 2002;3:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linding V., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Schilling O., Wenzel N., Naylor M., Vogel A., Crowder M., Makaroff C., Meyer-Klaucke W. Flexible metal binding of the metallo-β-lactamase domain: glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry. 2003;42:11777–11786. doi: 10.1021/bi034672o. [DOI] [PubMed] [Google Scholar]
- 34.Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 35.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 36.Rost B., Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobanov A.V., Kryukov G.V., Hatfield D.L., Gladyshev V.N. Is there a 23rd amino acid in the genetic code? Trends in Genet. 2006;22:357–360. doi: 10.1016/j.tig.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Snyder R.M., Mirabelli C.K., Crooke S.T. Cellular association, intracellular distribution, and efflux of auranofin via sequential ligand exchange reactions. Biochem. Pharmacol. 1986;35:923–932. doi: 10.1016/0006-2952(86)90078-x. [DOI] [PubMed] [Google Scholar]
- 41.Müller S., Liebau E., Walter R.D., Krauth-Siegel R.L. Thiol-based redox metabolism of protozoan parasites. Trends Parasitol. 2003;19:320–328. doi: 10.1016/s1471-4922(03)00141-7. [DOI] [PubMed] [Google Scholar]