Abstract
GTP hydrolysis catalyzed in the ribosome by a complex of two polypeptide release factors, eRF1 and eRF3, is required for fast and efficient termination of translation in eukaryotes. Here, isothermal titration calorimetry is used for the quantitative thermodynamic characterization of eRF3 interactions with guanine nucleotides, eRF1 and Mg2+. We show that (i) eRF3 binds GDP (Kd = 1.9 μM) and this interaction depends only minimally on the Mg2+ concentration; (ii) GTP binds to eRF3 (Kd = 0.5 μM) only in the presence of eRF1 and this interaction depends on the Mg2+ concentration; (iii) GTP displaces GDP from the eRF1•eRF3•GDP complex, and vice versa; (iv) eRF3 in the GDP-bound form improves its ability to bind eRF1; (v) the eRF1•eRF3 complex binds GDP as efficiently as free eRF3; (vi) the eRF1•eRF3 complex is efficiently formed in the absence of GDP/GTP but requires the presence of the C-terminus of eRF1 for complex formation. Our results show that eRF1 mediates GDP/GTP displacement on eRF3. We suggest that after formation of eRF1•eRF3•GTP•Mg2+, this quaternary complex binds to the ribosomal pretermination complex containing P-site-bound peptidyl-tRNA and the A-site-bound stop codon. The guanine nucleotide binding properties of eRF3 and of the eRF3•eRF1 complex profoundly differ from those of prokaryotic RF3.
INTRODUCTION
Termination of protein synthesis in eukaryotes is governed by three stop codons, UAA, UAG or UGA, at the ribosomal A-site and by two translation termination factors, designated eRF1 (1) and eRF3 (2). eRF1 recognizes all three stop codons in the decoding center via its N-terminal domain (3–6) in which the highly conversed YxCxxxF motif plays a pivotal role in discriminating purine bases in the second and third stop codon positions (7,8). The major function of eRF1 is to transfer a signal from the small to the large ribosomal subunit and to trigger peptidyl-tRNA hydrolysis at the peptidyl-transferase center [reviewed in (9)]. The key role in the termination reaction is played by the invariant GGQ motif located at the tip of the M domain of eRF1 (10–12). The C-terminal domain is not mandatory for eRF1 function in vitro in a simplified model system (13) but in a completely reconstituted in vitro protein synthesizing machinery it is essential (14).
eRF3 is a GTPase whose activity depends entirely on the ribosome and eRF1 (15). Binding of eRF3 to eRF1 as revealed in vivo and in vitro (2,16,17) is mediated by the C-terminal domains of both proteins (18–20).
Several hypotheses have been suggested regarding the possible biological function of eRF3, encoded by an essential gene [reviewed in (21)]. It is proposed that eRF3 helps eRF1 bind to the stop codon-charged ribosome similarly to prokaryotic elongation factor EF-Tu which increases the affinity of the aminoacyl-tRNA to the A-site programmed with a sense codon (22). This hypothesis is based on two observations: (i) eRF3 possesses a significant degree of homology with eEF1A, an analog of prokaryotic EF-Tu (23,24) and (ii) it stimulates peptidyl-tRNA hydrolysis triggered by eRF1 at low mRNA concentrations (15). In early studies on translation termination in eukaryotes (25,26), it was shown that a dimeric RF (in view of later data this was most likely an eRF1•eRF3 complex) bound to the ribosome and in the presence of GTP stimulated peptidyl-tRNA hydrolysis coupled with GTP hydrolysis. It has also been suggested that eRF3 possesses a proofreading function in stop codon decoding by eRF1 (15,21,27). Furthermore, it has been speculated that eRF3 could be a functional analog of RF3 known to facilitate RF1/RF2 release from the ribosome after peptidyl-tRNA hydrolysis (28).
The primary structure of yeast eRF3 has been subdivided into N, M and C regions and it was demonstrated that the variable N region is not mandatory for the eRF3 function in translation termination, while the C region which is highly conserved is functionally essential (23). The structure of the C-terminal part (positions 196–662) of Schizosaccharomyces pombe eRF3 was resolved by X-ray analysis (24). In this sequence truncated of its N-terminus, domain 1 (positions 237–467) binds guanine nucleotides and probably Mg2+, and domain 3 (positions 555–662) binds to the C-terminus of eRF1 as shown earlier (18–20). The function of domain 2 remains unknown.
Mutations of yeast eRF3 that reduce GTPase activity cause an increase in readthrough efficiency in vivo indicating a decrease in translation termination efficiency (27). GTP influences the interaction of eRF1 with eRF3 at physiological Mg2+ concentrations (29). In a completely reconstituted in vitro protein synthesizing system (14) eRF3 together with GTP considerably increase the rate of peptidyl-tRNA hydrolysis promoted by eRF1 which proceeds after GTP hydrolysis within the ribosome. The factors act cooperatively in the termination reaction and it is likely (14) that eRF3 increases the affinity of eRF1 for the ribosome as suggested previously (22).
Lack of sufficient information concerning the interaction between eRF3, eRF1 and guanine nucleotides in vitro hampers deeper understanding of the GTP/GDP cycle involved in translation termination. For this reason, the aim of the present study has been to use isothermal titration calorimetry (ITC) to follow quantitatively complex formation between eRF3 and its ligands comparing the thermodynamic parameters of these complexes. The principal advantage of the ITC approach stems from the fact that it does not require any kind of modifications of the interacting molecules under investigation and experiments are performed at equilibrium avoiding artifacts caused by possible dissociation of the complex in non-equilibrium conditions.
We have shown here that eRF3 binds GDP independently of the presence or absence of eRF1 whereas GTP binds only to the eRF1•eRF3 complex. We suggest that the quaternary eRF1•eRF3•GTP•Mg2+ complex binds to the ribosomal pretermination complex; owing to the action of the ribosomal GTPase center, the GTPase activity is thus induced which in turn is followed by peptidyl-tRNA hydrolysis. Based on these results, we conclude that the biological role of prokaryotic RF3 and eukaryotic eRF3 is fundamentally different in agreement with earlier conclusions that translation termination mechanisms in general are profoundly distinct in eukaryotes and prokaryotes (14).
MATERIALS AND METHODS
Human eRF3 and eRF1 preparations
The human eRF1 [wild-type and truncated eRF1 (amino acids 1–275) without the C domain] and truncated eRF3 (amino acids 139–637), all of them tagged with a C-terminal hexahistidine, were overproduced in Escherichia coli BL21(DE3) for eRF1 and C41(DE3) for eRF3. After affinity chromatography on Ni-NTA agarose (Qiagen) the resulting preparations were applied on the HiTrap Q HP 16/25 5 ml ion-exchange column (Amersham Pharmacia Biotech) in 50 mM Tris–HCl (pH 7.5), 0.1 M KCl, 10 mM β-mercaptoethanol, 0.2 mM imidazole and 15% glycerol for eRF3 and in 50 mM Tris–HCl (pH 7.5), 0.2 M KCl, 3 mM DTT, 0.035% Tween-20, 0.15 M imidazole and 10% glycerol for eRF1. The columns were washed with 0.1 M KCl, 50 mM Tris–HCl (pH 7.5), 10% glycerol and 5 mM β-mercaptoethanol for eRF3 and with 0.24 M KCl, 50 mM Tris–HCl (pH 8.0) and 0.1 mM EDTA for eRF1. Peak fractions of eRF3 eluted at 0.21 M KCl; they were collected and dialyzed against phosphate [0.1 M KCl, 25 mM potassium phosphate (pH 7.5), 10% glycerol and 1 mM DTT] or Tris [0.15 M KCl, 50 mM Tris–HCl (pH 7.5), 10% glycerol and 5 mM β-mercaptoethanol] buffers and varying concentrations of MgCl2. eRF1 was eluted with 255 mM KCl, 50 mM Tris–HCl (pH 8.0), 0.1 mM EDTA and dialyzed against either phosphate or Tris buffers as for eRF3. After dialysis the samples were used in ITC experiments. If necessary, the samples were concentrated by ion-exchange chromatography on HiTrap Q HP 7/25 1 ml column or using centricon UFV4BGC25 tubes (Millipore). The eRF1 and eRF3 preparations produced single bands after denaturing gel electrophoresis. The molecular masses of the proteins (55 and 50 kDa for eRF3 and eRF1, respectively) corresponded to the values deduced from the amino acid sequences. The protein concentrations were determined spectrophotometrically using a molar extinction coefficient ɛ280 = 39 770 M−1 cm−1 for eRF3 and ɛ280 = 29 780 M−1 cm−1 for eRF1.
Purification of guanine nucleotides
Commercial preparations of GDP and GTP (ICN) were purified using a high-performance liquid chromatography (Gilson) and a Lichrosorb RP-18 column. The purity of the GDP and GTP preparations estimated by NMR was not lower than 99%.
Isothermal titration calorimetry
The thermodynamic parameters of eRF3 binding to eRF1, GDP and GTP were measured using a MicroCal VP-ITC instrument (MicroCal, Northampton, MA). Experiments were carried out at 25°C in phosphate (25 mM K2HPO4, 10% glycerol, 1 mM DTT and 0.1 M KCl) or in Tris (50 mM Tris–HCl, 10% glycerol, 5 mM β-mercaptoethanol and 0.15 M KCl) buffers, at pH 7.5. The MgCl2 concentration varied from 0 to 10 mM. All solutions were degassed under vacuum for 5 min immediately before measurements. Aliquots (10 μl) of ligands were injected from a 296 μl syringe into the 1.42 ml cell containing the protein solution to achieve a complete binding isotherm. The protein concentration in the cell ranged from 6 to 15 μM and ligand concentration in the syringe ranged from 50 to 300 μM. The heat of dilution was measured by injecting the ligand into the buffer solution or by additional injections of ligand after saturation; the values obtained were subtracted from the heat of the reaction to obtain the effective heat of binding. The resulting titration curves were fitted using the MicroCal Origin software, assuming one set of sites. Affinity constants (Ka) and enthalpy variations (ΔH) were determined. Consequently, the Gibbs energy (ΔG) and the entropy variations (ΔS) were calculated from the relation: ΔG = −RTlnKa = ΔH − TΔS.
The stoichiometry of eRF3 binding to eRF1 and guanine nucleotides, calculated using ITC data, was below one. At physiological concentration of Mg2+ (2.0 mM) the binding stoichiometry was equal to 0.47 ± 0.15. One eRF3 molecule binds one eRF1 molecule (17), and bears a single site for binding to the guanine nucleotide (24); the binding stoichiometry observed shows that the efficient concentration of eRF3 and/or eRF1 is below 100% (30). This may be caused by partial inactivation or oligomerization of eRF3 and/or eRF1 during purification or measurements.
When the signal was too low to be measured directly, a displacement titration calorimetry method (31) was applied that consists in measuring an apparent binding constant (Kapp) corresponding to the displacement of one ligand by another. In this case, eRF3 was saturated with ligand L1 (GDP or GTP) placed into the calorimeter cell and then titrated with 10 μl aliquots of ligand L2 (GTP or GDP). An apparent binding constant, corresponding to the displacement of L1 by L2, was determined. Then, knowing the binding constant K1 of L1, we determined the value of the binding constant K2 of L2 for eRF3 from the equation K2 = Kapp(1 + K1[L1]) (31,32). All experiments were carried out two to four times.
RESULTS
ITC was used for direct determination of the thermodynamic parameters for binding of eRF3 and its complexes to eRF1, GDP and GTP. ITC experiments were performed in phosphate or Tris buffers (commonly used in biochemical experiments with translation factors) in order to examine the contribution of the heat of buffer ionization to complex formation. Table 1 presents the experiments performed in phosphate buffer. The data in Tris buffer were in most cases similar to those in phosphate buffer. Whenever the results depend on the buffer composition, this is mentioned in the text.
Table 1.
Thermodynamic parameters of eRF3 binding to eRF1, GDP and GTP at 25°C and pH 7.5 determined by ITCa
| Sample | Ligand | MgCl2 (mM) | Kab(M−1) | Kdc (μM) | ΔHd(kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|---|
| eRF3 | GDP | 0 | 9.1 × 105 | 1.1 | −9.8 | −1.7 | −8.1 |
| eRF3 | GDP | 2 | 5.6 × 105 | 1.9 | −9.2 | −1.4 | −7.8 |
| eRF3 | GDP | 10 | 3.6 × 105 | 2.8 | −2.1 | 5.5 | −7.6 |
| eRF3 | eRF1 | 0 | 6.0 × 105 | 1.7 | −7.4e | −0.5 | −7.9 |
| eRF3 | eRF1 | 2 | 1.4 × 106 | 0.7 | −7.2e | 1.2 | −8.4 |
| eRF3•GDP | eRF1 | 2 | 4.9 × 106 | 0.2 | −3.1e | 6.0 | −9.1 |
| eRF1•eRF3 | GDP | 2 | 5.1 × 105 | 2.0 | −11.8 | −4.0 | −7.8 |
| eRF1•eRF3 | GTP | 0 | 3.6 × 105 | 2.8 | 1.1 | 8.7 | −7.6 |
| eRF1•eRF3 | GTP | 2 | 2.0 × 106 | 0.5 | −2.2 | 6.4 | −8.6 |
| eRF1•eRF3•GDPf | GTP | 2 | 3.1 × 106 | 0.3 | −8.9 | ||
| eRF1•eRF3•GTPf | GDP | 2 | 4.6 × 105 | 2.2 | −7.7 |
aAll measurements were performed in phosphate (25 mM K2HPO4, 10% glycerol, 1 mM DTT and 0.1 M KCl) buffer.
bThe standard deviation did not exceed ±20%.
cCalculated as 1/Ka.
dThe standard deviation did not exceed ±8%.
eΔH was calculated taking into account the effect of protonation (for details see text).
fThe model of competitive ligand binding was used (Materials and Methods).
eRF3 binds GDP but not GTP
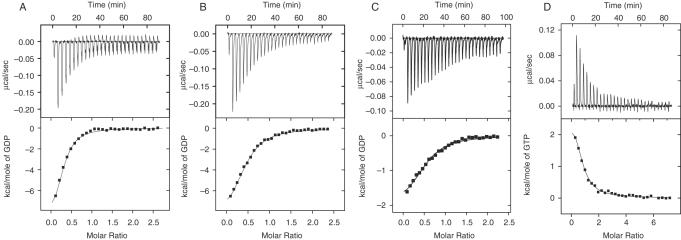
A typical set of ITC data for GDP binding to eRF3 in phosphate buffer in the presence or absence of Mg2+ is shown in Figure 1A–C. The upper panel presents the raw calorimetric data for the titration of the protein by ligand and the lower panel presents the binding isotherms. In both phosphate and Tris buffers elevation of Mg2+ concentration caused a slight decrease in GDP affinity for eRF3 (Table 1). Indeed, in the absence of Mg2+, the dissociation constant Kd is equal to 1.1 μM, while at physiological (2 mM) Mg2+ it increases 1.7-fold in both buffers. As is clear from the thermodynamic parameters (Table 1), the binding of GDP to eRF3 is enthalpy-driven. However, at non-physiological (10 mM) Mg2+ concentration the enthalpic contribution to overall free energy diminishes while the enthropic contribution increases. Thus, despite the fact that Mg2+ is not essential for GDP binding to eRF3, at high concentrations it dramatically changes the thermodynamic profile of the interaction (Table 1 and Figure 1). We observed no Mg2+ binding to eRF3. In contrast to GDP binding, we failed to reveal any measurable GTP binding to eRF3 in either phosphate or Tris in the absence and presence of 2 mM Mg2+. It should be noted that binding constants below 103 M−1 cannot be measured by the ITC method.
Figure 1.
ITC curves (upper panel) and binding isotherms (lower panel) of eRF3 interaction with GDP in the absence (A) or presence of 2 mM (B) or 10 mM (C) MgCl2; (D) interaction of the eRF1•eRF3•GDP complex with GTP at 2 mM MgCl2, 25°C, in phosphate buffer (pH 7.5).
Energetics of complex formation between eRF3 and eRF1
As mentioned above, eRF3 and eRF1 form an equimolar complex in vitro but the binding constant and thermodynamic parameters of this interaction are unknown. Complex formation between eRF1 and eRF3 is an enthalpy-favorable process (Table 1). In both phosphate and Tris buffers at 2 mM Mg2+ the association constant (Ka) is ∼106 M−1. This value decreases 2-fold in the absence of Mg2+. In Tris buffer the alteration in the observed enthalpy [ΔHobs(Tris) = −19.2 kcal/mol] is significantly higher than in phosphate buffer [ΔHobs(Pi) = −8.2 kcal/mol]. This difference is caused by changes in the protonation state of protein amino acid residues upon binding, because the enthalpy of ionization of Tris buffer is significantly higher than that of phosphate buffer (11 kcal/mol versus 1 kcal/mol) (33). The enthalpy of ionization (ΔHi) is a unique property of different buffer systems and when protonation accompanies complex formation, it can be used to determine the number of interacting protons (30). The ΔHobs from the titration experiment at a given pH is thus composed of the heat of binding and the protonation effect: ΔHobs = ΔHb + (Δn)ΔHi, where ΔHb is the true intrinsic heat of binding and Δn is the number of protons released or taken by the buffer upon binding.
Since ΔHobs in phosphate buffer is less exothermic, we suggest that protons are released at the association step. It cannot be determined whether protons are released from eRF3 or from eRF1 but the total number of released protons is determined by the following relation (33):
where ΔHi(Pi) is the enthalpy of phosphate buffer ionization and ΔHi(Tris) is the enthalpy of Tris buffer ionization. For eRF1•eRF3 binding Δn = −1.1. Hence, association of the factors is accompanied by proton release from the complex into solution. This is probably induced by conformational changes of one or of both factors (17), leading to alteration of the pK of an amino acid residue in the complex.
In the GDP-bound state, eRF3 binds to eRF1 with 3.5 times higher affinity than does free eRF3 (Table 1). As in the case of complex formation between eRF3 and eRF1, when the eRF3•GDP complex binds eRF1 in Tris buffer, ΔHobs is much higher (−13.5 kcal/mol) than in phosphate buffer (−4.1 kcal/mol). For the eRF3•GDP association with eRF1 Δn = −0.8, also indicating release of one proton into the solution.
It is known that C-terminal truncation of eRF1s from fission yeast and human largely or completely abolishes interaction of eRF1 with eRF3 (13,18,19,34). Human eRF1 deprived of its C domain loses its ability to bind eRF3 as determined by ITC experiments (data not shown), in entire agreement with the above mentioned biochemical observations.
The eRF1•eRF3 complex binds GTP
The eRF1•eRF3 complex binds GDP as efficiently as free eRF3 at 2 mM Mg2+ and the thermodynamic profile of GDP binding to the eRF1•eRF3 complex is virtually the same as to free eRF3 (Table 1). In contrast to free eRF3, eRF3 complexed with eRF1 acquires the ability to bind GTP (Table 1). Moreover, the constant of GTP binding to the eRF1•eRF3 complex is four times higher than that of GDP binding and is equal to 2 × 106 M−1 at 2.0 mM Mg2+ (Table 1). In contrast to the enthalply-driven process of GDP binding to the eRF1•eRF3 complex, GTP binding has a reduced enthalpic contribution and a larger entropic component. An increase of the entropic component during complex formation is frequently associated with a conformational change in a protein molecule that leads to burying hydrophobic residues located on the surface of the protein globule (30). Mg2+ increases the GTP affinity to the eRF1•eRF3 complex 6-fold (Table 1) in contrast to the eRF3•GDP complex, suggesting that Mg2+ is implicated in GTP coordination in the binding site, as shown for other GTP-binding proteins (35). GTP binding to the eRF1•eRF3 complex is observed only in phosphate buffer whereas in Tris buffer the signal is too low to be measured directly.
Nucleotide displacements in the ternary complex
The eRF1-dependent GTPase activity of eRF3 in the ribosome (15) implies that the substrate of this reaction should be a ternary eRF1•eRF3•GTP complex, which has been described in the previous section. Since the eRF1•eRF3 complex retained its ability to bind GDP, we attempted to displace GDP from the ternary complex by GTP, at 2 mM Mg2+. The eRF1•eRF3 complex was saturated with GDP and then titrated with GTP (Figure 1D). The model of competitive ligand binding (31) was applied. The GTP binding constant in phosphate buffer, determined by this method, is virtually the same as the constant measured for GTP binding to the eRF1•eRF3 complex (Table 1). As shown in Figure 1D, GDP displacement by GTP is an endothermic process. This could be explained by the fact that complex formation between GTP and the eRF1•eRF3 complex is an entropically driven process and the enthalpic contribution is lower than in the case of GDP binding (Table 1). Applying a competitive ligand binding model we determined the GTP binding constant (1.8 × 106 M−1) for the eRF1•eRF3 complex in Tris buffer, which appears to be similar to that in phosphate buffer (Table 1).
By analogy, the ternary eRF1•eRF3•GTP complex was titrated with GDP (data not shown). In contrast to the GDP/GTP displacement, the GTP/GDP exchange is an exothermic process. The GDP association constant in phosphate buffer determined using the displacement method is similar to the value determined for GDP binding to the eRF1•eRF3 complex (Table 1). In the absence of eRF1, if the eRF3•GDP complex was titrated with GTP, no binding was detected as shown for free eRF3 titrated with GTP.
DISCUSSION
It is becoming clear from recent data in vivo (27) and from results obtained in a fully reconstituted eukaryotic translation system (14) that eRF3 ensures a fast and high fidelity translation termination reaction in contrast to what has been shown for its prokaryotic analog RF3 (28). This explains why eRF3 is encoded by an essential gene while RF3 is dispensable in bacterial cells. Here, we measured the thermodynamic parameters of guanine nucleotide binding to human eRF3, eRF1 and to the complex of these factors in both phosphate and Tris buffer. We have shown that (i) eRF3 binds GDP with a rather weak dependence on Mg2+ concentration (Table 1 and Figure 1A–C); (ii) eRF1 and eRF3 bind to one another irrespective of the presence or absence of guanine nucleotides and of Mg2+ (Table 1); (iii) the eRF1•eRF3 complex binds both GDP and GTP with a preference for the latter; (iv) in the eRF1•eRF3•GDP complex, GDP can be exchanged for GTP (Figure 1D) and vice versa; GDP displacement by GTP proceeds more easily than the reverse reaction (Table 1).
ITC results compared to earlier data
Our data contradict the results obtained in a yeast system in which GTP and Mg2+ are required for complex formation between eRF1 and eRF3 (29). As shown here, neither GTP nor GDP is essential for complex formation and binding is also tolerant to Mg2+ concentration. One possible reason for such discrepancies may stem from the fact that eRF1 and eRF3 were tagged with different proteins (Myc and protein A) and complex formation was visualized using an indirect immunoblotting assay (29). Bulky tags may interfere with eRF1•eRF3 complex formation whereas GTP coordinated with Mg2+ can partially weaken this effect. The observation (29) that mutations in the GTP-binding site of eRF3 impair the ability of the mutated factor to bind eRF1 is in a sharp contradiction with other data showing that the eRF1•eRF3 interaction is mediated solely by the C domains of both factors without involvement of GTP-binding sites (18–20,34).
It has been shown for truncated eRF3 from the fission yeast S.pombe that Mg2+ is not required for GDP binding (24) in complete agreement with the data obtained here with total human eRF3 (Table 1 and Figure 1A–C). Our Kd values for the eRF3•GDP complex (Table 1) are similar to those for the S.pombe eRF3•GDP measured in the absence of Mg2+ by the ITC assay. However, at 2.0–2.5 mM Mg2+, the affinity of GDP for yeast eRF3 is lost (24). We assume that this remarkable difference between the data with human eRF3 and that with S.pombe eRF3 may be associated with the fact that yeast eRF3 was depleted of the N-terminal amino acid sequence (positions 1–195) and additionally, that the S.pombe eRF3 differs considerably in amino acid sequence from human eRF3. A hypothesis (24) that Mg2+ plays a role of guanine exchange factor (GEF) with respect to eRF3 is inconsistent with our observation that eRF3•GDP complex formation insignificantly depends on the Mg2+ concentration (Table 1).
For the human eRF3•GDP complex Kd =1.9 μM at 2 mM Mg2+ (Table 1). This is in sharp contrast to the Kd = 5.5 nM at 5 mM Mg2+ for the prokaryotic RF3•GDP complex (28). The three orders of magnitude difference in dissociation constants points to a profound difference between the structure of the GDP-binding sites of eRF3 and RF3. Since GDP binding to eRF3 is a one-step process (Table 1 and Figure 1), no conformational transformation of eRF3 is required to bind GDP to the factor. GDP binding to eRF3 is not affected by eRF1 (Table 1) in agreement with the data showing that eRF1•eRF3 complex formation is mediated by the C-termini of both factors and does not involve the GTP/GDP-binding center of eRF3 (18–20,34).
eRF1 as affector of guanine nucleotide binding to eRF3
In mammalian cells, the GTP concentration is one order of magnitude higher than the GDP concentration (36) and in yeast cells the eRF1 and eRF3 concentrations are roughly equal (37). However, the >2 orders of magnitude difference between the Kd's for GDP and GTP implies that in vivo in the absence of eRF1, eRF3 exists predominantly in a GDP-bound form as shown for bacterial RF3 (28). When eRF3 is complexed with eRF1, the binary complex possesses higher affinity for GTP than for GDP (Table 1) and displaces GDP from the complex, if it has been bound before eRF3. The most plausible explanation of the effect of eRF1 stems from the suggestion (17) that in a binary complex either eRF3 or eRF1 or both factors undergo significant conformational changes. This structural alteration can induce the appearance of a site in the nucleotide binding center of eRF3 able to bind the γ-phosphate moiety of GTP which is absent in the eRF3•GDP complex. Therefore, eRF1 does not disturb GDP binding to eRF3 as shown above but modifies the nucleotide binding center in such a way that GTP binding becomes more favorable than GDP binding. In accord with this interpretation, we may speculate that the function of eRF1 with respect to eRF3 resembles that of a GAP (GTPase-activating protein).
Surprisingly, the influence of Mg2+ on the GDP binding constant is very weak (Table 1 and Figure 1A–C) implying that Mg2+ is not involved in coordination with GDP as reported for some other GDP/GTP-binding proteins [reviewed in (35)]. Since most GEFs act by distorting the GDP/GTP-binding site stabilized by Mg2+ (38), because of Mg2+-independence in the case of eRF3, there is no need to distort the eRF3-binding site. Therefore, the absence of Mg2+ effect on eRF3•GDP association suggests that eRF3 does not require a special GEF as do other GTPases (35). For instance, the translation factors IF2 and EF-G do not require GEFs in the form of external proteins for GDP/GTP exchange and the ribosome itself performs this function toward these factors (39).
Since Mg2+ increases the affinity of GTP for the eRF1•eRF3 complex (Table 1) in contrast to GDP binding to eRF3, Mg2+ is most likely coordinated in the GTP-binding site stabilizing the appropriate conformation of the switch 1 and switch 2 elements that are disordered in free eRF3 as shown by structural analyses (24).
A model of eRF3 interaction with eRF1 and guanine nucleotides
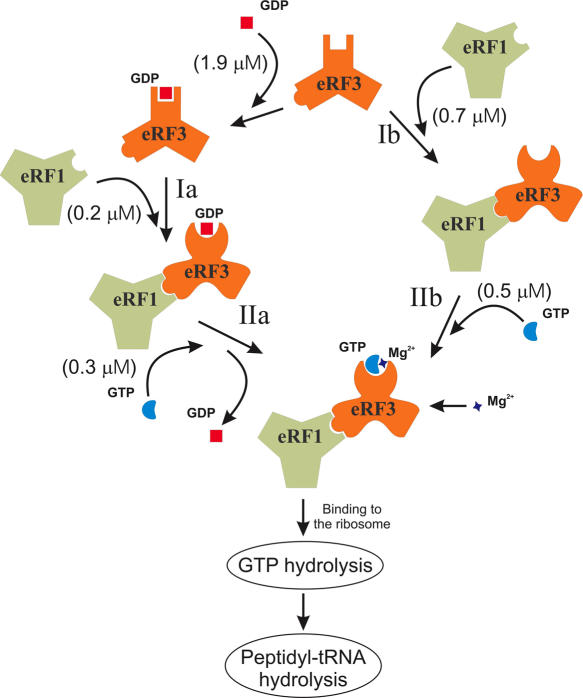
Based on the results described above (Table 1 and Figure 1) we suggest a sequence of interactions of eRF3 with its ligands that is consistent with the data available. Routes (a) and (b) can be considered (Figure 2). If eRF3 follows route (a), it binds GDP in a Mg2+-independent manner. Since eRF3 binds GDP but not GTP it seems possible that eRF3 similar to prokaryotic RF3 (28) is present in the eukaryotic cytosol in a GDP-bound state. At step Ia, eRF1 binds to the eRF3•GDP complex and probably changes the conformation of eRF3. As eRF3 is most likely in a GDP-bound form and the binding constant for reaction Ia is 3.5 times higher than for association of eRF3 with eRF1 (step Ib), the ternary eRF1•eRF3•GDP complex could be formed more easily than the eRF1•eRF3 complex. GDP in the eRF1•eRF3•GDPcomplex would then be displaced by GTP (step IIa). Owing to conformational changes induced by eRF1 upon binding as proposed earlier (17), eRF3 could acquire the ability to bind GTP and to displace GDP. This displacement/exchange reaction becomes possible not because of decrease of the eRF3-binding constant to GDP, which is not altered as shown here (Table 1). Since the constant of GTP binding to the eRF1•eRF3 complex is four times higher than that to GDP (Table 1) and the GTP concentration in mammalian cells is at least 10 times higher than the GDP concentration (36), this induces the displacement reaction. At physiological (0.5–2.0 mM) Mg2+ concentration (40) the affinity of GTP for eRF1•eRF3 is 6-fold higher than in the absence of Mg2+. In contrast for GDP binding the Mg2+ effect is absent.
Figure 2.
A model illustrating an interplay of eRF3, eRF1 guaninenucleotides and Mg2+ at termination of translation. The Kd values of the corresponding reactions (Table 1) are in parentheses. For details see Discussion. The results on the role of eRF3 in translation termination were taken into consideration (14).
Route (b) is in principle also possible (Figure 2). If eRF3 binds first to eRF1 and not to GDP as in route (a), it acquires the ability to bind directly to GTP (IIb). Although the binding constant of eRF3 with eRF1 is two times higher than that of eRF3 with GDP, route (b) appears less likely because the concentration of eRF1 seems to be lower than that of GDP.
Irrespective of whether the routes (a) or (b) or both take place, the resulting product is formation of the ternary eRF1•eRF3•GTP complex. Given that Mg2+ stabilizes the ternary complex, we assume that the structure is rather a quaternary complex which includes Mg2+ coordinated with GTP and eRF3. Quaternary eRF1•eRF3•GTP•Mg2+ complex formation can proceed in the cytosol. In view of our results, the functionally active quaternary complex requires neither an additional external GEF, nor the GEF activity of the ribosome known to catalyze guanine nucleotide exchange in prokaryotic ribosomes toward translation factor GTPases (39). If so, the role of the ribosome toward the quaternary complex is to catalyze GTPase hydrolysis which is entirely ribosome-dependent (15). Therefore, at termination of translation not only is the function of eRF3 and RF3 entirely different (14,41) but also it seems likely that the ribosomes in prokaryotes and eukaryotes are also distinct with regard to translation termination process.
The present study was carried out in parallel with investigations (42) based on entirely different tools, but dealt with the same problem. The results appear in most cases similar although some differences in interpretation exist.
Acknowledgments
We thank L. Frolova for valuable advice with regard to isolation and purification of proteins and P. Tsvetkov for fruitful discussions. We are very grateful to A.-L. Haenni for kind help in preparing this manuscript for publication. This study was supported by the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences, by the program Leading Scientific Schools of the RF via the Ministry of Education and Science and by the Russian Foundation for Basic Research (projects 05-04-49385, 03-0448943 and 05-04-49392). Funding to pay the Open Access publication charges for this article was provided by University of Oslo, Center for Medical Studies at Moscow.
Conflict of interest statement. None declared.
REFERENCES
- 1.Frolova L., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.-L., Celis J.E., Philippe M., et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 2.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolova L., Seit-Nebi A., Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito K., Frolova L., Seit-Nebi A., Karamyshev A., Kisselev L., Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl Acad. Sci. USA. 2002;25:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolosov P., Frolova L., Seit-Nebi A., Dubovaya V., Kononenko A., Oparina N., Justesen J., Efimov A., Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kisselev L.L., Ehrenberg M., Frolova L., Yu. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 12.Seit-Nebi A., Frolova L., Justesen J., Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolova L.Y., Merkulova T.I., Kisselev L.L. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Frolova L., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 16.Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolova L., Simonsen J., Merkulova T., Litvinov D., Martensen P., Rechinsky V., Camonis J., Kisselev L., Justesen J. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 1998;256:36–44. doi: 10.1046/j.1432-1327.1998.2560036.x. [DOI] [PubMed] [Google Scholar]
- 18.Ebihara K., Nakamura Y. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA. 1999;5:739–750. doi: 10.1017/s135583829998216x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkulova T.I., Frolova L.Y., Lazar M., Camonis J., Kisselev L.L. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- 20.Eurwilaichitr L., Graves F.M., Stansfield I., Tuite M.F. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- 21.Buckingham R.H., Grentzmann G., Kisselev L. Polypeptide chain release factors. Mol. Microbiol. 1997;24:449–456. doi: 10.1046/j.1365-2958.1997.3711734.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura Y., Ito K., Isaksson L.A. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 23.Ter-Avanesyan M.D., Kushnirov V.V., Dagkesamanskaya A.R., Didichenko S.A., Chernoff Y.O., Inge-Vechtomov S.G., Smirnov V.N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 24.Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S.pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 25.Konecki D., Aune K., Tate W., Caskey C. Characterization of reticulocyte release factor. J. Biol. Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 26.Tate W., Beaudet A., Caskey C. Influence of guanine nucleotides and elongation factors on interaction of release factors with the ribosome. Proc. Natl Acad. Sci. USA. 1973;70:2350–2355. doi: 10.1073/pnas.70.8.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salas-Marco J., Bedwell D.M. GTP Hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zavialov A.V., Buchingham R.H., Ehrenberg M.A. Posttermination ribosomal complex is the guanine exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi T., Funakoshi Y., Shin-ichi H., Katada T. The GTP-binding Release Factor eRF3 as a Key Mediator coupling translation termination to mRNA decay. J. Biol. Chem. 2004;279:45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- 30.Ladbury J.E., Doyle M.L. Biocalorimetry 2. Applications Calorimetry In The Biological Sciences. The Sussex John Wiley & Sons, Ltd; 2004. p. 259. [Google Scholar]
- 31.Sigurskjold B.W. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 32.Lafitte D., Lamour V., Tsvetkov P.O., Makarov A.A., Klich M., Deprez P., Moras D., Briand C., Gilli R. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry. 2002;41:7217–7223. doi: 10.1021/bi0159837. [DOI] [PubMed] [Google Scholar]
- 33.Pierce M.M., Raman C.S., Nall B.T. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 34.Ito K., Ebihara K., Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sprang S.R., Coleman D.E. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell. 1998;95:155–158. doi: 10.1016/s0092-8674(00)81746-8. [DOI] [PubMed] [Google Scholar]
- 36.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y., Wessling-Resnick M. GEF-mediated GDP/GTP exchange by monomeric GTPases: a regulatory role for Mg2+? BioEssays. 1998;20:516–521. doi: 10.1002/(SICI)1521-1878(199806)20:6<516::AID-BIES11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of The Cell. 4th edn. NY: Taylor & Francis Group Press; 2002. p. 616. [Google Scholar]
- 41.Kisselev L.L., Buckingham R.H. Translation termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 42.Hauryliuk V., Zavialov A., Kisselev L., Ehrenberg M. Biochimie. 2006. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. in press, PMID: 16797113. [DOI] [PubMed] [Google Scholar]