Abstract
We have characterized elongation complexes (ECs) of RNA polymerase from the extremely thermophilic bacterium, Thermus thermophilus. We found that complexes assembled on nucleic acid scaffolds are transcriptionally competent at high temperature (50–80°C) and, depending upon the organization of the scaffold, possess distinct translocation conformations. ECs assembled on scaffolds with a 9 bp RNA:DNA hybrid are highly stable, resistant to pyrophosphorolysis, and are in the posttranslocated state. ECs with an RNA:DNA hybrid longer or shorter than 9 bp appear to be in a pretranslocated state, as evidenced by their sensitivity to pyrophosphorolysis, GreA-induced cleavage, and exonuclease footprinting. Both pretranslocated (8 bp RNA:DNA hybrid) and posttranslocated (9 bp RNA:DNA hybrid) complexes were crystallized in distinct crystal forms, supporting the homogeneity of the conformational states in these complexes. Crystals of a posttranslocated complex were used to collect diffraction data at atomic resolution.
INTRODUCTION
Studies of bacterial transcription have traditionally involved Escherichia coli RNAP. While a large body of biochemical and genetic data are available for this enzyme, high-resolution structural data are still lacking. On the other hand, RNAPs from thermophilic bacteria, namely Thermus thermophilus and Thermus aquaticus, have emerged as excellent models for structural studies. In the past few years, structures of the holo enzyme (α2ββ′ωσ) of Thermus RNAPs in the presence or absence of various ligands and antibiotics have become available (1–8). However, crystallographic characterization of transcription complexes in association with nucleic acids or transcription factors is still lagging. Previous efforts have shown that such studies require an extensive prior biochemical characterization to define the nature of the complexes and to provide conditions for successful crystallization (9). This work represents a significant step in this direction.
During elongation, RNAP alternates between different conformational states that are defined by the position of 3′ end of the transcript relative to the enzyme catalytic site (10,11). Following formation of the phosphodiester bond, the 3′ end of the RNA occupies the insertion site, and the complex is in the pretranslocated state. Subsequent translocation of polymerase along the DNA template is required for the next round of the transcription cycle. This transition results in the posttranslocated state of the complex, in which the 3′ end of the RNA is transferred to the product site, and the template acceptor base is available for substrate binding and selection (12,13). The manner in which RNAP oscillates between pre- and posttranslocated states during the nucleotide addition cycle, and what controls translocation, are poorly understood. Several models have been proposed to explain RNAP translocation. In the Brownian ratchet model, binding of the incoming substrate NTP stabilizes the oscillating RNAP in the posttranslocated state and serves as a pawl to prevent backward movement, as suggested by footprinting and single molecule studies (14–16). An alternative model (called the power stroke mechanism) suggests that motor proteins convert energy that derives from NTP hydrolysis into mechanical energy through conformational changes of the protein (17). An example of such a mechanism has been reported in the T7 RNAP system, where release of pyrophosphate after formation of the phosphodiester bond triggers a relaxation of a strained protein intermediate, which causes the transition from the pre- to posttranslocated state (11). In the latter case, it is the conformation of polymerase, not NTP-binding that determines the most stable position of the 3′ end of the RNA.
Elongation complexes (ECs) of bacterial RNAP halted downstream from a promoter differ significantly in their translocation conformations (18–22) which complicates studies of the mechanisms of transcription elongation. In this work, we demonstrate that ECs of Tth RNAP assembled on nucleic acid scaffolds that resemble the organization of the components of the EC have distinct translocation conformations. Such complexes will be useful for functional and structural studies of ECs at different stages of the nucleotide addition cycle.
MATERIALS AND METHODS
Purification of RNAP and polymerase activity assay
WT Tth core and holo RNAP were purified from cell biomass obtained from HB8 strain as described in (23). WT E.coli core polymerase was purified from cell biomass obtained from MRE600 strain as described in (24). The purity of these RNAPs was greater than 99.8% as judged by SDS–PAGE. Polymerase activity was measured by the ability to extend a 32P-labeled RNA primer by 1 nt in a reaction where the concentrations of nucleic acid scaffold (R8/TS35/NT35) and RNAP were equimolar (see transcription conditions below). Only RNAP preps that extended the RNA primer with close to 100% efficiency within 1 min of incubation at 37°C (E.coli core ) or at 60°C (Tth core) were used.
RNA and DNA oligonucleotides
The following synthetic oligonucleotides were used (all sequences are 5′ to 3′).
RNA oligomers (Dharmacon): (R7) GCGGCGA, (R8) GCGGCGAU, (R9) GCGGCGAUA, (R10) GCGGCGAUAU, (R13) GAGUCUGCGGCGA, (R14) GAGUCUGCGGCGAU, (R15) GAGUCUGCGGCGAUA.
DNA oligomers (IDT DNA): (TS35) CCTGTCTGAATCGATATCGCCGC, (TS42) CCCTGTCTGAATCTCTATCGCCGG, (TS11) GGGTCCTGTCTGAAATCGACATCGCCGCTCAACATACG, (TS38 or TS38b, biotinylated) CCAACATACGGCTCGGACAGAGGTCCTGTCTGAATCGATATCGCCGC, (NT35) TCGATTCAGACAGG, (TS351) CCTGTCTGAATCGCTATCGCCGC, (TS352) CCTGTCTGAATCGCAATCGCCGC, (NT36) CTGTCCGAGGAGATGACACTCGATTCAGACAGG, (NT38) ATCGATTCAGACAGG, (NT380) CGATTCAGACAGGACCTCTGTCCGAGCCGTATGTTGG, (NT05, biotinylated) CGTATGTTGAGCGGCGATGTCGATTTCAGACAGGACCC, (NT351) GCGATTCAGACAGG, (NT01, biotinylated) CGATTTCAGACAGGACCC.
All DNA and RNA oligomers were high-performance liquid chromatography (HPLC) purified and had better than 95% purity.
Assembly of ECs and transcription conditions
Nucleic acid scaffolds were assembled by annealing together appropriate oligomers (10 μM each) as described previously (25). Where indicated, RNA and DNA oligomers were labeled at their 5′ ends using [γ-32P]ATP and polynucleotide kinase (NEB) prior to assembly. To assemble ECs, core RNAP (1 μM) was incubated with an equimolar concentration of scaffold for 5 min at RT in 10 μl of transcription buffer [40 mM Tris (pH 7.9 at 25°C), 100 mM NaCl, 10 mM MgCl2 and 5 mM 2-mercaptoethanol]. Primer extension was achieved by incubation of complexes with substrate NTPs (10 μM) for 1 min at 60°C (Tth RNAP) or 37°C (E.coli RNAP). All walking experiments were performed according to (26) except biotinylated template and streptavidin agarose beads (Pierce) were used. Where indicated, all incubations and wash steps were performed at 60°C using pre-heated transcription buffer. Reactions were stopped by the addition of formamide/EDTA containing buffer and products of the reaction were resolved by 20% PAGE in the presence of 6 M urea and visualized by Phosphoimager™ (GE Health).
Pyrophosphorolysis and nucleolytic activity assays
ECs were incubated with 50 μM PPi at 60°C (Tth RNAP) or 37°C (E.coli) for the times indicated. GreA was added to the Tth ECs at an equimolar ratio, and reactions were allowed to continue for 1–20 min at 60°C. Reactions were stopped by the addition of formamide/EDTA containing buffer and samples were resolved by electrophoresis in 20% PAGE in the presence of 6 M urea and visualized by Phosphoimager™ (GE Health). Experiments on the intrinsic exo/endonuclease activity of RNAP were performed in transcription buffers containing 40 mM Tris (pH 7.9 at 25°C) or 40 mM HEPES (pH 8.0 at 25°C).
Footprinting assay
EC14 (R14/TS38/NT380) that contained a 32P-labeled non-template (NT) strand was assembled as described above. EC15 was obtained by incubation of EC14 with substrate ATP for 2 min at 60°C. Exonuclease III (Exo III) (NEB, 0.1 U/μl) was added for 5–10 min at 37°C. Reactions were stopped by the addition of formamide/EDTA containing buffer and products of the reaction were analyzed as described above.
Photo cross-linking
Scaffold complexes R13/TS351/NT351 and R14/TS352/NT351, assembled as described above were incubated with 50 μM of 4-thio UTP (Trilink Biotech) for 2 min at 60°C to allow incorporation of this reagent at the 3′ end of 32P-labeled RNA and to form EC14 and EC15, correspondingly. Efficiency of 4-thio UTP incorporation was monitored by running the aliquot of the samples in 20% PAGE and exceeded 95%. Cross-linking was then initiated by the exposure of the complexes to ultraviolet (UV) light (312 nm) for 5 min at 60°C. After cross-linking, samples were loaded onto 4–12% NuPAGE MES gel (Invitrogen) and visualized by Phosphoimager™ (GE Health).
Crystallization, data collection and processing
EC14 (R14/TS35/NT35) and EC15 (R15/TS35/NT35) were prepared as described above by incubating Tth RNAP (10 mg/ml) with an equimolar concentration of nucleic acid scaffold for 10 min at RT. Crystallization was carried out by the sitting drop vapor-diffusion technique at 20°C using screening solutions (Hampton Research). Initial crystallization conditions for EC14 were 6% polyethylene glycol (PEG) 8000, 0.1 M sodium cacodylate and 0.2 M (NH4)2SO4. Hexagonal crystals (0.5 × 0.07 × 0.07 mm) appeared within 5–7 days. Crystals of EC15 were obtained in solution containing 30% 2-methyl-2,4-pentandiol, 0.1 M sodium cacodylate (pH 6.5) and 0.2 M magnesium acetate. The crystals had bi-pyramidal shape and grew within 6–8 weeks to a final size of about 0.6–0.8 mm in the longest dimension.
The quality of the crystals was probed using a laboratory X-ray generator (Rigaku FR-D). Diffraction was measured using a Rigaku R-AXIS IV++ imaging-plate detector. For data collection at cryo temperature, the crystals were soaked in a well solution in which the concentration of cryoprotectant was gradually increased. The cryoprotectant solutions contained 10% PEG 8000, 30% glycerol (EC14 crystals) or reservoir solution (EC15 crystals). Complete diffraction data were collected using synchrotron radiation at beam line BL5 at Photon Factory (Tsukuba, Japan) and at the SERCAT beamline (Argonne, Chicago). The diffraction data were processed using the HKL2000 program package (27).
RESULTS
Core Tth RNAP can be assembled into functional ECs on nucleic acid scaffolds
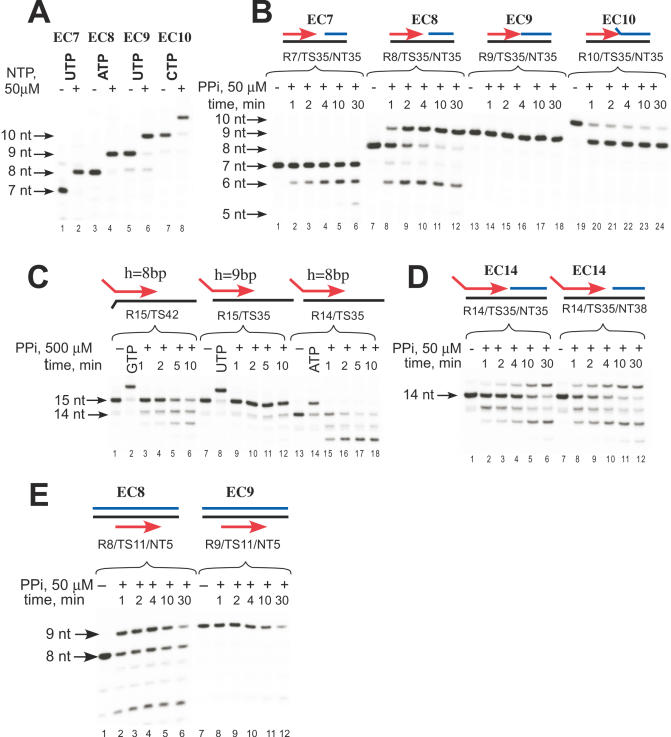
To assemble Tth RNAP EC, we used RNA primers (7–16 nt) pre-annealed to a complementary DNA that had 14–18 bp of downstream duplex DNA (Figure 1 and Supplementary Figure S1). Such complexes have previously been shown to posses many of the properties of ECs of bacterial and phage RNAPs (25,28). The Tth core RNAP (α2ββ′ω) can bind the RNA:DNA scaffold and efficiently extend the RNA primer (Figure 1A) in a similar manner to E.coli RNAP (28). The efficiency of the RNA primer extension did not depend on the length of the RNA:DNA hybrid itself, as we observed nearly 100% conversion of the initial 5′ 32P-labeled primers from 7–10 nt within 5 min of incubation with the substrate NTP (Figure 1A). In agreement with the data on E.coli RNAP, Tth ECs having an 8–9 bp RNA:DNA hybrid were equally stable (28) and withstood incubation with upto 0.8 M NaCl, as revealed by salt-challenge experiments (Supplementary Figure S2).
Figure 1.
Sensitivity of different Tth RNAP ECs to pyrophosphorolysis. (A) Tth RNAP ECs (EC7, EC8, EC9, EC10, R7–10/TS35/NT35) were incubated with the substrate NTP (50 μM) for 5 min at RT as described in Materials and Methods. (B–D) Effect of the length of the RNA:DNA hybrid (B) and scaffold topology (C and D) on sensitivity to pyrophosphorolysis. Scaffold ECs were obtained as noted and then incubated with 50 (B, D and E) or 500 μM (C) PPi at 60°C for the times indicated. (E) Pyrophosphorolysis in complexes having fully complementary NT strand. EC9 was obtained by incubation of EC8 (R8/TS11/NT05b) with GTP for 2 min at 60°C followed by the removal of the NTP by washing with transcription buffer. The complexes were then incubated with 50 μM PPi at 60°C for the times indicated. Note that in EC8 (B and E) and EC14 (D) the RNA primer is extended by incorporation of substrate NTP released during pyrophosphorolysis. Color scheme: RNA-red, DNA T strand-black and DNA NT strand-blue.
Scaffold ECs can be assembled in pre- or posttranslocated conformations
The length of the RNA:DNA hybrid in bacterial polymerase ECs halted downstream from a promoter has been probed by various techniques, and has been determined to be 8–9 bp (21,29). In this work, we probed the sensitivity of the assembled ECs to pyrophosphate in a reaction, which is the reverse of substrate incorporation and is characteristic of the pretranslocated state of EC (Figure 1B). We found, that the Tth RNAP exhibited notably different pyrophosphorolytic activity, which depended upon the length of the RNA:DNA hybrid in the assembled EC. Thus, incubation with 50 μM PPi resulted in pyrophosphorolysis in all complexes except EC9 (Figure 1B). While EC7 was readily converted into EC6, further cleavage of 6mer RNA was not observed, likely due to dissociation of the EC6 at 60°C (Figure 1B, lanes 1–6). Progressive pyrophosphorolysis of EC8 resulted in the appearance of 6 and 7mer RNAs and, at the same time, accumulation of a stable 9mer RNA due to incorporation of UTP that was released during conversion from 7mer to 6mer (Figure 1B, lanes 7–12). Complexes with a 9 bp hybrid remained intact even during extended incubation (Figure 1B, lanes 13–18) or when a high (up to 1 mM) concentration of PPi was used (data not shown). EC10 was highly sensitive to pyrophosphorolysis and was rapidly converted to a stable EC9 complex (Figure 1B, lanes 19–24). Similar PPi cleavage patterns were obtained when EC8, EC9 and EC10 were obtained by extension of EC7 (data not shown).
Changes in the topology of the scaffold (i.e. the presence of a single-stranded RNA ‘tail’ and the lack of a complementary non-template strand downstream or upstream) did not change the trend in PPi sensitivity, suggesting a primary role for the length of RNA:DNA hybrid in complex translocation conformation (Figure 1C–E). Thus, EC14 and EC15, which have an RNA ‘tail’ of 6 nt and a 8 or 9 bp RNA:DNA hybrid, respectively, behaved like EC8 and EC9. That the 9th RNA:DNA base pair confers stabilization of the posttranslocated state (and thus resistance to PPi), and not the presence of upstream DNA, becomes evident from comparison of pyrophosphorolysis in scaffolds R15/TS35 and R15/TS42 (Figure 1C). Similarly, 1 nt gap between the 3′ end of the RNA and the 5′ end of the NT DNA strand did not affect sensitivity to PPi (Figure 1D).
It is known that NT DNA strand contributes to the EC integrity and may effect its conformation and stability (30,31). To monitor whether the presence of NT strand affects pyrophosphorolysis, we assembled complexes on scaffolds having fully complementary DNA strands and thus most closely resembling promoter originating ECs, using an approach first described by Kashlev and co-workers (28). In this assay, a 5′ end labeled RNA primer was first pre-annealed with template (T) strand DNA and incubated with RNAP, followed by addition of a 10-fold excess of biotin-labeled NT strand DNA (Figure 1E). The ECs were then immobilized on streptavidin agarose beads and washed several times with transcription buffer to ensure that all assembled complexes contained an NT strand. As in the previous experiment with scaffolds lacking an upstream DNA duplex, we observed that only ECs with a hybrid of 9 bp were resistant to PPi cleavage (Figure 1E).
In an other experiment, we repeated the PPi sensitivity assay on the scaffolds described above using E.coli core RNAP, and found the same trend in PPi sensitivity in complexes with different RNA:DNA hybrid lengths (Supplementary Figure S3).
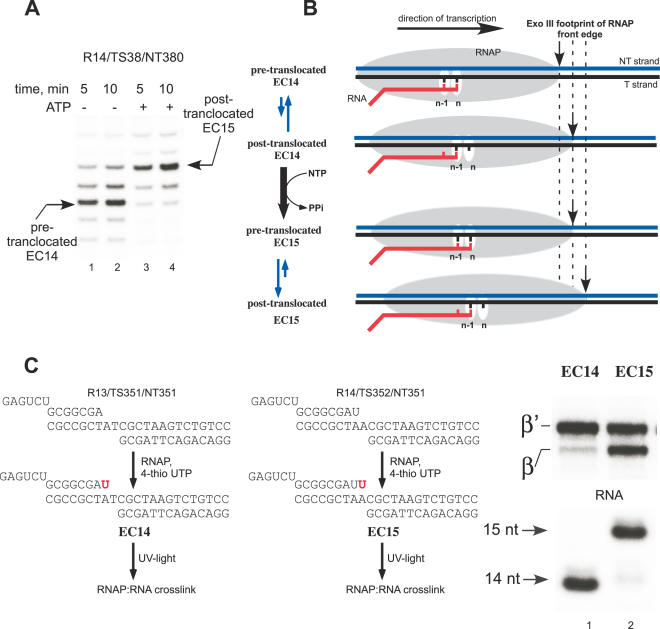
The differences in PPi sensitivity may be interpreted as resulting from the relative partitioning between pre and posttranslocated states of the EC. To examine this, we employed an Exo III footprinting assay, in which we labeled the 5′ end of the NT DNA strand with [γ32P]ATP and treated ECs with Exo III for 5 min at 37°C. Consistent with footprinting data for E.coli RNAP (32), in the Tth EC14 complex about 14 nt of downstream DNA is protected (Figure 2A, lanes 1 and 2). Upon addition of ATP, EC14 was converted to EC15, which shifted the position of the front edge of RNAP downstream by 2 bp (Figure 2A, lanes 3 and 4). Whereas the footprinting assay shows that some other conformations may be present, these results, together with the PPi cleavage data described above, suggest that EC14 is mostly in the pretranslocated state, while EC15 is mostly in the posttranslocated state (Figure 2B).
Figure 2.
Probing of the EC translocation conformations. (A) Exo III footprints of EC14 and EC15. Tth EC14 (lanes 1 and 2) was assembled as described in Materials and Methods. EC15 (lanes 3 and 4) was obtained by incubation of EC14 with substrate ATP for 2 min at 60°C. Exo III (0.02 U/μl) was added for 5 (lanes 1 and 3) or 10 (lanes 2 and 4) min at 37°C. (B) Schematics of RNAP front edge oscillations in EC14 and EC15. (C) Photo cross-linking patterns of Tth EC14 and EC15, EC14 and EC15 containing 5′ 32P-labeled RNA primers were prepared as illustrated (left). The photo cross-linking analog 4-thio UTP (50 μM) was incorporated into the transcript for 2 min at 60°C followed UV light irradiation for 5 min at RT. The cross-linked species were separated using gel electrophoresis (right).
Lastly, we examined the difference in position of the 3′ end of RNA in ECs by photo cross-linking method in which we incorporated 4-thio UTP at the 3′ end of RNA and UV-irradiated the complexes. While both β and β′ subunits became cross-linked in EC14 and EC15 Tth complexes, the distribution of the cross-link in these complexes was notably different (Figure 2C). In EC14, most of the cross-link was associated with the β′ subunit, while in EC15 the 3′ end of the RNA cross-linked to both β′ and β subunits. Although the 3′ end RNA cross-linking does not by itself attest to the conformation of the complex, the observed distribution of the cross-link in EC14 and EC15 likely reflects changes in the protein environment between the product and the substrate site, and thus supports our findings of distinct EC conformations. The substrate site, which is occupied by the 3′ end of the RNA in EC14, may involve strong interactions with several structural elements in β′ subunit, (such as the bridge helix and trigger loop) important for substrate binding and formation of the closed complex (6,7,15) and thus yield an efficient protein:RNA cross-link exclusive to this subunit.
Exo- and endo- nucleolytic activity of Tth RNAP
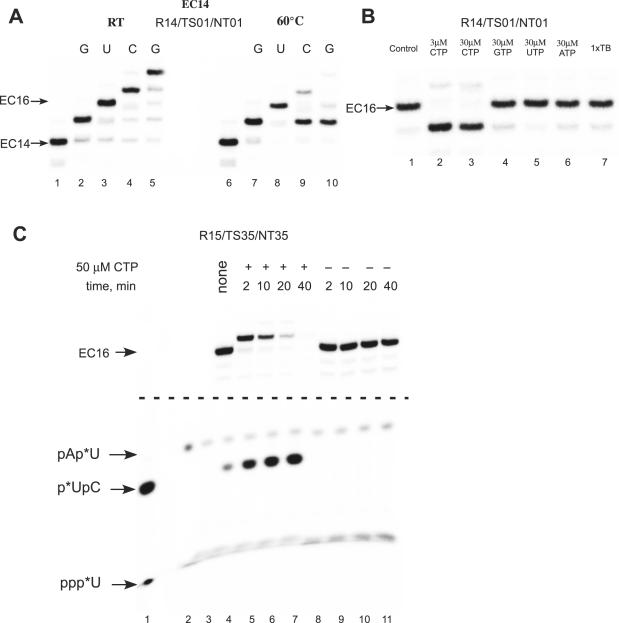
Besides pyrophosphorolysis, bacterial RNAP is also capable of 3′ to 5′ exo- and endo-RNA hydrolysis, activities that require the pretranslocated or the backtracked state of RNAP (33). To monitor intrinsic cleavage in scaffold ECs, we incubated EC14 (R14/TS35/NT35), EC15 (R15/TS35/NT35) and complexes obtained by extension of RNA primer in EC15 by 1 nt (EC16), without addition of any nucleotides for several hours at 60°C. Independent on the pH of the buffer, the RNAP intrinsic cleavage activity was weak in these and all aforementioned complexes (data not shown), with somewhat more efficient endonucleolytic activity observed in EC16 (see below). As we noted above, core Tth RNAP is able to efficiently extend an RNA primer in the EC at RT. Interestingly, when we compared the results of walking experiments performed at different temperatures, we found that instead of extension of EC16 in the presence of the next incoming substrate, CMP, the RNA was trimmed to 15 nt within 2 min at 60°C (Figure 3A). Addition of non-cognate NTPs or incubation with transcription buffer alone did not result in the cleavage of the transcript, suggesting that EC16 by itself is not prone to the endonucleolytic cleavage within the time of the experiment (Figure 3B). To test whether the cognate NTP ‘stimulated’ cleavage of the RNA in EC16 at 60°C, we labeled the RNA at its 3′ end by incorporation of [α-32P]UTP and incubated the complex with or without substrates (Figure 3C). Because of RNAP endonucleolytic activity, di-nucleotide product (pApU) appeared upon incubation of EC16 in transcription buffer (Figure 3C, lane 8–11). When substrate CTP was added to EC16, we observed accumulation of labeled di-nucleotide product pUpC (Figure 3C, lanes 4–7). Thus, the RNA hydrolysis observed during extension of EC16, was due to hypersensitivity of EC17 to the intrinsic endonucleolytic activity of polymerase.
Figure 3.
Intrinsic endonuclease activity in Tth RNAP EC. (A) Walking of Tth RNAP on an immobilized DNA template. EC14, formed using biotinylated NT DNA strand (NT01) and a 5′ 32P-labeled RNA primer was immobilized on streptavidin agarose beads, and the RNA primer was extended by several subsequent steps of NTP incorporation and washing at RT (lanes 1–5) and 60°C (lanes 6–10). (B) Nucleolytic activity in Tth RNAP EC16. Tth EC16 was obtained by extension of the RNA primer in EC14 using the walking technique at 60°C (lane 1) and incubated with the CTP (lanes 2 and 3), non-cognate NTPs (lanes 4–6) or without substrates (lane 7) for 5 min at 60°C. (C) EC16 (lane 3) was obtained by extension of the RNA primer in R15/TS35/NT35 by incorporation of [α-32P]UTP at the 3′ end of the RNA. The complex was then incubated in the presence (lanes 4–7) or absence (lanes 8–11) of 50 μM CTP for 2–40 min at 60°C, and the products of the reaction were analyzed by 23% PAGE electrophoresis. The central portion of the gel was removed for clarity. The position of 32P-labeled markers—pUpC (lane 1) and pApU (lane 2) are indicated by arrows; * denotes position of the labeled phosphate.
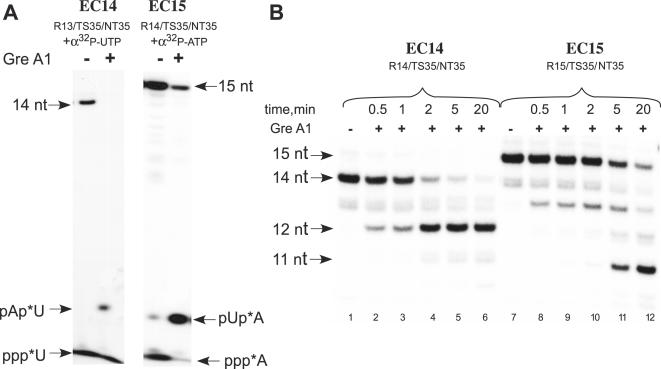
GreA-induced nucleolytic activity
Transcript cleavage factor GreA greatly stimulates endonucleolytic activity of Tth RNAP (34,35). We examined the sensitivity of scaffold ECs having 8 and 9 bp RNA:DNA hybrids to GreA-induced cleavage, as it may also attest to the conformation state of the polymerase in EC. To monitor the products of the reaction, we labeled the 3′ end of the RNA by incorporation of [α-32P]NTP in pre-assembled complexes (Figure 4A). GreA-induced cleavage of both EC14 and EC15 resulted in accumulation of di-nucleotide products (Figure 4A) as was shown previously for Tth RNAP complexes (34,35). When the pretranslocated EC14 having a 5′-labeled RNA was incubated with GreA, the initial 14mer RNA was completely converted into 12mer RNA within 5 min (Figure 4B, lanes 1–6). It is likely that shortening of the RNA:DNA hybrid in EC12 (to 6 bp) results in an unstable complex and precludes further RNA cleavage. Interestingly, posttranslocated EC15 was also GreA- sensitive; however, RNA hydrolysis was notably slower, as more than 20 min were required to generate EC13 that was, in turn, rapidly converted into EC11 (Figure 4B, lanes 7–12).
Figure 4.
Sensitivity of Tth RNAP ECs to GreA stimulated nucleolytic activity. (A) Complexes R13/TS35/NT35 and R14/TS35/NT35 were labeled by incorporation of [α-32P]UTP and ATP, respectively, and incubated with an equimolar amount of Tth GreA (1 μM) for 10 min at 60°C. The products of the reaction were resolved in 23% PAGE. (B) EC14 and EC15, containing 5′ 32P-labeled RNA primers were incubated with equimolar amount of Tth GreA at 60°C for the times indicated.
Crystallization of Tth RNAP ECs
One of the goals of this work was to establish conditions for crystallization of functional EC of Tth RNAP. We selected EC14 and EC15 for crystallization trials as the nucleic acid components in these complexes most closely represent the minimal system required for assembly of a stable and functional EC and likely would not be involved in crystal contacts (29,36). Thus, the downstream DNA region (14 bp) is completely protected by polymerase from Exo III cleavage and the single-stranded portion of RNA (6 nt) must be located within the RNA exit channel (31).
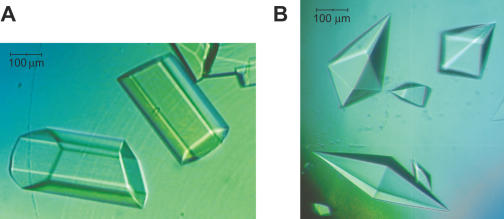
Using a sitting drop vapor-diffusion technique, we performed extensive screening for crystallization conditions, avoiding solutions containing buffers with extreme pH and/or high ionic strength precipitants in order to prevent dissociation of the elongation complex. We found crystallization conditions for both pretranslocated EC14 and posttranslocated EC15. The pretranslocated EC14 crystallized in the presence of a number of precipitating agents most of which included PEG 4000 or PEG 8000, and produced large hexagonal crystals (Figure 5A). Remarkably, the conditions used to crystallize EC14 did not produce any crystals with EC15, for which conditions were searched independently. We found that EC15 formed large pyramid-like crystals in the presence of alcohols, such as iso-propanol, 1,6-hexanediol and MPD (2-methyl-2,4-pentandiol) (Figure 5B). The latter crystals appeared to be of a much better quality compared to poorly diffracting (8–9 Å) crystals of EC14 and were used to collect a complete diffraction data at 2.5 Å resolution using synchrotron radiation (Table 1).
Figure 5.
Crystals of Tth RNAP ECs. (A) Hexagonal crystals of pretranslocated Tth RNAP EC14 (B) Bi-pyramidal crystals of posttranslocated Tth RNAP EC15.
Table 1.
Data collection statistics for the Tth RNAP elongation complex
| Unit-cell parameters | ||
|---|---|---|
| a (Å) | 145 | |
| b (Å) | 145 | |
| c (Å) | 498 | |
| α (°) | 90 | |
| β (°) | 90 | |
| γ (°) | 90 | |
| Temperature (K) | 100 | |
| Space group | P41212 | |
| Molecules in asymmetric unit | 1 | |
| Solvent content (%) | ∼73 | |
| Resolution (Å) | 2.5 (2.59–52.5) | |
| Observations | 749 175 | |
| Unique reflections | 193 488 | |
| Rmergea | 0.102 (0.432) | |
| Completeness (%) | 91.3 (83.5) | |
| I/σ(I) | 14.4 (2.8) |
Values in parentheses are for highest resolution shell.
aRmerge = , where Ij is the intensity of reflection j and (Ij) is the average intensity of reflection j.
DISCUSSION
During each catalytic cycle, transcription complexes must move along the DNA template, a process that is accompanied by conformational changes in the RNAP and the organization of the EC (11,13,37,38). Thus, the position of the active site of the enzyme relative to the 3′ end of the nascent RNA (pre and posttranslocated states) is variable, and, as shown in this study, is subject to various influences. These factors and their effects on the equilibrium between the two states complicate the interpretation of a number of biochemical and structural studies. For example, a number of E.coli ECs exhibited different sensitivity to exonuclease cleavage and pyrophosphorolysis and produced quite distinct patterns of cross-linking when reactive analogs were placed at the 3′ end of RNA (39). Thus, biochemical experiments have suggested that halted E.coli ECs represent a mixed population of pre and posttranslocated complexes (40). The heterogeneity of ECs has also been revealed in single molecule experiments (19).
Successful crystallization of ECs, however, requires homogeneous populations of complexes. One of the important goals of this work, therefore, was to determine conditions that would allow the formation of homogeneous populations of ECs. Characterization of ECs assembled on nucleic acid scaffolds, as described in this work, offers a number of advantages over promoter-originated ECs in terms of manipulation, convenience and, particularly, homogeneity. Interestingly, Tth RNAP ECs assembled on a 8 bp RNA:DNA hybrid scaffold appear to be in a pretranslocated form, while ECs that were assembled on a 9 bp RNA:DNA scaffold, or obtained by extension of EC8 by 1 nt, were in a posttranslocated state. This difference most clearly observed during pyrophosphorolysis—posttranslocated EC9 are extremely resistant to PPi (Figure 1). EC10 obtained by extension of EC8 by 2 nt (or by direct assembly) is highly sensitive to PPi and rapidly cleaves RNA to 9 nt. An immediate conclusion is that RNAP in the EC10 failed to translocate upon substrate incorporation, likely due to inability to displace the 5′ end of the RNA in the hybrid in the absence of a fully complementary NT strand (41). This suggests that the length of RNA:DNA hybrid that gives rise to the most stable conformation for Tth RNAP is 9 bp (in the posttranslocated state) as both 8 and 10 bp scaffold ECs exhibit high sensitivity to pyrophosphorolysis. This conclusion is in agreement with cross-linking data on the length of the RNA:DNA hybrid in a bacterial EC (21) and the fact, that additional interaction(s) with the transcript in the EC involve contacts with the RNA base that is 9 nt away from RNAP active site, as revealed by photo cross-linking studies (42).
In all assays used in this work assembled ECs having the same length RNA:DNA hybrid behaved similarly, regardless of scaffold topology, however, we cannot exclude that the absence of a particular DNA component affects complex conformation. Thus, pol II EC assembled on scaffold having an 8 bp RNA:DNA hybrid appeared to be in the posttranslocated state (12), whereas the 8 bp Tth ECs described here are in the pretranslocated state. While this discrepancy might be attributed simply to differences in the nature of the two proteins, it may also reflect differences in scaffold design (especially in view of the high conservation of the RNA:DNA hybrid binding site in eukaryotic and prokaryotic RNAPs). In contrast to scaffolds employed for Tth ECs, the pol II EC was assembled on a ‘bubble’ scaffold template that contained 11 bp of artificially melted DNA in the region of the RNA:DNA hybrid and 14 bp of upstream DNA duplex (12). As in the previous crystallization studies with T7 RNAP EC (43), a significant portion of the upstream DNA duplex was missing from the electron density in the pol II EC, suggesting limited protein–nucleic acid interactions in that region. Since the lack of upstream DNA duplex does not compromise the stability of bacterial ECs (31), we used such ‘minimal’ scaffolds for both biochemical and structural experiments.
The results of exonuclease footprinting experiments suggest that the tested complexes are mostly in pre (EC14) and posttranslocated (EC15) states. Intriguingly, we observed GreA stimulated endonuclease activity in a highly stable, posttranslocated EC15 (Figure 4A and B). Our data indicate that GreA-induced RNAP nuclease activity cleavage produced exclusively di-nucleotide products (Figure 4A). It is unclear, however, why during progressive cleavage the EC would backtrack by exactly 2 nt. It is tempting to speculate that GreA can shift the equilibrium towards backtracked conformation, e.g. by inducing large conformational changes in the RNAP. Such changes were observed in the case of a functional homolog of GreA, TFIIS, as revealed by the crystal structure of this transcription factor in complex with pol II EC (12). In that case, binding of TFIIS resulted in conformational changes in RNAP that led to realignment of the RNA in the active center and stabilization of an alternative conformation of the EC (12). Interestingly, dinucleotides are also the predominant products of TFIIS-stimulated cleavage (44,45). An alternative, more parsimonious explanation of sensitivity of EC15 to GreA is that it may serve as a more powerful (and thus more revealing) probe, compared to PPi, in monitoring oscillations of the EC between pre and posttranslocated conformations.
Interestingly, in contrast to E.coli RNAP (33), we did not find significant intrinsic cleavage activity in most of the Tth RNAP scaffold ECs and promoter complexes (though we tested only a limited number of such complexes). However, when the length of the RNA:DNA hybrid in the EC was increased to 10–11 bp we observed efficient endonucleolytic activity (Figure 3). It is likely that complexes assembled on scaffolds lacking the NT strand in the region of the transcription bubble, (such as EC16 and EC17, Figure 3) cannot efficiently displace the 5′ end of the transcript (41). Thus the formation of the overextended RNA:DNA hybrid results in a complex that is prone to endonucleolytic cleavage (Figure 3C).
All together, the results of biochemical assays indicate that ECs assembled on nucleic acid scaffolds with different structures (topology) possess quite distinct conformations. In the absence of NTPs, one conformation is greatly stabilized and preferred over another. Our data suggest that the length of the RNA:DNA hybrid plays a primary role not only in stabilization of EC (21,28) but also shifting the equilibrium between pre and posttranslocated conformations. Interestingly, the pretranslocated state of the EC (as observed in EC14) appears to be stable and preferred even in the absence of NTP, since very distinct patterns of footprinting and cross-links were observed (Figure 2). This observation argues against the power stroke mechanism of transcription elongation, which suggests that PPi release is required for the transition from a pre to a posttranslocated state in multi-subunit RNAPs (11). However, in contrast to bacterial ECs (which can assume different translocation conformations), halted T7 RNAP ECs appear to exist preferentially in a posttranslocated state (9,11,13,43,46). Moreover, ECs of T7 RNAP assembled on scaffolds similar or identical to those used in the current work are highly resistant to pyrophosphorolysis (25,47). Hence, it is possible that the single subunit T7-like RNAPs and bacterial RNAPs utilize different mechanisms of translocation.
The results of biochemical experiments suggesting that Tth RNAP ECs possess distinct conformations are in a good agreement with finding that pretranslocated EC14 and posttranslocated EC15 crystallized under different conditions. Since scaffold components of EC are buried inside of the polymerase, they are unlikely to affect contacts in the crystal lattice. Instead, yet to be identified conformational changes in the RNAP itself are likely to result in packing of the molecules into distinct crystal forms. Thus, it will be possible to use scaffold ECs to monitor different intermediates of transcription complexes of bacterial polymerases during the nucleotide addition cycle by structural analysis. Model building of the posttranslocated EC15 and refinement of the structure are under way.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Dr William T. McAllister for fruitful discussions and the critical reading of the manuscript, and Dr Anna Perederina for the expert technical assistance. This work was supported by National Institutes of Health grants GM74252 and GM74840 (to D.G.V.), UMDNJ Foundation Grant (to D.T.) and by RIKEN to (D.G.V.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vassylyev D.G., Svetlov V., Vassylyeva M.N., Perederina A., Igarashi N., Matsugaki N., Wakatsuki S., Artsimovitch I. Structural basis for transcription inhibition by tagetitoxin. Nature Struct. Mol. Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell E.A., Korzheva N., Mustaev A., Murakami K., Nair S., Goldfarb A., Darst S.A. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 3.Murakami K.S., Masuda S., Darst S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 4.Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 5.Artsimovitch I., Patlan V., Sekine S., Vassylyeva M.N., Hosaka T., Ochi K., Yokoyama S., Vassylyev D.G. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 6.Temiakov D., Zenkin N., Vassylyeva M.N., Perederina A., Tahirov T.H., Kashkina E., Savkina M., Zorov S., Nikiforov V., Igarashi N., et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 8.Symersky J., Perederina A., Vassylyeva M.N., Svetlov V., Artsimovitch I., Vassylyev D.G. Regulation through the RNA polymerase secondary channel. Structural and functional variability of the coiled-coil transcription factors. J. Biol. Chem. 2006;281:1309–1312. doi: 10.1074/jbc.C500405200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahirov T.H., Temiakov D., Anikin M., Patlan V., McAllister W.T., Vassylyev D.G., Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 10.Landick R. Active-site dynamics in RNA polymerases. Cell. 2004;116:351–353. doi: 10.1016/s0092-8674(04)00121-7. [DOI] [PubMed] [Google Scholar]
- 11.Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in t7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 12.Kettenberger H., Armache K.J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 13.Temiakov D., Patlan V., Anikin M., McAllister W.T., Yokoyama S., Vassylyev D.G. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Guajardo R., Sousa R. A model for the mechanism of polymerase translocation. J. Mol. Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 15.Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 16.Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang M.Y., Sheetz M.P. Mechanics of myosin motor: force and step size. Bioessays. 1994;16:531–532. doi: 10.1002/bies.950160803. [DOI] [PubMed] [Google Scholar]
- 18.Komissarova N., Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuman K.C., Abbondanzieri E.A., Landick R., Gelles J., Block S.M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 20.Shaevitz J.W., Abbondanzieri E.A., Landick R., Block S.M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nudler E., Mustaev A., Lukhtanov E., Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 22.Sosunov V., Sosunova E., Mustaev A., Bass I., Nikiforov V., Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassylyeva M.N., Lee J., Sekine S.I., Laptenko O., Kuramitsu S., Shibata T., Inoue Y., Borukhov S., Vassylyev D.G., Yokoyama S. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta. Crystallogr. 2002;D58:1497–1500. doi: 10.1107/S0907444902011770. [DOI] [PubMed] [Google Scholar]
- 24.Nudler E., Gusarov I., Bar-Nahum G. Methods of walking with the RNA polymerase. Meth. Enzymol. 2003;371:160–169. doi: 10.1016/S0076-6879(03)71011-8. [DOI] [PubMed] [Google Scholar]
- 25.Temiakov D., Anikin M., McAllister W.T. Characterization of T7 RNA polymerase transcription complexes assembled on nucleic acid scaffolds. J. Biol. Chem. 2002;277:47035–47043. doi: 10.1074/jbc.M208923200. [DOI] [PubMed] [Google Scholar]
- 26.Kashlev M., Martin E., Polyakov A., Severinov K., Nikiforov V., Goldfarb A. Histidine-tagged RNA polymerase: dissection of the transcription cycle using immobilized enzyme. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. Processing X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Sidorenkov I., Komissarova N., Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 29.Korzheva N., Mustaev A., Nudler E., Nikiforov V., Goldfarb A. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harb. Symp. Quant. Biol. 1998;63:337–345. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- 30.Ryder A.M., Roberts J.W. Role of the non-template strand of the elongation bubble in intrinsic transcription termination. J. Mol. Biol. 2003;334:205–213. doi: 10.1016/j.jmb.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 31.Komissarova N., Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korzheva N., Mustaev A. Transcription elongation complex: structure and function. Curr. Opin. MicroBiol. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- 33.Sosunov V., Zorov S., Sosunova E., Nikolaev A., Zakeyeva I., Bass I., Goldfarb A., Nikiforov V., Severinov K., Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laptenko O., Borukhov S. Biochemical assays of Gre factors of Thermus thermophilus. Meth. Enzymol. 2003;371:219–232. doi: 10.1016/S0076-6879(03)71016-7. [DOI] [PubMed] [Google Scholar]
- 35.Hogan B.P., Hartsch T., Erie D.A. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J. Biol. Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- 36.Temiakov D., Tahirov T.H., Anikin M., McAllister W.T., Vassylyev D.G., Yokoyama S. Crystallization and preliminary crystallographic analysis of T7 RNA polymerase elongation complex. Acta. Crystallogr. D. Biol. Crystallogr. 2003;59:185–187. doi: 10.1107/s0907444902019777. [DOI] [PubMed] [Google Scholar]
- 37.Cramer P. RNA polymerase II structure: from core to functional complexes. Curr. Opin. Genet. Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 39.Markovtsov V., Mustaev A., Goldfarb A. Protein–RNA interactions in the active center of transcription elongation complex. Proc. Natl Acad. Sci. USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erie D.A. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim. Biophys. Acta. 2002;1577:224–239. doi: 10.1016/s0167-4781(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 41.Kireeva M.L., Komissarova N., Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J. Mol. Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 42.Korzheva N., Mustaev A., Kozlov M., Malhotra A., Nikiforov V., Goldfarb A., Darst S.A. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 44.Izban M.G., Luse D.S. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J. Biol. Chem. 1993;268:12864–12873. [PubMed] [Google Scholar]
- 45.Gu W., Reines D. Variation in the size of nascent RNA cleavage products as a function of transcript length and elongation competence. J. Biol. Chem. 1995;270:30441–30447. doi: 10.1074/jbc.270.51.30441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheetham G.M., Steitz T.A. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 47.Temiakov D., Mentesana P.E., Ma K., Mustaev A., Borukhov S., McAllister W.T. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl Acad. Sci. USA. 2000;97:14109–14114. doi: 10.1073/pnas.250473197. [DOI] [PMC free article] [PubMed] [Google Scholar]