Short abstract
Two contrasting views of the relation of the metabolic cycle to the cell cycle in budding yeast are compared.
Abstract
Slowly growing budding yeast store carbohydrate, then liquidate it in late G1 phase of the cell cycle, superimposing a metabolic cycle on the cell cycle. This metabolic cycle may separate biochemically incompatible processes. Alternatively it may provide a burst of energy and material for commitment to the cell cycle. Stored carbohydrate could explain the size requirement for cells passing the Start point.
Yeast, like my children, are at their most vibrant in high concentrations of sugar. The budding yeast Saccharomyces cerevisiae extracts just 2 moles of ATP per mole of glucose via fermentation (that is, glycolysis to pyruvate, then reduction of the pyruvate to ethanol), but it grows rapidly with a doubling time of about one and a half hours. In low concentrations of glucose (or in non-fermentable carbon sources) yeast grow via oxidative respiration, extracting more than 30 moles of ATP per mole of glucose - but now their doubling time increases to 3 hours or longer.
Yeast growing oxidatively in limited glucose use this glucose in three major ways. First, of course, they use it as an energy source; glucose flows through glycolysis to generate ATP, NADH and pyruvate, and the pyruvate flows through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation to generate even more ATP. Second, they use glucose as a raw material for building the cell wall. Third, another large portion of the glucose is stored, some in the polysaccharide glycogen and some in the disaccharide trehalose. So, even though these respiring cells are in some sense starved for glucose (as they could grow faster if more glucose were available), they nevertheless store a fair portion of the glucose. About 16% of the dry weight of a respiring cell is stored carbohydrate (that is, glycogen plus trehalose), whereas a cell growing via fermentation on abundant glucose has virtually no stored carbohydrate [1,2].
Storing and burning, storing and burning
The fate of this stored carbohydrate is remarkable. The story is old [3-5] but complex. And recent studies [6,7] showing the oscillation of many genes as a function of the metabolic cycle have added another level of complexity, as discussed later. In the long G1 phase of a slowly growing, glucose-limited cell, cells oxidize glucose to grow by respiration, but they also store glucose as glycogen and trehalose. But in late G1, some event, possibly a spike in the level of cyclic AMP [8], changes all this. Storage ceases. The cell's stores of glycogen and trehalose are suddenly liquidated to glucose. The released glucose now floods through glycolysis into oxidative respiration, greatly increasing the rate of respiration. The sudden wave of glucose is, however, too much to be absorbed by the respiratory pathway, and a good deal of the glucose is simply fermented to ethanol. Amazingly, at this point in the cycle, glucose-limited cells are actually excreting ethanol from overflow glycolysis into the medium [8]. Thus, briefly, these cells are obtaining some of their energy from fermentation, by suddenly burning their stores of carbohydrate, and they greatly increase their production of ATP. The cells express mRNAs for the cyclins Cln1 and Cln2, commit to passage through the cell cycle by passing the point known as Start, and enter S phase, in which the DNA is replicated [7]. Shortly afterwards, having exhausted their stores of carbohydrate, the cells stop fermentation, respire at a low rate as permitted by the small amounts of glucose (and now ethanol) available from the medium, and begin the arduous process of storing carbohydrate for the next cell cycle (see Figure 1). Similar events also occur in yeast cells limited for other carbon sources.
Figure 1.
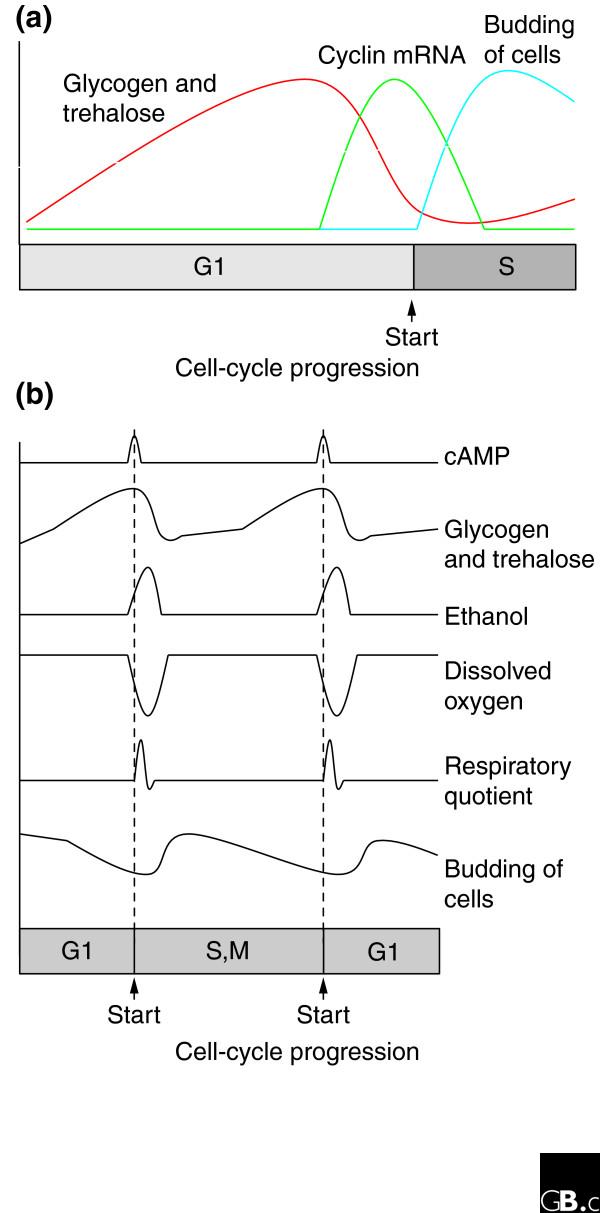
The metabolic cycle in slowly growing yeast cells. (a) The cycle of stored carbohydrate. In slowly growing cells, glycogen and trehalose build up during G1, then are suddenly liquidated in late G1. Shortly after liquidation, the mRNA levels of the G1 cyclins Cln1 and Cln2 reach a peak, Start is passed, and then budding and DNA synthesis occur. Adapted from the data of Sillje et al. [9], who studied cells in which the length of G1 was 500 to 600 minutes. (b) The metabolism of spontaneously synchronized cells in limiting glucose. The cyclic changesin the levels of various indicators of metabolism are shown. A small spike of cyclic AMP is seen in mid or late G1. Almost immediately afterwards, glycogen and trehalose are liquidated. Ethanol appears in the medium, presumably the result of fermentation of the freed glucose. The amount of dissolved oxygen in the medium plunges at the same time that stored carbohydrate is disappearing, and the same time as ethanol is appearing. The disappearance of oxygen suggests that glucose from stored carbohydrate isbeing metabolized by respiration as well as by fermentation. The respiratory quotient spikes from just below 1 to about 1.2, signifying a shift from nearly pure respiration (which would give a respiratory quotient of 1.0) tometabolism involving some fermentation. Budding follows shortly afterwards. Part (b) is adapted from a study by Muller et al. [8], in which cyclic AMP varies from about 6 nmol/g dry weight to about 12 nmol/g dry weight; stored carbohydrate varies from 175 mg/g dry weight to about 80 mg/gdry weight; ethanol varies from 0 to 125 mg/l, dissolved oxygen varies from 80-65% saturation (in Muller et al. [8]) or 60-20% saturation (in Tu et al., see Figure 1 in [7]); the respiratory quotient variesfrom 0.85 to 1.2 at the top of the spike; and budding index varies from 5% to 40%.
These peculiar cycles of storing and then burning carbohydrate can have an even more peculiar effect on the yeast culture as a whole. Depending on factors such as dilution rate, oxygen levels and pH, a culture of yeast grown in a chemostat in limiting glucose can become spontaneously synchronized with respect to the cell cycle: all the cells in the culture align at the same place in the cell cycle and then progress through the cycle together, an effect first seen more than 35 years ago [9-13]. Again the full story is complex (see [14] and references therein), but a simplified explanation is that the cells that first liquidate their stored carbohydrate and secrete ethanol are thus feeding ethanol to other cells in the culture. The cells receiving the ethanol can therefore grow faster, and so can catch up to the more advanced cells. Once they catch up, they too become feeders rather than receivers. To say the same thing in a different way, each cell has an internal oscillation of storing, then burning, carbohydrate, and these oscillations can be synchronized through a whole culture by the cross-feeding effects of released ethanol and perhaps other metabolites (for instance, hydrogen sulfide and acetaldehyde have been suggested [15]). Thus, Figure 1 can be viewed as the events happening in a single cell in an asynchronous culture or, as in the studies by Muller et al. [8] and Sillje et al. [9], as the events happening to all the cells in a synchronous culture.
The excretion of ethanol into the medium is energetically wasteful from the cell's point of view. But yeast grow as clonal cultures, so most of the excreted ethanol is likely to be taken up by genetically identical cells, minimizing the cost from the clone's point of view.
Many genes oscillate as a function of the metabolic cycle
Recently, two groups have used microarrays to analyze gene expression throughout the cell cycle of spontaneously synchronized cells [6,7]: that is, cells synchronized by growth in limiting glucose and experiencing synchronous waves of storing and then burning carbohydrate. Up to half of all genes showed at least a weak oscillation. Of course, some of these are typical cell-cycle-linked genes such as the histone genes, as noted in previous studies [16]. But there were many strongly oscillating genes that are intimately associated with the metabolic oscillation described above and that do not significantly oscillate in a cell cycle in high glucose. These oscillating metabolic genes form a number of functionally and temporally related clusters. For instance, in mid-G1, at about the time that stored carbohydrate is being liquidated and ATP production is maximal, there is a large cluster of genes involved in protein synthesis and ribosomal biogenesis, including the ribosomal proteins themselves [6,7] (a similar cluster was also noted by [17]). Other genes involved in protein synthesis, and genes involved in sulfur metabolism, amino-acid synthesis, and RNA processing, also peak at this time. The combination of high ATP production, ribosome biogenesis and amino-acid synthesis and related functions suggests that cells in mid-G1 may have an especially high rate of protein synthesis. Cells pass through Start near the end of this period. Interestingly, a similar phenomenon of a peak in ribosome biogenesis and protein synthesis occurs in the fission yeast Schizosaccharomyces pombe just before commitment [17], even though in S. pombe this commitment occurs in the G2 phase.
At about the time that stored carbohydrate has been exhausted and the amount of dissolved oxygen in the medium begins to climb (indicating a lack of substrate for respiration), the microarray analysis shows expression of histones [6,7], indicating on-going DNA replication. Genes for spindle-pole components are also expressed, consistent with the idea that the cells are in S phase. At this time, cells have budded, again consistent with S phase. Finally, many nuclear genes for mitochondrial proteins, such as mitochondrial ribosomal proteins, peak at this time [6,7]. The reason for this peak is not obvious, as the highest rate of respiration has by now passed. Tu et al. [7] suggest that cells are either rebuilding or duplicating their mitochondria at this time. Many variations on this theme are possible; for instance, it might be a time when mitochondrial import is particularly favored, and so mitochondrial proteins are synthesized to meet a window of opportunity for import.
Finally, late in the cell cycle, many peroxisomal proteins and certain other classes of proteins are upregulated [7]. Peroxisomes are the site of β-oxidation of fatty acids, yielding acetyl-CoA, which is the starting point for the generation of ATP via respiration. Thus, just as the cell is getting an extra energy boost from stored carbohydrate in mid-G1, it could be getting an energy boost from stored lipid in G2/M.
Compartmentalization versus the finishing kick
So, why does the respiring yeast cell have these metabolic cycles and the associated oscillations of hundreds or thousands of genes? There are two views, not mutually exclusive, which I will call the 'compartment' hypothesis, and the 'finishing kick' hypothesis. These are built on work from two distinct groups of researchers, publishing in distinct journals, and in some cases possibly not aware of each other's results. The compartment hypothesis is championed by Tu et al. [7], and it states that some of the different metabolic processes in the cell are incompatible with one another and therefore they are compartmentalized in time. This view is built on earlier work by Murray, Kuriyama, and colleagues [6,15,18], who viewed the metabolic oscillation shown in Figure 1b as primarily an oscillation in redox potential, shown by a strong oscillation in NADH [18]. As an example of compartmentalization, Tu et al. [7] suggest that respiration is incompatible with glycolysis, and therefore that the two processes are carried out at two different times of the cell cycle (this particular suggestion contrasts with the view given above where peak respiration occurs simultaneously with peak glycolysis). As a second example, both Klevecz et al. [6] and Tu et al. [7] suggest that respiration might be incompatible with DNA synthesis, as respiration might be mutagenic. Furthermore, there might be many circumstances under which two pathways might interact to cause futile cycles unless the pathways were separated in either space or time.
The idea that respiration and glycolysis happen at different times cannot be completely correct: for one thing, respiration requires glycolysis to produce pyruvate and acetyl-CoA. But the idea that they might occur largely at different times arises from interpretations of dissolved oxygen measurements. All investigators agree that in spontaneously synchronized respiring cultures, there is a period when the concentration of dissolved oxygen in the chemostat medium falls sharply (see Figure 1b). This decrease in dissolved oxygen is a sign of increased respiration. Tu et al. [7] call this period the 'Ox' period, and they interpret it as a brief window in the cycle during which respiration can occur; they feel that respiration is relatively insignificant outside of the Ox period. Later, the concentration of dissolved oxygen in the medium rises sharply (see Figure 1b), indicative of a decreased rate of respiration. Tu et al. [7] interpret this as the cessation of respiration, and name this period of time the 'R/B' period (for reductive, building period). They do not specify how cells obtain ATP during this period. Finally, there is a period when the concentration of dissolved oxygen is high and stable; this is named the 'R/C' period (the reductive, charging period) and Tu et al. [7] suggest that during this period, ATP is obtained by glycolysis, but not (or not significantly) by respiration. In summary, Tu et al. [7] suggest that respiration is more-or-less confined to the Ox period of the metabolic cycle, while glycolysis is more-or-less confined to the R/C period of the metabolic cycle, and this temporal separation minimizes conflicts between different modes of metabolism.
More detailed metabolic measurements throw serious doubt on this interpretation, however. Although there are quite a number of relevant papers, I will focus on the example of Muller et al. [8], who made extensive measurements in very comparable cells, which were also spontaneously synchronized in low glucose (see Figure 1b). They measured not only dissolved oxygen, but also the respiratory quotient (which is the ratio of the volume of carbon dioxide produced to the volume of oxygen consumed: for pure respiration, this ratio is 1, whereas for pure fermentation, the ratio is infinitely high, as no oxygen is consumed). They also measured the amount of stored carbohydrate, ethanol and acetate in the medium, and other parameters. The measurements of Muller et al. [8] show that during the Ox phase, both respiration and glycolysis are occurring at a high rate; a high rate of respiration is shown by a high rate of oxygen consumption, whereas a high rate of glycolysis is shown by a respiratory quotient that spikes above 1, and by the loss of storage carbohydrate and its reappearance as ethanol (the end-product of yeast fermentation) in the medium. Indeed, Tu et al. [7] also find ethanol in the medium during their Ox phase, a sign that in their experiments glycolysis and fermentation must also be extremely active at this time. In the R/B and R/C phases, the measurements of Muller et al. [8] indicate almost pure respiration; the respiratory quotient is stable, just less than 1 (as expected if respiration is occurring but some carbon is retained by the cell for anabolism). In summary, the measurements of Muller et al. [8] suggest that there is no temporal compartmentalization of respiration from glycolysis; rather, almost the opposite is true: respiration is occurring all the time (though certainly at a higher rate during the Ox period than at other times), and glycolysis is most intense during the Ox period, exactly when respiration is most intense. According to these results the intense respiration and the simultaneous glycolysis could be the consequences of the liquidation of stored carbohydrate occurring at this time.
A related proposal is that oxidative and reductive processes are compartmentalized; this is based in part on an oscillation in NADH [18]. This oscillation is tricky to interpret, however. First, oxidative and reductive steps through glycolysis, the TCA cycle and oxidative respiration usually are coupled in time, so while a portion of the yeast metabolic cycle may provide an exception to this coupling, it is nevertheless difficult to argue that the processes must occur at different times. Second, NADH is in flux through pathways, and a measurement of instantaneous concentration is not sufficient to describe the flux. Nevertheless, the oscillation of NADH is very clear [18] and cries out for an explanation. It may be significant that the peak of NADH occurs just as respiration is slowing down, and just as nuclear genes for mitochondrial functions are reaching peak expression. Perhaps mitochondria are taking a break from oxidative phosphorylation to import new protein and reorganize themselves; this might be an example of compartmentalization.
Another interesting possibility is that the oscillation in NADH (and the opposite-phase oscillation in NAD) could affect the activity of Sir2, an NAD-dependent histone deacetylase [19]. NADH peaks at about S phase, and the lower NAD concentration at this time could decrease activity of Sir2 [19,20], and so increase the acetylation of histones and perhaps other proteins. Conceivably, this could be somehow connected to the fact that re-establishment of gene silencing in yeast requires passage through S phase, but not actual DNA replication [21,22].
Even if respiration and glycolysis are not temporally compartmentalized, many other processes in the metabolic cycle certainly are, and so the compartment hypothesis remains a powerful idea that may explain other aspects of the metabolic oscillation. We still lack a clear example of temporally compartmentalized processes known to be mutually incompatible, however.
An alternative view which I propose here, the 'finishing-kick' hypothesis, is an outgrowth of the work of Muller et al. [8], Sillje et al. [9], Schneider et al. [23], and many other authors. This hypothesis focuses on the requirements for Start, the commitment to the cell cycle that takes place at the G1/S transition. Start depends on three G1 cyclins, Cln1, Cln2 and Cln3, which bind and activate the cyclin-dependent kinase (CDK) Cdc28; the kinase activity of the resulting complexes then catalyzes Start. The three G1 cyclins are all very unstable proteins (even at very low growth rates [23]) encoded by very unstable mRNAs. The finishing-kick hypothesis states that at low rates of protein synthesis, cells will not pass through Start. One mechanism for preventing Start at low protein synthesis rates is that G1 cyclins cannot be synthesized to the requisite level, because they turn over too quickly. This rapid turnover can only be overcome by high rates of protein synthesis. Therefore, the slowly growing cell organizes its metabolism to store sufficient carbohydrate, then suddenly burns it to provide a burst of ATP and protein synthesis in late G1. This metabolic burst provides enough G1 cyclin and other materials (glucans for the wall of the new bud, deoxynucleotides for DNA synthesis, and so on) for this key event of the cell cycle. That is, there is a finishing kick to Start and, exactly like the finishing kick of an Olympic 10,000-meter runner, it involves a lot of glycolysis.
Although the instability of G1 cyclin provides the mechanism by which Start is delayed, providing G1 cyclin is not the point, or at least not the whole point, of the metabolic burst. If it were, the cell would evolve a more stable G1 cyclin and be done with it. Rather, the unstable G1 cyclin is a gating device that limits Start to times when carbohydrate, other materials and protein synthesis rates are sufficiently high for all needs.
The finishing kick and critical size
It has been known for many years that slowly growing cells have a long G1, and only when these cells have grown to 'critical size' can they pass through Start. In the finishing kick hypothesis, critical size is equivalent to stored carbohydrate; that is, the hypothesis predicts that the size-correlated parameter being measured by the cell is glycogen plus trehalose. When enough carbohydrate is stored for successful passage through this energy- and material-intensive part of the cell cycle, this is somehow sensed (perhaps via some glucose-related metabolite such as glucose 6-phosphate and the cyclic AMP pathway), a signal is sent (again, perhaps via the cyclic AMP pathway), the carbohydrate is liquidated, ATP is produced, and a burst of metabolism, nucleotide synthesis, protein synthesis and all the other events of Start ensue. The late-G1 peak in expression of ribosome and protein synthesis genes noted by both Klevecz et al. [6] and Tu et al. [7] is explained by the need for a burst of protein synthesis. Interestingly, most of the mutations affecting critical cell size either affect the synthesis of G1 cyclins (for example, CLN3-1, whi3 and whi5) or are related to (though not actually in) the cyclic AMP pathway (sch9 and sfp1) [24]. Oscillations in other metabolites and sets of genes would be explained as downstream effects of the oscillation in stored carbohydrate and of the metabolic burst.
The finishing-kick hypothesis can only explain critical size and Start in slowly growing cells. Cells growing rapidly on abundant glucose have little or no stored carbohydrate, and in any case no need for a metabolic burst, and the finishing-kick hypothesis is irrelevant to such cells. But there is also evidence that cells use multiple mechanisms for controlling the time of Start [23], and the mechanisms that apply in fast-growing cells may be quite different from those in slow-growing cells [25].
It is clear that glucose-limited yeast do undergo a metabolic oscillation superimposed on their cell-cycle oscillation. Whether this metabolic oscillation is primarily for temporal compartmentalizing of different metabolic process, or primarily for managing Start under difficult circumstances, or whether both hypotheses are true, remains to be seen. One promising avenue for distinguishing the hypotheses is the study of mutants that do not store any glycogen or trehalose [2,26]. Such mutants are alive, but with aberrant cell cycles. At present, phenotypic analysis is not detailed enough to distinguish the two hypotheses, but in principle, mutants that lack storage carbohydrate should allow some interesting experiments.
Acknowledgments
Acknowledgements
I thank Benjamin Tu for helpful discussions; Steve Oliver for acquainting me with the work of Muller et al. [8], Adam Rosebrock, Aaron Neiman and Janet Leatherwood for comments on the manuscript, and Bob Halti-wanger for expert discussions of yeast metabolism. This work was supported by the NIH, RO1 GM39978.
References
- Francois J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Guillou V, Plourde-Owobi L, Parrou JL, Goma G, Francois J. Role of reserve carbohydrates in the growth dynamics of Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:773–787. doi: 10.1016/j.femsyr.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Panek A. Function of trehalose in baker's yeast (Saccharomyces cerevisiae). Arech Biochem Biophys. 1963;100:422–425. doi: 10.1016/0003-9861(63)90107-3. [DOI] [PubMed] [Google Scholar]
- Kuenzi MT, Fiechter A. Changes in carbohydrate composition and trehalase-activity during the budding cycle of Saccharomyces cerevisiae. Arch Mikrobiol. 1969;64:396–407. doi: 10.1007/BF00417021. [DOI] [PubMed] [Google Scholar]
- Kuenzi MT, Fiechter A. Regulation of carbohydrate composition of Saccharomyces cerevisiae under growth limitation. Arch Mikrobiol. 1972;84:254–265. doi: 10.1007/BF00425203. [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Muller D, Exler S, Aguilera-Vazquez L, Guerrero-Martin E, Reuss M. Cyclic AMP mediates the cell cycle dynamics of energy metabolism in Saccharomyces cerevisiae. Yeast. 2003;20:351–367. doi: 10.1002/yea.967. [DOI] [PubMed] [Google Scholar]
- Sillje HH, ter Schure EG, Rommens AJ, Huls PG, Woldringh CL, Verkleij AJ, Boonstra J, Verrips CT. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiechter A, von Meyenburg HK. Regulatory properties of growing cell populations of Saccharomyces cerevisiae in a continuousculture system. In: Kockova-Kratochrilova A, editor. Proceedings of the 2nd Symposium on Yeast, Bratislava. Slovenskej; 1968. pp. 387–398. [Google Scholar]
- von Meyenburg HK. Stable synchrony oscillations in continuous cultures of Saccharomyces cerevisiae under glucose limitation. In: Chance B, Pye EK, Ghosh TK, Hess B, editor. Biological and Biochemical Oscillators. New York: Academic Press; 1973. pp. 411–417. [Google Scholar]
- Parulekar SJ, Seamons GB, Rolf MJ, Lim HC. Induction and elimination of oscillations in continuous cultures of Saccharomyces cerevisiae. Biotechnol Bioeng. 1986;28:700–710. doi: 10.1002/bit.260280509. [DOI] [PubMed] [Google Scholar]
- Porro D, Martegani E, Ranzi BM, Alberghina L. Oscillations in continuous cultures of budding yeast: a segregated parameter analysis. Biotech Bioeng. 1988;32:411–417. doi: 10.1002/bit.260320402. [DOI] [PubMed] [Google Scholar]
- Jones KD, Kompala DS. Cybernetic model of the growthdynamics of Saccharomyces cerevisiae in batch and continuous cultures. J Biotechnol. 1999;71:105–131. doi: 10.1016/S0168-1656(99)00017-6. [DOI] [PubMed] [Google Scholar]
- Murray DB, Klevecz RR, Lloyd D. Generation and maintenance of synchrony in Saccharomyces cerevisiae continuous culture. Exp Cell Res. 2003;287:10–15. doi: 10.1016/S0014-4827(03)00068-5. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DB, Engelen F, Lloyd D, Kuriyama H. Involvement of glutathione in the regulation of respiratory oscillation during a continuous culture of Saccharomyces cerevisiae. Microbiology. 1999;145:2739–2745. doi: 10.1099/00221287-145-10-2739. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- Kirchmaier AL, Rine J. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:852–862. doi: 10.1128/MCB.26.3.852-862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AM, Nasmyth KA. Role of DNA replication in therepression of silent mating type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Zhang J, Markwardt J, Tokiwa G, Volpe T, Honey S, Futcher B. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start. Mol Cell Biol. 2004;24:10802–10813. doi: 10.1128/MCB.24.24.10802-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Laabs TL, Markwardt DD, Slattery MG, Newcomb LL, Stillman DJ, Heideman W. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:10275–10280. doi: 10.1073/pnas.1833999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


