Short abstract
A high-throughput luciferase-based assay for circadian gene expression gives a boost to the study of circadian rhythms in Chlamydomonas.
Abstract
Recent work on the circadian clock of the unicellular green alga Chlamydomonas reinhardtii strengthens its standing as a convenient model system for circadian study. It was shown to be amenable to molecular engineering using a luciferase-based real-time reporter for circadian rhythms. Together with the completed draft genomic sequence, the new system opens the door for genome-scale forward and reverse genetic analysis.
Chlamydomonas reinhardtii is often referred to as 'green yeast' to convey the usefulness of this eukaryotic unicellular green alga as a model organism for plant research [1]. Like a plant cell, Chlamydomonas possesses a cell wall and a chloroplast, but like animal sperm cells, it has a flagellum. This structure enables it to carry out phototaxis, moving towards or away from light and so maximizing light perception for photosynthesis and minimizing photodamage. When given a light source, C. reinhardtii can be grown in large quantities in a simple medium containing only inorganic salts. In the dark, it can also grow non-photosynthetically with acetate as its sole carbon source. As most of its life cycle is in the haploid phase, C. reinhardtii is amenable to genetic screens, which have led to major discoveries in the fields of chloroplast biogenesis and the structure of eukaryotic flagella and the basal bodies from which they derive [2].
In the past decade, the C. reinhardtii community has embraced the genomic revolution [3]. In addition to the mitochondrial and chloroplast genomes, the 110 Mb nuclear genome has been sequenced and is currently being annotated using the information generated by two large-scale expressed sequence tag (EST) projects. More than 15,000 genes have now been identified and this information is accessible through the C. reinhardtii genome portal [4]. Genome-scale analyses of protein sequences have described the repertoire of cell-cycle regulatory proteins, tyrosine kinases and flagella and basal body genes [5-8]. The EST sequencing projects have also been instrumental in designing cDNA macroarrays and oligonucleotide arrays [3,9,10]. In addition, the generation of a sequence database has facilitated the data-mining processes needed to identify large protein complexes by mass spectrometry [11,12]. Finally, progress made toward facilitating map-based cloning, and insertional mutagenesis has turned this little green eukaryotic cell into a key model system for the 21st century [13,14]. Its usefulness as a model system is well illustrated by a recent article from Matsuo et al. [15], which describes the application of genetic engineering and high-throughput technology to provide a new and convenient system for studying circadian rhythms in C. reinhardtii.
To provide a real-time readout of circadian rhythms in C. reinhardtii, Matsuo and colleagues [15] generated a strain expressing luciferase driven by the circadian-regulated psbD gene promoter. Luciferases are naturally occurring enzymes capable of emitting light after substrate catalysis. The most widely used luciferase comes from the firefly, which uses the substrate luciferin [16]. This enzyme is better suited than green fluorescent protein (GFP) as a reporter for gene-expression analyses as it has a half-life of a few hours and does not require external illumination, thus avoiding the problem of autofluorescence and photodamage to the cells. In the newly developed strain, Matsuo et al. [15] re-engineered the single-chain firefly luciferase with optimized codons for high-level expression in C. reinhardtii and integrated the reporter construct into the chloroplast genome.
There have been previous reports of C. reinhardtii engineered to express luciferases from bacteria or the sea pansy Renilla, a coelenterate, but Matsuo et al. [15] are the first to integrate firefly luciferase and to demonstrate the feasibility of measuring circadian rhythms in real time continuously for several days [15,17-19]. The advantage of using the firefly luciferase over Renilla or bacterial luciferase is that the substrate luciferin is more affordable and soluble than their substrates (coelenterate luciferin and decanal, respectively), an important factor to consider when developing a high-throughput system. To be able to monitor several hundred individual cultures, they created an automated high-throughput platform composed of a 96-well plate-conveying system, a measuring unit, a controller, and an analyzing computer that accumulates and analyzes bioluminescence data [20,21]. In the course of a 4-day experiment, they can measure the level of the reporter at hourly intervals by detecting and quantifying the light produced by the enzyme acting on the exogenously provided substrate luciferin.
The capacity to monitor circadian rhythms in a non-invasive way using such techniques has proved a tremendous step forward in the molecular dissection of the circadian clock in cyanobacteria, plants, Drosophila, and mouse [16]. Even if the emitted bioluminescence readout displays a 24-hour periodicity, the first step that has to be taken following the development of such a tool is to prove that the pattern is indeed generated by a circadian clock. Matsuo and colleagues [15] did this by showing firstly that, after a few days of entrainment, the emitted signal maintains a circadian period under constant light and constant darkness, secondly that the signal is temperature compensated (an innate property of all circadian clocks) as it maintains an approximately 24-hour period over a wide range of temperatures, and thirdly that the circadian phase is sensitive to light pulses.
Opening up the clock
C. reinhardtii was chosen as a model for circadian clock studies in the 1970s, when Victor Bruce [22] showed that C. reinhardtii phototaxis was circadian regulated. Since then, other physiological processes, as well as the expression of several chloroplast- and nuclear-encoded genes, have been shown to be under circadian control, but clock components have yet to be identified in this species [23]. Pioneer studies have identified several mutants with circadian-period phenotypes, but the associated genes remain unknown [24].
In the plant Arabidopsis thaliana, forward genetic screens using a circadian-driven luciferase reporter followed by map-based cloning identified three clock genes: TOC1, ZTL, and LUX [25]. Initial circadian reporter measurements revealed that mutation of these genes results in a short period (about 21 hours), long period (about 28 hours), or arrhythmic phenotype, respectively, after being entrained under 24-hour light-dark cycles. As mentioned earlier, a forward genetic screen in C. reinhardtii has led to the isolation of mutants displaying circadian-period defects. In their final but critical experiment, Matsuo and colleagues [15] showed that their system can accurately monitor the period difference identified in the classical period mutants per-1 and per-4 isolated by Victor Bruce in the 1970s [24]. Along with validating the methodology, this result also demonstrates that the period length of the chloroplast-integrated reporter depends upon the genotype of the nucleus, suggesting that it is under the control of a nucleus-encoded circadian oscillator. With their high-throughput system, Matsuo et al. now have the capability to undertake a genomic clone complementation strategy to isolate those mutations and identify the per-1 and per-4 genes. In addition, the high-throughput system also offers the possibility of conducting mutant screens that do not rely on a physiological phenotype such as phototaxis, as it is now possible to directly monitor rhythmicity with the reporter.
The C. reinhardtii strain developed by Matsuo et al. [15] is also suitable for use in reverse genetic screening. It is possible to find potential clock genes in a homology-based genome survey and then test their roles in the C. reinhardtii clock using a newly developed collection of insertion mutants and analyzing insertion lines containing the reporter [14]. Such an approach has been very successful in A. thaliana in ascribing clock function to genes with sequence similarity to TOC1 (for example, PRR3, PRR5, PRR7 and PRR9) [25]. As no core clock component has yet been identified in C. reinhardtii, an attractive strategy would be to identify genes showing sequence homology to clock genes in other species. Comparison of fly and mouse genomes has been critical in the identification of clock genes in mouse and humans [26]. To satisfy our curiosity, we and others have already peeked at the C. reinhardtii genome draft sequence to identify potential clock gene sequences ([27] and G.B. and S.A.K., unpublished observations).
Looking for clock parts in the Chlamydomonas genome
Because C. reinhardtii is a photosynthetic eukaryotic organism, it is likely that it has homologs to some A. thaliana clock genes. In higher plants and animals, the central clock mechanism is an interlocking feedback loop. In A. thaliana, light signaling through the phytochrome and cryptochrome families of protein pigments induces the entrainment to the circadian cycle of two Myb-related transcription factors, CCA1 and LHY, which regulate the 'evening' element (EE)-containing genes PRR9, TOC1, and LUX, so-called as the genes first associated with the element were found to be genes expressed in the evening [25,28] (Figure 1). The TOC1, PRR7 and PRR9 proteins contain a pseudo-response regulator and a CCT domain specific to plants whose function still needs to be identified. The other core clock component, LUX, is a novel Myb-related transcription factor from the Golden 2, Arabidopsis response regulator B, Phosphate starvation response 1 (GARP) subfamily and its target site in DNA is currently unknown. These core proteins might be part of a larger complex involved in controlling the expression of CCA1 and LHY throughout the day. In addition, other genes encoding the so-called 'pioneer' clock proteins ELF3, ELF4, and GI were identified. The loss-of-function phenotypes of these genes displayed clock phenotypes ranging from compromised to arrhythmic behavior, but their functional role in the Arabidopsis clock is currently unknown [25,28]. BLAST searches against the C. reinhardtii genome sequence versions 2.0 and 3.0 revealed that it encodes small Myb-related transcription factors with homology to the DNA-binding domains of LUX, CCA1, and LHY. One ELF4 homolog and several PRR-like genes containing a response-regulator domain and a CCT domain were also found ([27] and G.B. and S.A.K., unpublished observations). All those genes are C. reinhardtii candidate clock components whose function could be assessed using insertion lines and the reporter strain developed by Matsuo et al. [15].
Figure 1.
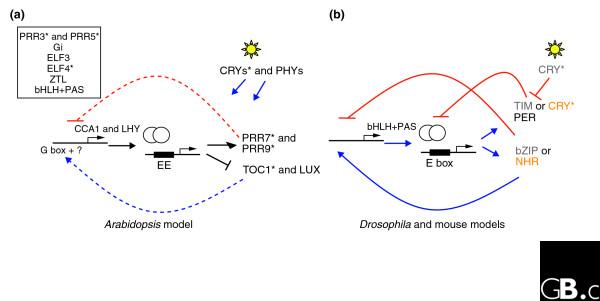
Similarities between the plant, Drosophila and mouse circadian clocks. (a) In Arabidopsis, light perceived through cryptochromes (CRYs) and phytochromes (PHYs) acts to entrain the circadian clock. The Myb-like transcription factors CCA1 and LHY (white circles) bind to the evening element (EE), forming a feedback loop that regulates genes that are thought to positively (blue dashed line) and negatively (red dashed line) regulate the expression of CCA1 and LHY; positive regulation probably occurs through binding of G box and possibly other elements. Forward and reverse genetic screens have identified pioneer genes whose function in the clock still needs to be defined (boxed). (b) In Drosophila and mouse, transcription factors with both bHLH and PAS domains bind the E-box element to regulate positive (blue line) and negative (red line) regulators of their own expression. Fly proteins are indicated in grey and mouse proteins in orange; proteins indicated in black are found in both. The regulators are bZIP type factors in fly and nuclear hormone receptor (NHR) proteins in mouse. In addition, the proteins containing the bHLH and PAS domains also regulate the expression of binding partners that will repress their action. The PER protein acts with TIM in the fly to inhibit the function of the bHLH proteins, whereas it acts with CRY in mouse. In the fly, the CRY protein is a photoreceptor involved in TIM degradation. Proteins marked with an asterisk represent potential clock genes for which homologs were detected in the C. reinhardtii genome sequence.
C. reinhardtii also has some animal-like features that are absent in plants, such as a flagellum and tyrosine kinases [6,7]. Thus, the C. reinhardtii genome could yield genes homologous to fly or mouse circadian clock genes. Despite the large evolutionary distance between mouse and Drosophila, their circadian clock mechanisms show some degree of similarity to each other (Figure 1) [26,29]. In each system, proteins with basic helix-loop-helix (bHLH) DNA-binding motifs and two PAS protein-protein interaction domains positively regulate genes with E-box control elements. Some of the targeted genes act as activators or repressors of the bHLH genes, creating a feedback loop (Figure 1). The resemblance is not perfect, as in mammals those activators and repressors are nuclear hormone receptor proteins, whereas in Drosophila they are basic leucine zipper (bZIP)-type factors. In addition to the similarity in E-box-binding proteins and feedback loop architecture, sequence similarity is also found among the proteins that directly interact with the bHLH proteins. In mammals, the bHLH dimer also regulates the transcription of a pair of proteins, the crytochrome-like chromophore-binding protein CRY and the PAS-domain Period (PER) protein, that directly interact with the bHLH dimer to inhibit its function (Figure 1). The scenario is different in Drosophila, where the bHLH interactors are the PAS-domain proteins PER and Timeless (TIM). CRY in the fly does not directly interact with the bHLH dimer; instead it is involved in light-induced targeted degradation of TIM [30].
The C. reinhardtii genome has two CRY proteins, one of which is more similar to the mouse CRY than to the Drosophila or Arabidopsis CRYs. This suggests that a mammalian-type CRY and a hitherto unidentified PER homolog may be part of a protein complex with a role in the C. reinhardtii clock [27]. The second C. reinhardtii CRY is closer to its higher-plant cryptochrome counterpart, suggesting that it may function as a photoreceptor. In regard to light sensing and the clock, C. reinhardtii may have an amalgam clock including ancestral mechanisms from before the evolutionary split between the plant and animal kingdoms. In this case, a combination of reverse genetics and the system developed by Matsuo and colleagues [15] may help define the C. reinhardtii clock mechanism, as well as helping us learn more about higher plant and animal clocks. For example, both plants and animals have bHLH transcription factors with PAS-like domains that can bind E-box-like elements (known as the G-box in plants). Unfortunately, in plants there is no indication yet that these proteins are as embedded in the core oscillator as are their mammalian and fly counterparts.
To sum up, the bioluminescence reporter strains developed by Matsuo and colleagues [15] can now be used in conjunction with new genomic resources to design new mutant screens as well as reverse genetics approaches to test C. reinhardtii genes homologous to genes involved in other circadian system. As plant biologists, we look forward to further developments and have begun guessing whether our favorite genes will be part of the Chlamydomonas clock or not. Given the information coming from the C. reinhardtii genome sequence, the excitement is likely to spread to the mammalian community as well. Is it a hybrid plant and animal clock, two parallel systems, or a novel system with as-yet unidentified players? Whatever the outcome, the luciferase lights are on and Chlamydomonas is ready to shine.
Acknowledgments
Acknowledgements
We thank members from the Kay lab for suggestions and critical reading of this manuscript.
References
- Gutman BL, Niyogi KK. Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol. 2004;135:607–610. doi: 10.1104/pp.104.041491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, Zhang Z. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell. 2003;2:1137–1150. doi: 10.1128/EC.2.6.1137-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlamydomonas Center http://www.chlamy.org
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/S0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/S0092-8674(04)00412-X. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol Biol Evol. 2004;21:828–840. doi: 10.1093/molbev/msh077. [DOI] [PubMed] [Google Scholar]
- Bisova K, Krylov DM, Umen JG. Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:475–491. doi: 10.1104/pp.104.054155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Okamoto K, Tabata S, Fukuzawa H, Ishiura M. Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol Biol. 2005;57:889–906. doi: 10.1007/s11103-005-3248-1. [DOI] [PubMed] [Google Scholar]
- Eberhard S, Jain M, Im CS, Pollock S, Shrager J, Lin Y, Peek AS, Grossman AR. Generation of an oligonucleotide array for analysis of gene expression in Chlamydomonas reinhardtii. Curr Genet. 2006;49:106–124. doi: 10.1007/s00294-005-0041-2. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR, 3rd, Mayfield SP. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70 S ribosome. J Biol Chem. 2003;278:33774–33785. doi: 10.1074/jbc.M301934200. [DOI] [PubMed] [Google Scholar]
- Wagner V, Gessner G, Mittag M. Functional proteomics: a promising approach to find novel components of the circadian system. Chronobiol Int. 2005;22:403–415. doi: 10.1081/CBI-200062348. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Handley JM, Thomas M, Stern DB. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:557–566. doi: 10.1104/pp.104.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Onai K, Okamoto K, Minagawa J, Ishiura M. Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol Cell Biol. 2006;26:863–870. doi: 10.1128/MCB.26.3.863-870.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Hausherr A, Ferbitz L, Schodl T, Heitzer M, Hegemann P. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol. 2004;55:869–881. doi: 10.1007/s11103-004-2150-6. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Schultz J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 2004;37:449–458. doi: 10.1046/j.1365-313X.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- Minko I, Holloway SP, Nikaido S, Carter M, Odom OW, Johnson CH, Herrin DL. Renilla luciferase as a vital reporter for chloroplast gene expression in Chlamydomonas. Mol Gen Genet. 1999;262:421–425. doi: 10.1007/s004380051101. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Onai K, Ezaki N, Ofuchi T, Ishiura M. An automated apparatus for the real-time monitoring of bioluminescence in plants. Anal Biochem. 2005;340:187–192. doi: 10.1016/j.ab.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Onai K, Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal Biochem. 2005;340:193–200. doi: 10.1016/j.ab.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Bruce VG. The biological clock in Chlamydomonas reinhardtii. J Protozool. 1970;17:328–334. [Google Scholar]
- Mittag M, Wagner V. The circadian clock of the unicellular eukaryotic model organism Chlamydomonas reinhardtii. Biol Chem. 2003;384:689–695. doi: 10.1515/BC.2003.077. [DOI] [PubMed] [Google Scholar]
- Bruce VG. Mutants of the biological clock in Chlamydomonas reinhardi. Genetics. 1972;70:537–548. doi: 10.1093/genetics/70.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Imaizumi T, Kay SA. The plant circadian clock: review of a clockwork Arabidopsis. In: Hall A, McWatters HG, editor. Endogenous Plant Rhythms. Vol. 21. Oxford: Blackwell; 2005. pp. 1–23. [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii What is it for? What is it similar to? Plant Physiol. 2005;137:399–409. doi: 10.1104/pp.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kay SA. Circadian clocks in daily and seasonal control of development. Science. 2003;301:326–328. doi: 10.1126/science.1085935. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sassone-Corsi P. Structural and functional features of transcription factors controlling the circadian clock. Curr Opin Genet Dev. 2005;15:548–556. doi: 10.1016/j.gde.2005.07.003. [DOI] [PubMed] [Google Scholar]


