Short abstract
An RNAi screen in Drosophila cells has identified about 100 TANGO proteins, which may regulate protein exocytosis or secretion.
Abstract
Although the organization and functions of the constitutive secretory pathway have been intensively studied for decades, a recent genome-wide RNAi screen in Drosophila cells has identified about 100 genes encoding novel so-called TANGO proteins (for transport and Golgi organization) that may be direct regulators of various aspects of protein exocytosis or secretion.
Soon after the phenomenon of RNA interference (RNAi) was described in animal cells [1], it became evident that RNAi not only represented an ancient mechanism to control viral infections and physiological developmental processes, but it would also provide researchers with a powerful tool for functional genetic analysis [2]. Because of its efficiency and specificity, RNAi has gained ground as a method of disrupting protein function over the traditional method of transiently expressing dominant-negative mutant proteins, which can yield misleading results [3]. In fact, downregula-tion or depletion of a protein with unknown function by RNAi has nowadays become an almost obligatory part of its characterization.
The availability of entire genome sequences of an increasing number of organisms has given RNAi an additional boost. The construction of double-stranded (ds) RNA libraries representing nearly all the genes in a genome provides the opportunity to identify proteins that regulate specific cellular and developmental processes. Several genome-wide RNAi screens have already been carried out in Caenorhabditis elegans and Drosophila cells, whereas only smaller-scale screens are feasible at the moment in mammalian cells [4]. High-throughput RNAi screens in Drosophila S2 cells have been used to unravel genes involved in cell growth and viability [5], cell-cycle progression [6], cytokinesis [7], and several signaling pathways such as Wnt/Wingless [8], JAK-STAT [9,10] and Hedgehog [11]. In this article, we will focus on a recent genome-wide RNAi screen in the S2 cell model system with which Bard et al. [12] have looked into another fundamental cellular process - protein secretion.
The secretory pathway
The secretory pathway is the compulsory route taken by most secreted and plasma membrane proteins from the site of their synthesis to their final destination. Thus, its main function is protein transport. The organization of the pathway was established by the pioneering work of George Palade [13]. It consists of several membrane-bounded compartments including the rough endoplasmic reticulum, where proteins enter the pathway, the transitional endoplasmic reticulum sites (tER sites) where newly synthesized proteins exit the endoplasmic reticulum via specialized transport vesicles called COPII-coated vesicles, the Golgi apparatus, the post-Golgi carriers and the plasma membrane (see [14] for an introduction to the molecular players in the pathway). Post-translational modifications and sorting take place as proteins travel through the pathway, and these are crucial for the proteins' proper localization and function.
For decades, the tractable genetics of budding yeast have made it a valuable tool for studying the secretory pathway. Screens of temperature-sensitive mutants in Saccharomyces cerevisiae have identified many genes for proteins involved in protein secretion (sec genes) [15,16]. The secretory pathway in S. cerevisiae lacks the complexity of that of higher eukaryotes, however, and has several differences. For instance, Golgi stacks are rarely observed under normal conditions and tER sites do not form a discrete compartment in yeast [17,18]. Instead, exit from the endoplasmic reticulum can take place throughout the membrane network [18].
Drosophila S2 cells are a good alternative model system to study exocytosis. They have secretory compartments with similar morphological and molecular characteristics to those of mammalian cells. For example, anterograde protein transport (transport away from the endoplasmic reticulum) is inhibited by brefeldin A or H-89 [19,20], and the Golgi stacks disassemble during mitosis [21]. The main difference between the two systems is in the organization of the Golgi apparatus. In mammalian cells, Golgi stacks are laterally interconnected, forming a 'Golgi ribbon' capping the nucleus, whereas in Drosophila cells they are independent of each other and are distributed throughout the cytoplasm in close proximity to tER sites, forming tER-Golgi units (Figure 1a) [19,21]. The number of tER-Golgi units in S2 cells is relatively small and constant and can be easily monitored, which is an additional advantage to using these cells to study the secretory pathway.
Figure 1.
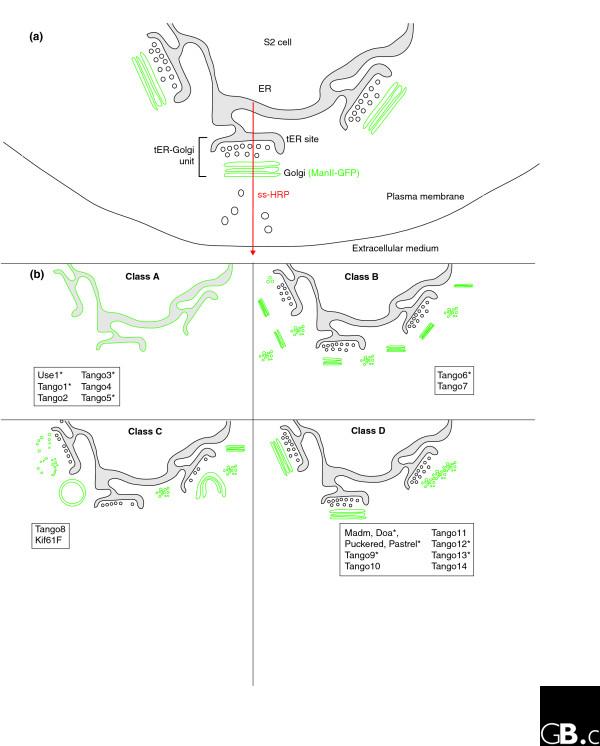
Schematic representation of the effects of depleting TANGO (transport and Golgi organization) genes on Golgi organization. (a) The constitutive protein secretory pathway in Drosophila S2 cells, showing its major compartments. The red arrow indicates the route through which the reporter signal sequence-HRP protein (ss-HRP) used in the RNAi screen by Bard et al. [12] is transported and secreted. The Golgi membranes marked by mannosidase II (ManII)-GFP are outlined in green. The lumen of the endoplasmic reticulum (ER) is shaded pale gray. tER, transitional endoplasmic reticulum; PM, plasma membrane. (b) The four phenotypes (classes A-D) that Bard et al. [12] identified as a result of the depletion of 130 TANGO genes are shown here; they were defined by the localization of ManII-GFP, which normally resides in the Golgi stacks. Depletion of class A genes led to a redistribution of ManII-GFP to the endoplasmic reticulum; depletion of class B genes caused fragmentation of Golgi membranes; knock-down of class C genes led to aggregation of ManII-GFP-positive membranes (and formation of ring-like structures); and depletion of class D genes inhibited protein secretion but did not affect the Golgi structure. The 20 TANGO proteins that were further characterized by Bard et al. [12] are listed under the diagram depicting the phenotype induced by their depletion. Proteins containing at least one putative transmembrane domain are marked with an asterisk.
Screening for proteins regulating constitutive protein secretion
Using Drosophila S2 cells, Bard et al. [12] carried out a genome-wide RNAi screen to identify novel proteins involved in constitutive protein secretion. The primary screen was performed in S2 cells stably transfected with the soluble protein horseradish peroxidase (HRP) fused to a signal sequence (ss-HRP; Figure 1a) to direct the protein to the classical secretory route. Expression of the HRP fusion protein was under the control of a metallothionein promoter, which was switched on after 5 days of RNAi. The efficiency of secretion was assessed by measuring HRP activity in the culture medium by chemiluminescence. As positive controls, depletion of β-COP (a COP I coat subunit) and syntaxin 5, a t-SNARE (soluble NSF attachment protein receptor) protein previously shown to block anterograde transport and cause Golgi stack vesiculation [19], were performed. As expected, depletion by RNAi led to significant inhibition of HRP secretion. Depletion of green fluorescent protein (GFP) served as a negative control.
The primary screen yielded 1,133 dsRNAs whose downregulation affected generic protein secretion. To narrow down this number, the authors excluded genes that were presumed to inhibit protein secretion indirectly, including genes previously implicated in transcription, translation, apoptosis and basic metabolism, as well as positive hits in screens examining cell growth and viability [5]. This first selection reduced the positive hits to 284 dsRNAs. Surprisingly, established components of the secretory pathway have not been picked up in a screen for genes involved in cell growth [5], although some key components for secretion are expected to be essential for growth, as are the yeast sec genes [15,16]. Of course, the RNAi method may produce hypomorphic phenotypes, where a small amount of the protein is still expressed. It is, however, anticipated that a 90% reduction in synthesis of an essential protein of the pathway would lead to a significant inhibition of anterograde transport and impede growth in cell size just by preventing plasma membrane growth. Furthermore, mutations in proteins involved in secretion, such as COG5 [22] and syntaxin 5 ([23] and our unpublished work), have been shown to impair cytokinesis. This should also lead to a reduction in cell numbers. Thus, the selection by Bard et al. [12] may have eliminated some regulators of the exocytic pathway that hinder cell growth rate directly or indirectly.
To exclude the possibility that the measured reduction in protein secretion was not an indirect effect of an arrest in cell growth or cell proliferation, the remaining 284 dsRNAs were tested again in the ss-HRP secretion assay in combination with cell counting (nuclear staining by Hoechst). This, plus discarding non-reproducible results, led to the further elimination of 154 hits.
Characterization of the TANGO genes
Of the remaining 130 genes, 100 were completely uncharacterized and have not been found to be directly involved in protein secretion before. All these genes were named by Bard et al. [12] as TANGO (transport and Golgi organisation) genes and their role in the organisation of the Golgi apparatus was characterized upon their depletion by RNAi. To investigate this, Bard et al. [12] used a stable S2 cell line expressing GFP-tagged α-mannosidase II (ManII-GFP), a transmembrane enzyme marker for Golgi membranes [24] (Figure 1a). RNAi for these 130 genes in ManII-GFP S2 cells produced four different phenotypic classes [12] (Figure 1b). Class A (23.8%) comprises genes whose depletion leads to a redistribution of ManII-GFP into the endoplasmic reticulum. This is likely to be due to impaired translocation into the endoplasmic reticulum or impaired transport from the endoplasmic reticulum to the Golgi, as this class includes proteins with established roles in these processes, such as a translocon subunit, Arf/Sar GTPases, vesicle docking proteins (SNAREs), and COPI and COPII subunits. Class B genes (15.3%) encode proteins that when knocked down cause a fragmentation of Golgi membranes into smaller elements (either small stacks or vesiculated membrane), whereas the depletion of class C gene products (4.6%) leads to aggregation and swelling of the ManII-GFP-positive membranes, including the formation of ring-like structures. Finally, class D genes represent the majority of the 130 selected genes (56.2%) that inhibit protein secretion, but their depletion did not significantly alter Golgi organization. It should be pointed out, however, that the fluorescent spots seen with this phenotype may not necessarily correspond to intact Golgi stacks, but to highly fragmented ones, a question that only electron microscopy can resolve [19]. Because the Golgi phenotypes B and C include genes previously associated with mitotic progression, Bard et al. [12] looked at the mitotic index after RNAi to see whether these phenotypes simply correspond to cells arrested in mitosis. They found no increase in the mitotic index, confirming the earlier Hoechst staining results that did not exclude these genes. In any case, phenotype C would be difficult to explain as a mitotic phenotype, as Golgi stacks are known to disassemble during metaphase in S2 cells [21].
As a proof of the RNAi screen's validity, several genes previously reported to control intracellular vesicle-trafficking steps were identified, such as the gene for the vesicle fusion protein NSF (N-ethylmaleimide-sensitive fusion protein). Not all anticipated genes were picked up in the screen, however - for example, not all COPI and COPII coat subunits were scored. This could reflect functional redundancies (for example, protein isoforms) or be an inherent drawback of such large-scale analyses.
Twenty representative TANGO proteins from all four phenotypic classes were further studied: six formerly annotated genes and 14 uncharacterized gene products (see Figure 1). These genes were cloned, tagged with a V5 epitope and then expressed to examine the localization of their gene products. Interestingly, 17 localize partially or exclusively in compartments of the early secretory pathway, consistent with a potential direct role in protein secretion. At least some of these proteins contain a predicted signal sequence (for example, TANGO9, TANGO13 and TANGO14), and half of them (marked with asterisks in Figure 1b) have one or more predicted transmembrane regions (predicted using PRED-TMR2 version 1.0 [25]), through which they could be targeted to the membranes of the exocytic pathway. Running a BLAST search to find sequence homologs indicates that these proteins may regulate protein secretion by carrying out very diverse functions, including membrane docking and fusion (the Golgin-like coiled-coil protein TANGO1 and the endoplasmic reticulum SNARE Use1), control of the pH in membrane compartments (the transmembrane transporter TANGO12), or protein modification (the sulfotransferase activity of TANGO13).
The big challenge now for researchers in the field is to determine the relevance of these novel gene products in protein secretion and the directness of their effects. First, the proteins that are directly involved in anterograde transport should be distinguished from those involved indirectly through inhibition of retrograde transport. Second, positive hits corresponding to uncharacterized proteins or to proteins unrelated to exocytosis might have indirect effects on the process. TANGO7, for example, contains a PINT domain that is found in proteasome subunits, and TANGO14 encodes a putative lipid synthase, raising the question of the role of proteasomes and lipid metabolism in protein secretion. The highly conserved TANGO2 protein, which contains a DUF833 domain (domain of unknown function), is another potential regulator requiring further characterization. Finally, TANGO5 bears significant similarity to vacuolar membrane protein1 (VMP1), a protein that normally localizes to the endoplasmic reticulum and Golgi membranes and is upregulated under stress conditions that induce vacuole formation and cell death [26]. The characterization of the role of TANGO5 under physiological conditions may reveal another novel player of the secretory pathway.
In conclusion, the screen performed by Bard et al. [12] represents a major effort and is the first of several to tackle various aspects of the exocytic pathway. The classes of genes identified in this screen will be of great use for comparisons with regulators of the anterograde transport of specific proteins, such as Wingless and Hedgehog, secreted proteins that stimulate two signaling pathways of crucial importance in development. Such comparisons are likely to reveal factors that control specialized trafficking pathways versus constitutive protein secretion. Furthermore, constitutive transport of transmembrane proteins may require different sets of genes compared with transport of soluble proteins. Thus, additional screens using transmembrane reporters will need to be performed. Lastly, proteins that disrupt Golgi stack architecture, such as Golgi matrix proteins dp115, dGM130 and dGRASP, are not in the list of the positive hits, because their depletion does not block anterograde transport [19,20]. Therefore, the identification of proteins that are involved in Golgi stack organization without affecting secretion will require yet another screening design.
The combination of high-throughput RNAi screens with other large-scale molecular datasets on genome-wide expression profiling and proteomics are likely to provide leads for cell biology research in the years to come. With this prospect, Drosophila as a model system will supply the means to transfer the knowledge we obtain from these genomic approaches to the more complex level of developmental biology, thus providing a more global view on many cellular processes.
Acknowledgments
Acknowledgements
We would like to thank Fulvio Reggiori and Madelon Maurice for their helpful comments. V.K. is supported by the Horizon Program from the National Regie-Orgaan Genomics (050-71-029).
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hudson DF, Morrison C, Ruchaud S, Earnshaw WC. Reverse genetics of essential genes in tissue-culture cells: 'dead cells talking'. Trends Cell Biol. 2002;12:281–287. doi: 10.1016/S0962-8924(02)02281-X. [DOI] [PubMed] [Google Scholar]
- Cullen LM, Arndt GM. Genome-wide screening for gene function using RNAi in mammalian cells. Immunol Cell Biol. 2005;83:217–223. doi: 10.1111/j.1440-1711.2005.01332.x. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HF, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Mellman I, Warren G. The road taken past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/S0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Schekman R. Lasker Basic Medical Research Award. SEC mutants and the secretory apparatus. Nat Med. 2002;8:1055–1058. doi: 10.1038/nm769. [DOI] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Spoorendonk KM, Rabouille C. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol Biol Cell. 2005;16:4061–4072. doi: 10.1091/mbc.E04-10-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci USA. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas RM, Giansanti MG, Gatti M, Fuller MT. The Drosophila Cog5 homologue is required for cytokinesis, cell elongation, and assembly of specialized Golgi architecture during spermatogenesis. Mol Biol Cell. 2003;14:190–200. doi: 10.1091/mbc.E02-06-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Brill JA, Hsien J, McBride R, Boulianne GL, Trimble WS. Syntaxin 5 is required for cytokinesis and spermatid differentiation in Drosophila. Dev Biol. 2002;251:294–306. doi: 10.1006/dbio.2002.0830. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kuntz DA, Lockyer A, Watson R, Signorelli T, Rose DR, van den Heuvel M, Roberts DB. The Drosophila GMII gene encodes a Golgi alphamannosidase II. J Cell Sci. 1999;112:3319–3330. doi: 10.1242/jcs.112.19.3319. [DOI] [PubMed] [Google Scholar]
- PRED-TMR2 version 1.0 http://o2.biol.uoa.gr/PRED-TMR2
- Dusetti NJ, Jiang Y, Vaccaro MI, Tomasini R, Azizi Samir A, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn JC, Iovanna JL. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun. 2002;290:641–649. doi: 10.1006/bbrc.2001.6244. [DOI] [PubMed] [Google Scholar]


