Abstract
During biosynthesis of bacteriochlorophyll or chlorophyll, three protein subunits of 140, 70, and 42 kDa interact to insert Mg2+ into protoporphyrin IX. The semidominant Chlorina-125, -157, and -161 mutants in barley are deficient in this step and accumulate protoporphyrin IX after feeding on 5-aminolevulinate. Chlorina-125, -157, and -161 are allelic to the recessive xantha-h mutants and contain G559A, G806A, and C271T mutations, respectively. These mutations cause single amino acid substitutions in residues that are conserved in all known primary structures of the 42-kDa subunit. In vitro complementation and reconstitution of Mg-chelatase activity show that the 42-kDa subunits are defective in the semidominant Chlorina mutants. A mutated protein is maintained in the Chlorina plastids, unlike in the xantha–h plastids. Heterozygous Chlorina seedlings have 25–50% of the Mg-chelatase activity of wild-type seedlings. Codominant expression of active and inactive 42-kDa subunits in heterozygous Chlorina seedlings is likely to produce two types of heterodimers between the strongly interacting 42-kDa and 70-kDa subunits. Reduced Mg-chelatase activity is explained by the capacity of heterodimers consisting of mutated 42-kDa and wild-type 70-kDa protein to bind to the 140-kDa subunit. The 42-kDa subunit is similar to chaperones that refold denatured polypeptides with respect to its ATP-to-ADP exchange activity and its ability to generate ATPase activity with the 70-kDa subunit. We hypothesize that the association of the 42-kDa subunit with the 70-kDa subunit allows them to form a specific complex with the 140-kDa subunit and that this complex inserts Mg2+ into protoporphyrin IX.
Keywords: chlorophyll synthesis, complementation assays, protoporphyrin IX, chlorina, aurea
Chelation of Mg2+ into protoporphyrin IX during the biosynthesis of bacteriochlorophyll or chlorophyll requires an enzyme, Mg-chelatase, consisting of three soluble subunits (1–6). The subunits show significant conservation of their primary amino acid sequences and molecular mass from Rhodobacter to cyanobacteria and higher plants, including barley, thale cress, and tobacco (for reviews, see refs. 7 and 8). The three subunits have nominal molecular masses of 140 kDa (range: 123–154 kDa), 70 kDa (range: 60–87 kDa), and 42 kDa (range: 37–46 kDa), respectively. Although the functional analysis of recombinantly expressed subunits is more advanced for Rhodobacter than for higher-plant Mg-chelatase, genetic studies and analyses of subunits from tobacco and barley thus far have indicated congruent characteristics. The largest subunit binds the substrate protoporphyrin IX. The metal-insertion step requires Mg2+ and ATP, in addition to the large subunit and protoporphyrin IX. It is preceded by an activation step, consisting of ATP hydrolysis, which is carried out by the combined action of the 70- and 42-kDa subunits (2, 9).
In barley, the 140-, 70-, and 42-kDa subunits are encoded by the Xantha-f, Xantha-g, and Xantha-h genes, respectively (4, 5). Mutations in these loci yielded the recessive, lethal genes xantha-f10, -f26, -f27, -f40, -f41, -f58, -f60, and -f68; xantha-g28, -g37, -g44, -g45, and -g65; and xantha-h30, -h38, -h56, and -h57 (10). These mutants lack Mg-chelatase activity and accumulate protoporphyrin IX and its monomethyl ester when fed the chlorophyll biosynthetic precursor 5-aminolevulinic acid. Some of the mutants are defective in transcription (e.g., xantha-h56 and -h57); others are defective in translation (e.g., xantha-h30 and -h38); and others are missense mutations (e.g., xantha-f10; ref. 4).
In amphidiploid tobacco, the gene of the semidominant aurea mutation Sulfur and its wild-type allele have been cloned by transposon tagging and sequenced (11, 12). Homozygotes of aurea mutations are yellow-seedling lethals, whereas the heterozygotes have a reduced amount of chlorophyll and are yellow-green. The sulfur gene encodes a 42-kDa Mg-chelatase subunit of tobacco, and the Sulfur mutation results in a single amino acid substitution from Asn to Ile in position 269 of the subunit inherited from the Nicotiana sylvestris parent. Nguyen (11) interpreted the dominant action of the single-mutated 42-kDa subunit by the formation of inactive Mg-chelatase, which thereby limits the synthesis of chlorophyll.
The three Mg-chelatase subunits of tobacco have been investigated by Gräfe (8) for expression in yeast and for protein–protein interaction in the GAL4/two-hybrid-yeast system. A strong interaction between the 42-kDa subunit (designated CHL I) and the 70-kDa subunit (designated CHL D) was observed in the activation of transcription of the reporter gene when the two subunits were fused to the activation and binding domains, respectively, of the GAL4 transcription factor. A weak interaction was observed between 70-kDa subunits and interpreted as homodimer formation. No interaction was registered with the 140-kDa subunit (designated CHL H), although this subunit was required for Mg-chelatase activity. Dissection of the 70-kDa subunit by expression of five peptide fragments in yeast, together with the cDNAs of the 140-kDa and 42-kDa tobacco subunits, indicated that the 312 N-terminal amino acid residues of the mature protein with significant homology to the 42-kDa subunit are dispensable for maximal Mg-chelatase activity. By coexpression of the proline-rich linker region and the two flanking helical domains at the center of the primary structure of the 70-kDa subunit with the two other subunits, 30% of maximal activity was achievable.
Recently, it has been shown that the purified 42-kDa subunit of Rhodobacter sphaeroides (designated BchI) catalyzes a phosphate-exchange reaction from one ATP molecule to an ADP, generating ADP and ATP, respectively, whereas ATP hydrolysis requires the combined presence of the 42-kDa (BchI) and 70-kDa (BchD) subunits (9). The 140-kDa subunit (BchH) by itself had weak ATPase activity. Recently, the unusual ATP-to-ADP phosphate-exchange activity has been found to be an intrinsic activity in the heat-shock protein Hsp70, which is thought to act as a molecular chaperone in protein folding (13). The semidominant action of the mutated 42-kDa tobacco Sulfur (CHL I) subunit and the possible importance of this subunit for correct folding of Mg-chelatase subunits led us to investigate three aurea mutants in barley, designated Chlorina-125, -157, and -161 (14, 15). They are allelic and segregate into a phenotypic ratio of one green wild-type leaf (clo/clo) to two viable light-green chlorina leaves (clo/Clo) to one lethal yellow xantha leaf (Clo/Clo) (Fig. 1).
Figure 1.
Leaves (6 days old) obtained from the seeds of a heterozygous barley Chlorina mutant. Because of the semidominant behavior of the Chlorina mutation, the leaves segregate into a phenotypic ratio of one normal green wild-type to two light-green heterozygotes to one yellow, chlorophyll-lacking homozygote. In this spike, the ratio is 3 normal green to 13 light-green to 5 yellow.
MATERIALS AND METHODS
Plant Material.
Grains of two barley lines were treated with the mutagen sodium azide (16) and sown in the field. The progeny grains (M2) from these plants were germinated; 4-week-old seedlings were screened visually for light-green (chlorina) color, and the viable mutants were propagated for further analyses. Among the 78 viable Chlorina mutants selected, there were 3 allelic aurea mutants. Mutant Chlorina-125 was induced in a wild-type line of cultivar Bonus, and Chlorina-157 and Chlorina-161 were induced in a wild-type line of Ca710516 (Triumph × Goldspear). Grains of the stock collection were planted in moist vermiculite and grown in darkness at 20°C for 6 days. Homozygous yellow and heterozygous light-green seedlings were distinguishable from green wild-type seedlings after 4 h of illumination with Osram Fluora fluorescent lights (Berlin) at 2.59 W/m2.
Cloning and Sequencing of the Xantha-h Gene Encoding the 42-kDa Subunit of Mg-Chelatase and Its Mutant Alleles.
Chromosomal DNA was extracted from barley leaves with cetyltrimethylammonium bromide (17). The relevant parts of the wild-type Xantha-h genes and their dominant mutant alleles in the Chlorina mutants were cloned by PCR by using the DNA from the Chlorina mutants and their two wild-type segregants as templates. The PCR consisted of 30 cycles of 45 s at 95°C, 60 s at 57°C, and 120 s at 72°C, and oligonucleotides with the sequences 5′-AAC GTC ATC GAC CCC AA-3′ and 5′-GCC AAA GAC TTC ATA GAA CTT-3′ were used as primers. Sequencing was performed directly on the PCR fragments by using an ABI377 sequencer and the BigDye Primer Cycle Sequencing Kit supplied by the manufacturer (Applied Biosystems).
Complementation Assays of Mg-Chelatase Activity with Mutant and Wild-Type Subunits.
Between 20 and 40 g (fresh weight) of leaves from the homozygous mutants were homogenized in grinding medium and processed according to the method of Kannangara et al. (5). Heterozygous Chlorina and normal green seedlings were also processed in this way. Briefly, the homogenate was squeezed through a single layer of nylon tissue, filtered through another layer, and then centrifuged at 5,000 × g at 4°C for 6 min. After removal of the supernatant, 1 ml of the resuspended chloroplasts was purified by centrifugation through a 5-ml Percoll cushion (Amersham Pharmacia; 35% Percoll in grinding medium) at 12,800 × g at 4°C for 10 min. The purified chloroplasts were lysed in 150 μl of 20 mM Tricine⋅NaOH {N-[tris(hydroxymethyl)methyl]glycine⋅NaOH} buffer (pH 9.0) containing 1 mM EDTA, 2 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. The membranes were removed by centrifugation at 21,000 × g for 15 min, and the supernatants were used for analysis of in vitro complementations.
Wild-type subunits were also prepared in large scale from 3 kg (fresh weight) of leaves. This allows the 42- and 70-kDa subunits of Mg-chelatase in the stromata of purified plastids to be separated into a supernatant and pellet fraction, respectively (5). The stroma supernatant obtained by lysis of the purified plastids, as described above, was centrifuged for 90 min at 272,000 × g, yielding a clear-as-glass pellet containing the ribosome-associated 70-kDa subunit and a supernatant containing the 42-kDa subunit. The 140-kDa subunit is present in both fractions.
Mg-chelatase activity was measured in the following way. From stock solutions of 1 mM deuteroporphyrin IX in 2% (vol/vol) ammonia and the other components, a mixture containing deuteroporphyrin (60 μM), ATP (50 mM), creatine phosphate (250 mM), and MgCl2 (250 mM) was prepared. The assays were started by adding 4.25 μl of this mixture to protein solutions adjusted to 45 μl with 20 mM Tricine⋅NaOH (pH 9.0) and then adding 1 μl (2.5 units) of creatine kinase. The reaction mixtures were incubated and shaken for 20 min at 30°C, and the reaction was stopped by adding 1 ml of acetone/water/25% ammonia (80:20:1, vol/vol/vol) and 200 μl of hexane. After mixing, the tubes were centrifuged briefly to separate the phases. The emission spectrum of the bottom acetone phase was recorded from 550 to 650 nm with an excitation wavelength of 408 nm. The amount of Mg-deuteroporphyrin formed was calculated from the fluorescence intensity at 585 nm by using a standard curve obtained with known amounts of authentic compound.
Other Methods.
Total leaf protein from mutants and wild type was prepared for Western blot analysis as described (18). SDS/PAGE was performed as described (19) with the Tris⋅Tricine buffer system (20). Proteins were electrotransferred to Immobilon P filters (Millipore) with a semidry electroblotter (Ancos, Copenhagen, Denmark). Antigens were visualized by chemiluminescence (CLONTECH). Optimal sensitivity of detection was obtained according to published protocols (18). Protein concentrations were determined with the Bio-Rad protein assay based on the binding of Coomassie brilliant blue dye.
RESULTS
The Chlorina Mutants Are Deficient in the Synthesis of Mg-Protoporphyrin IX.
To identify the block in the biosynthetic pathway, the Chlorina-125, -157, and -161 mutants were analyzed after feeding 5-aminolevulinic acid to primary leaves. The primary leaves of normal green seedlings (clo/clo), heterozygous light-green seedlings (Clo/clo), and yellow seedlings (Clo/Clo) were detached and placed for 15 h in 20 mM 5-aminolevulinic acid and 10 mM sodium phosphate buffer (pH 7.0) in the dark. A red pigment accumulated in all homozygous (Clo/Clo) and heterozygous (Clo/clo) mutant leaves, whereas the wild type accumulated green pigment. The leaves were ground in liquid nitrogen and suspended in acetone/water/25% ammonia (80:20:1, vol/vol/vol). Carotenoids were removed with hexane, and the absorption spectra were determined. All three homozygous (Clo/Clo) mutants, whether grown in the dark or in the light, accumulated protoporphyrin IX. The wild-type seedlings (clo/clo) accumulated protochlorophyllide under these conditions, whereas the heterozygotes accumulated both protoporphyrin IX and protochlorophyllide. Accumulation of protoporphyrin IX after feeding with 5-aminolevulinic acid is also characteristic for the xantha-f, -g, and -h mutants; however, in these cases, the mutations are recessive—the heterozygotes are normal green and accumulate only protochlorophyllide (21). We conclude that the semidominant Chlorina mutants are deficient in Mg-chelatase.
The Semidominant Chlorina Mutations Are Allelic to the Recessive xantha-h Mutations.
Heterozygous xantha-h57 plants (Xan/xan) were emasculated and pollinated with pollen from heterozygous Chlorina-161 (Clo/clo) plants. The F1 progeny segregated into 25 green, 8 light-green, and 10 yellow seedlings, which is in agreement with the expected 2:1:1 ratio for allelism (χ2 = 0.782; P ≈ 0.6). In crosses of heterozygous xantha-f26 and xantha-g28 with Chlorina-161, only green and light-green seedlings segregated in a 1:1 ratio.
The Chlorina-125, -157, and -161 Mutations Result in Single Amino Acid Changes in the 42-kDa Mg-Chelatase Subunit.
Oligonucleotide primers for the gene encoding the 42-kDa subunit were synthesized according to the known cDNA sequence of the Xantha-h gene (4). From comparisons between the deduced barley Xantha-h gene product with the 42-kDa Mg-chelatase subunit of other organisms, it can be estimated that approximately 20 amino acid residues of the sequence of the mature XANTHA-H protein are undetermined (Fig. 2). The known part of the Xantha-h gene was sequenced from chromosomal DNA of the wild-type lines Bonus and Ca710516 (Triumph × Goldspear) and the homozygous Chlorina-125, -157, and -161 mutants. All three Chlorina mutations were found to be point mutations resulting in changes of single amino acid residues that are conserved strictly in the 14 sequences, 6 of which are compared in Fig. 2. When numbering the nucleotides according to the incomplete barley Xantha-h sequence, the Chlorina-125, -157, and -161 mutations are G559A (resulting in amino acid exchange D187N), G806A (R269K), and C271T (L91F), respectively (indicated by arrows in Fig. 2). The original Xantha-h sequence (4) very likely contains a sequencing mistake, which appears as a CTA insertion at positions 831 to 833. The extra codon adds a tyrosine residue to the polypeptide sequence at position 278 (barley numbering). This tyrosyl has no homology in the amino acid sequences of the 42-kDa subunits of other species and causes a gap in polypeptide alignments (Fig. 2). The tobacco Sulfur mutation is an amino acid substitution (N269I; ref. 11). This asparagine residue is also a completely conserved amino acid residue and corresponds to N175 in the barley sequence (indicated by an arrow in Fig. 2). As the N-terminal half of the 70-kDa subunit shows substantial similarity to the complete 42-kDa subunit, it is interesting to note that the four mutations change amino acid residues that are conserved in an alignment between the two subunits.
Figure 2.
Polypeptide sequence alignment of 42-kDa subunits of Mg-chelatase. The barley sequence and 5 of the 14 sequences available in the GenBank Database: Hvu (Hordeum vulgare; barley; GenBank accession no. U26545), Gma (Glycine max; soybean; D45857), Nta (N. tabacum; tobacco; AF014053), Ath (Arabidopsis thaliana; thale cress; X91411), Rca (Rhodobacter capsulatus; Z11165), and Rsp [R. sphaeroides; AF017642 (AJ001690)]. The N-terminal sequence of the soybean, tobacco, and thale cress polypeptides is the same as that of the unprocessed proteins. Conserved amino acid residues in all 14 sequences are marked by asterisks. The species of origin of the 42-kDa subunits (not displayed) are Porphyra purpurea (U38804), Odontella sinensis (Z67753), Cryptomonas phi (Z21976), Anabaena variabilis (D49426), Synechocystis PCC6803 (U35144), Olisthodiscus luteus (Z21959), Chlorobium vibrioforme (Z83933), and Heliobacillus mobilis (AF080002). Arrows indicate the amino acid residues changed in mutants: L91F, D187N, and R269K. The tobacco Sulfur mutation is N269I.
The Chlorina-125, -151, and -161 Mutants Contain the 42-kDa Subunit Protein.
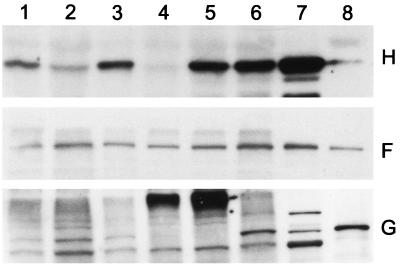
It has been shown that the recessive xantha-h mutants devoid of Mg-chelatase activity lack the 42-kDa but not the 140-kDa subunit (4). Therefore, it was of interest to determine whether three semidominant missense mutations in the same gene result in stable Mg-chelatase subunits. Western blot analyses with antibodies specific for the 42-, 70-, and 140-kDa subunits were performed according to the method of Hansson et al. (18). The homozygous Chlorina mutants contained all three subunits (Fig. 3). The recessive xantha-f10 mutant also contains all three subunits and has been diagnosed as having a missense mutation in the 140-kDa subunit. In contrast, the xantha-g28 mutant lacked the 70-kDa subunit, and the xantha-h56 mutant lacked both the 42- and 70-kDa subunits.
Figure 3.
Western blot analysis of the barley Mg-chelatase subunits XANTHA-H (Top), XANTHA-F (Middle), and XANTHA-G (Bottom). Lanes: 1, Chlorina-125; 2, Chlorina-157; 3, Chlorina-161; 4, xantha-h57; 5, xantha-g28; 6, xantha-f10; 7, wild-type chloroplast high-speed supernatant; and 8, wild-type chloroplast high-speed pellet. The molecular masses of the standard bands labeled in lanes 7 and 8 are 42 kDa for H, 150 kDa for F, and 76 kDa for G.
In Vitro Complementation and Reconstitution of Mg-Chelatase Activity with Mutant Extracts.
In vitro complementation of barley Mg-chelatase activity takes place when stroma fractions of the xantha-f, -g, and -h mutants are combined pairwise (5). Analogous complementation experiments were performed with the stroma proteins of Chlorina-161 (Table 1). The wild-type stroma had a Mg-chelatase activity that yielded 188 pmol of Mg-deuteroporphyrin IX in 20 min. The homozygous mutant had no activity, whereas the stroma proteins of the heterozygous seedlings had 21.6% of wild-type activity (per mg of protein). Soluble stroma of homozygous Chlorina-161 was mixed with soluble stroma proteins from homozygous xantha-f10, -g28, or -h56 mutants and assayed for Mg-chelatase activity. Stroma of Chlorina-161 mixed with stroma from xantha-f10 showed slight activity. As expected, the addition of the xantha-h56 extract did not reconstitute Mg-chelatase activity of stroma from Chlorina-161, a result which is in agreement with the lack of the 42-kDa XANTHA-H subunit in the xantha-h56 cell extract (Fig. 3). The extract of xantha-g28 did not restore activity, probably because the protein concentrations attained were not high enough. The combinations of the mutant with the wild-type preparations in assays indicate the absence of inhibitors of Mg-chelatase in the mutant extracts.
Table 1.
In vitro complementation of Mg-chelatase activity with barley chloroplast extracts of wild-type and Chlorina and xantha mutants.
| Plastid stroma assayed | Activity, pmol/20 min | Subunits present |
|---|---|---|
| Wild-type | 188 | F, G, H |
| Chlorina-161 (Clo/Clo) | 0.0 | F, G, h |
| Chlorina-161 (clo/Clo) | 3.0 | F, G, H, h |
| xantha-h56 (xan/xan) | 0.0 | F |
| xantha-f10 (xan/xan) | 0.0 | f, G, H |
| xantha-g28 (xan/xan) | 0.0 | F, G |
| Chlorina-161 (Clo/Clo) + xantha-h56 | 0.0 | F, G, h |
| Chlorina-161 (Clo/Clo) + xantha-f10 | 4.1 | F, f, G, H, h |
| Chlorina-161 (Clo/Clo) + xantha-g28 | 0.0 | F, G, H, h |
| Chlorina-161 + wild-type | 181 | F, G, H, h |
| xantha-h56 + wild-type | 185 | F, G, H |
| xantha-f10 + wild-type | 176 | F, f, G, H |
| xantha-g28 + wild-type | 146 | F, G, H |
Subunits: F = 140 kDa, G = 70 kDa, H = 42 kDa, h = inactive 42 kDa, f = inactive 140 kDa. The amounts of protein used in 50-μl assays were 725 μg of wild-type plastid stroma, 138 μg of Clorina-161 (Clo/Clo), 250 μg of Chlorina-161 (clo/Clo), 275 μg of xantha-h56, 200 μg of xantha-f10, and 150 μg of xantha-g28.
The Mg-chelatase subunits of barley chloroplasts can be separated by high-speed centrifugation into a clear supernatant containing the 42-kDa XANTHA-H subunit and a pellet containing the 70-kDa XANTHA-G subunit (5). Both fractions contain the 140-kDa XANTHA-F subunit. The supernatant and pellet fractions individually showed little and no activity, respectively (Table 2, lines 4 and 5). Reconstitution of the pellet with the supernatant restored high activity (Table 2, lines 1–3). The stromata of the three chlorina wild types (clo/clo) had high Mg-chelatase activity, whether tested alone or with fractions containing XANTHA-H or XANTHA-G (Table 2). Activity expressed on a protein basis could indicate some reduction in activity when the pellet fraction was added. Mg-chelatase activity in extracts of the homozygous mutants Chlorina-125, -157, and -161 (Clo/Clo) was reconstituted only when the 42-kDa XANTHA-H subunit in the supernatant fraction of wild type was supplied (Table 2). This result shows that the 42-kDa subunits present in the Chlorina mutants are functionally defective. The assays of the stroma preparations of the heterozygous Chlorina-125 and -161 genotypes (Clo/clo) showed a reduced Mg-chelatase activity, ranging from 25 to 50% of the activity present in the wild type, which could be normalized by supplying wild-type 42-kDa subunits but not by providing the 70-kDa subunits. The stroma preparation of the Chlorina-157 heterozygotes had an unexpectedly high Mg-chelatase activity, which might be caused by misclassification of the seedlings extracted (i.e., an inadvertent admixture of some wild-type leaves).
Table 2.
Reconstitution assays of Mg-chelatase activity in soluble chloroplast extracts from barley Chlorina mutants
| Plastid stroma | Wild-type subunits supplied | Mg-chelatase
activity
|
|
|---|---|---|---|
| pmol/30 min | pmol/min/mg protein | ||
| Wild-type | None | 164 ± 64 | 17.6 ± 3.0 |
| F, H | 318 ± 11 | 19.9 ± 2.7 | |
| F, G | 148 ± 66 | 4.78 ± 3.43 | |
| None | F, H | 18.6 | 2.38 |
| F, G | 0.0 | 0.0 | |
| F, G, H | 307 | 10.9 | |
| Chlorina-125 (Clo/Clo) | None | 0.0 | 0.0 |
| F, H | 49.7 | 4.04 | |
| F, G | 0.0 | 0.0 | |
| Chlorina-157 (Clo/Clo) | None | 0.0 | 0.0 |
| F, H | 64.4 | 6.71 | |
| F, G | 0.0 | 0.0 | |
| Chlorina-161 (Clo/Clo) | None | 0.0 | 0.0 |
| F, H | 141 | 9.15 | |
| F, G | 0.0 | 0.0 | |
| Chlorina-125 (Clo/clo) | None | 40.6 | 4.51 |
| F, H | 317 | 18.9 | |
| F, G | 35.5 | 1.21 | |
| Chlorina-157 (Clo/clo) | None | 201 | 16.7 |
| F, H | 294 | 14.9 | |
| F, G | 152 | 4.70 | |
| Chlorina-161 (Clo/clo) | None | 81.2 | 6.59 |
| F, H | 314 | 15.6 | |
| F, G | 71.0 | 2.16 | |
Wild-type 42-kDa subunits (H) were separated into a high-speed supernatant from pelleted wild-type 70-kDa subunits (G). The 140-kDa subunit (F) was present in both fractions. These wild-type subunits were supplied to reconstitution assays of Mg-chelatase activity. The amounts of protein used in 50-μl assays were 260 μg of supernatant (F, H), 680 μg of pellet (F, G), 150 μg of Chlorina-125 (Clo/Clo), 300 μg of Chlorina-125 (Clo/clo), 159 μg of Chlorina-125 (clo/clo), 60 μg of Chlorina-157 (Clo/Clo), 399 μg of Chlorina-157 (Clo/clo), 420 μg of Chlorina-157 (clo/clo), 255 μg of Chlorina-161 (Clo/Clo), 411 μg of Chlorina-161 (Clo/clo), and 300 μg of Chlorina-161 (clo/clo).
DISCUSSION
In prokaryotic and eukaryotic photosynthetic organisms, the insertion of Mg2+ into the natural substrate protoporphyrin IX or the more water-soluble in vitro substrate deuteroporphyrin IX requires the interaction of three protein subunits with nominal molecular masses of 140, 70, and 42 kDa. The reaction has an absolute requirement for ATP and proceeds in two steps (22, 23). In a first activation step, the 70- and 42-kDa subunits interact, resulting in ATP hydrolysis as well as synthesis of ATP from ADP by the intrinsic phosphate-exchange activity of the 42-kDa subunit (9).
The mutations in the three semidominant barley mutants Chlorina-125, -157, and -161 and in the tobacco Sulfur mutation cause four different amino acid substitutions in amino acid residues that are conserved in all 14 known primary structures of the 42-kDa subunit. Western blot analyses of the three barley mutants indicate that the mutated protein is maintained stably in the plastids, and the absence of Mg-chelatase activity in the yellow homozygous Chlorina (Clo/Clo) mutant seedlings proves that the 42-kDa XANTHA-H subunit is mutated into an inactive form. The heterozygous barley seedlings have only 25–50% of the Mg-chelatase activity observed in the wild type. The expression of the active and the inactive 42-kDa subunit in the heterozygous barley seedlings can lead to two types of heterodimers between the strongly interacting 42-kDa and 70-kDa subunits. One heterodimer will consist of wild-type 42-kDa and 70-kDa subunits (H + G), and the other will consist of mutated 42-kDa and wild-type 70-kDa subunits (h + G). Therefore, the reduced Mg-chelatase activity would be explained by the capacity of the heterodimers consisting of mutated 42-kDa protein and wild-type 70-kDa protein to bind to the 140-kDa subunit. However, because these heterodimers cannot carry out the activation step, their association with the third subunits (h + G + F) leads to inactive Mg-chelatase. If the 42-kDa and 70-kDa subunits were to dissociate after the activation step and an activated 70-kDa subunit alone is to interact with the 140-kDa subunit as suggested (8), we would not expect a codominant action of the inactive 42-kDa subunit, a reduction in Mg-chelatase activity, and a diminished chlorophyll content. Under this hypothesis, the heterodimers of an inactive 42-kDa with a 70-kDa subunit would fail to be activated and to dissociate. In heterozygotes, this failure would leave only the activated 70-kDa subunits that had been associated with wild-type 42-kDa subunits to interact with the 140-kDa subunit. Such a recessive action actually occurs in heterozygotes containing mutated 140-kDa subunits and in heterozygous Xantha-h mutants that fail to produce a 42-kDa subunit protein.
In contrast to the Chlorina mutants, the originally isolated and analyzed xantha-h mutants are recessive. The heterozygotes of the recessive mutants xantha-h30, -h38, -h56, and -h57 are normal green, are indistinguishable from wild type, and have no reduced Mg-chelatase activity. The homozygous lethal mutants lack the 42-kDa subunits, as determined by Western blot analysis. Of the three recessive xantha-h mutants analyzed by Northern blotting, two were devoid of transcripts of the gene, and the third had a truncated transcript. In the absence of a nonfunctional 42-kDa protein in the heterozygotes, only wild-type 42-kDa subunits are present, and these interact in the standard way with the 70-kDa and 140-kDa subunits. (In view of these results, the three Chlorina mutants should be renamed Xantha-hclo 125, -hclo 157, and -hclo 161.)
Mutations in the Xantha-f gene like xantha-f10 that produce an inactive but stable 140-kDa subunit (Fig. 3; ref. 4) are recessive and thus show no interference with association of wild-type subunits in the heterozygotes. Considering the observation that major deletions of 70-kDa protein domains still can support functional Mg-chelatase activity, we expect some mutated 70-kDa subunits to show recessive behavior (8). However, mutations in the 70-kDa subunit that affect the interaction with the 42-kDa subunit are likely to be codominant. The recessive xantha-g28 mutant investigated here lacked only the 70-kDa subunit (Fig. 3), and Mg-chelatase activity can be reconstituted by supplying this subunit (5). The xantha-h56 mutant preparation analyzed here seems to lack both the 42- and 70-kDa protein, although exclusively supplying the 42-kDa protein readily compensated for the lack of activity in the stroma of xantha-h57 (5). We interpret this inconsistency to indicate that, in the absence of a 42-kDa subunit, the 70-kDa subunit may have reduced stability and may be sensitive to proteolytic degradation in certain mutants and during certain developmental stages.
A nucleotide phosphate binding site of the type GX4GKS has been recognized in the 42-kDa subunit (4). This subunit has unique properties. By itself, the subunit catalyzes the exchange of one phosphate group of ATP to an ADP molecule, generating ADP and ATP, respectively (9). The binding of the 42- and 70-kDa subunits elicits a strong ATPase activity, hydrolyzing ATP to ADP plus free phosphate. This hydrolysis is characteristic of the activation reaction. Inhibitor studies indicate that both subunits contribute to the active site. The 140-kDa subunit that binds the protoporphyrin IX substrate has a weak ATPase activity by itself and seems to have two ATP binding sites that react differently to NaF inhibition (9).
An enhancement of ATPase activity is characteristic for the association in Escherichia coli of the 70-kDa DnaK heat-shock protein with the 41-kDa DnaJ heat-shock protein. These proteins are used as chaperones in refolding denatured polypeptides (24, 25) but also function in the conversion of, for example, the 32-kDa RepA initiator protein from a dimer to a monomer that binds to the origin of replication of the P1 plasmid (26, 27). In both cases, a third heat-shock protein, the 22-kDa GrpE protein, interacts with the other two. The three proteins are also essential for the replication of λ-phage DNA (28). In this case, ATPase activity of DnaK is stimulated up to 50-fold in the simultaneous presence of the DnaJ and GrpE proteins. The GrpE protein alone increases the rate of release of bound ATP or ADP without affecting the rate of hydrolysis and is thought to expedite the exchange of ADP to ATP on DnaK. In eukaryotic chaperones, the required phosphate-exchange activity postulated in E. coli for the GrpE protein has been identified recently as an intrinsic activity in the bovine heat-shock protein Hsp70 (13). In Hsp70, the interaction with protein substrate is regulated by ATP hydrolysis and ADP-to-ATP exchange. When bound to ADP, Hsp70 binds tightly to protein substrates, but when Hsp is bound to ATP, the peptides are released (29). ATP binding, but not ATP hydrolysis per se, is essential for substrate dissociation (30). In the binding and release cycle, ATP hydrolysis and ADP-to-ATP exchange are the methods by which Hsp70 cycles between the ADP⋅Hsp70 and ATP⋅Hsp70 complexes.
Although there is no obvious homology between the primary structure of Hsp70 and the 42-kDa subunit of Mg-chelatase, the similar use of ATP and ADP by these proteins provides an incentive to explore the functions of the 42-kDa subunit in controlling the folding and conformation of its own and the other two subunits. The result of the codominant action on the Mg-chelatase activity in heterozygotes of wild-type and mutant 42-kDa subunits with single amino acid changes leads us to the following working hypothesis. The ATP-to-ADP exchange reaction promotes the binding of the 42-kDa subunit to the 70-kDa subunit in a manner similar to the binding of Hsp70 with an unfolded protein. Once the two subunits are bound, the ATPase activity makes it possible for these subunits to bind the 140-kDa subunit. All three subunits bound together insert Mg2+ into protoporphyrin with ATP hydrolysis.
Acknowledgments
This work was made possible thanks to generous support from The Royal Swedish Academy of Agriculture and Forestry, The Swedish Council for Forestry and Agricultural Research, and the Carl Trygger Foundation. This is scientific paper 9912-16 from the College of Agriculture and Home Economics Research Center, Washington State University.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U26545).
References
- 1.Gibson L C, Willows R D, Kannangara C G, von Wettstein D, Hunter C N. Proc Natl Acad Sci USA. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willows R D, Gibson L C D, Kannangara C G, Hunter C N, von Wettstein D. Eur J Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]
- 3.Jensen P E, Gibson L C D, Henningsen K W, Hunter C N. J Biol Chem. 1996;271:16662–16667. doi: 10.1074/jbc.271.28.16662. [DOI] [PubMed] [Google Scholar]
- 4.Jensen P E, Willows R D, Petersen B L, Vothknecht U C, Stumann B M, Kannangara C G, Henningsen K W, von Wettstein D, Henningsen K W. Mol Gen Genet. 1996;250:383–394. doi: 10.1007/BF02174026. [DOI] [PubMed] [Google Scholar]
- 5.Kannangara C G, Vothknecht U C, Hansson M, von Wettstein D. Mol Gen Genet. 1997;254:85–92. doi: 10.1007/s004380050394. [DOI] [PubMed] [Google Scholar]
- 6.Papenbrock J, Gräfe S, Kruse E, Hänel F, Grimm B. Plant J. 1997;12:981–990. doi: 10.1046/j.1365-313x.1997.12050981.x. [DOI] [PubMed] [Google Scholar]
- 7.Walker C J, Willows R D. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gräfe, S., Saluz, H.-P., Grimm, B. & Hänel, F. (1999) Proc. Natl. Acad. Sci. USA96, in press. [DOI] [PMC free article] [PubMed]
- 9.Hansson M, Kannangara C G. Proc Natl Acad Sci USA. 1997;94:13351–13356. doi: 10.1073/pnas.94.24.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henningsen K W, Boynton J E, von Wettstein D. R Dan Acad Sci Lett Biol Skr. 1993;42:1–349. [Google Scholar]
- 11.Nguyen L V. Ph.D. thesis. Raleigh, NC: North Carolina State Univ.; 1995. [Google Scholar]
- 12.Kjemtrup S, Sampson K S, Peele C G, Nguyen L V, Conkling M A, Thompson W F, Robertson D. Plant J. 1998;14:91–100. doi: 10.1046/j.1365-313X.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 13.Hiromura M, Yano M, Mori H, Inoue M, Kido H. J Biol Chem. 1998;273:5435–5438. doi: 10.1074/jbc.273.10.5435. [DOI] [PubMed] [Google Scholar]
- 14.Simpson D J, Machold O, Høyer-Hansen G, von Wettstein D. Carlsberg Res Commun. 1985;50:223–238. [Google Scholar]
- 15.Simpson D J, von Wettstein D. Barley Genet Newsletter. 1993;21:102–108. [Google Scholar]
- 16.Jende-Strid B. Barley Genet Newsletter. 1978;8:55–57. [Google Scholar]
- 17.Murray H G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansson M, Gough S P, Kannangara C G, von Wettstein D. Plant Physiol Biochem. 1997;35:827–836. [Google Scholar]
- 19.Fling S P, Gregerson D S. Anal Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- 20.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.von Wettstein D, Kahn A, Nielsen O F, Gough S P. Science. 1974;184:800–802. doi: 10.1126/science.184.4138.800. [DOI] [PubMed] [Google Scholar]
- 22.Walker C J, Hupp L R, Weinstein J D. Plant Physiol Biochem. 1992;30:263–269. [Google Scholar]
- 23.Walker C J, Weinstein J D. Biochem J. 1994;299:277–284. doi: 10.1042/bj2990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrick J P, Hartl F-U. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 25.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl F-U. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickner S H. Proc Natl Acad Sci USA. 1990;87:2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis M J, Pelham H R B. EMBO J. 1985;4:3137–3143. doi: 10.1002/j.1460-2075.1985.tb04056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palleros D R, Reid K L, Shi L, Welch W J, Fink A L. Nature (London) 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]