Abstract
It is difficult to observe human tumor progression as precursor lesions are systematically removed. Alternatives to direct observations, commonly used to reveal the hidden past of species and populations, are sequence comparisons or molecular clocks. Noncoding microsatellite (MS) loci were employed as molecular tumor clocks in 13 human mutator phenotype (MSI+) colorectal tumors. Quantitative analysis revealed that specific patterns of somatic MS mutations accumulate with division after loss of mismatch repair (MMR). Tumors had unique patterns of MS mutation, and, therefore, based on this model, each tumor had its own unique history. Loss of MMR occurred very early relative to terminal clonal expansion, with an estimated average of 2,300 divisions since loss of MMR and 280 divisions since expansion. Contrary to the classical adenoma-cancer sequence, MSI+ adenomas were nearly as old as cancers (2,000 versus 2,400 divisions since loss of MMR). Negative clinical examinations preceded six tumors, independently documenting an absence of visible precursors during early MSI+ adenoma or cancer progression. These findings further extend a window beyond visible progression since loss of MMR appears to start a genetic phase involving clone sizes or phenotypes below a threshold of clinical detection. This previously occult prologue before visible neoplasia is longer and therefore likely more important than generally appreciated.
The mutations present in colorectal tumors are diverse, suggesting a variety of pathways to cancer (1–3). Which of the many potential pathways do individual tumors follow to cancer? Tumor histories based on direct observations are problematic. Adenomas are routinely removed, and surveillance intervals necessary to observe the entire progression to cancer may span decades. Adenomas can persist for years, with years before the appearance of cancers (4–6).
The precursors of mutator phenotype (MSI+) cancers have been extremely difficult to study because adenomas are not markedly increased in hereditary nonpolyposis colorectal cancer patients (3, 7). MSI+ colorectal cancers are deficient in DNA mismatch repair (MMR) and have greatly elevated mutation rates, especially at microsatellite (MS) loci (3). MSI+ tumors appear to progress more rapidly than repair-proficient cancer (3, 7). Of note, mice and rare humans with inherited MMR deficiencies are tumor prone but otherwise phenotypically normal and accumulate somatic MS mutations in histologically normal cells (8–11). Therefore, it is possible that at least some of the mutations present in MSI+ tumors accumulate before visible neoplasia.
To overcome limitations imposed by direct observations, we hypothesize that the unique histories of individual tumors are recorded by their somatic mutations. Progression is thought to occur through successions of selection and clonal expansion (1–3). The final tumor eventually arises from a single cell that represents the last bottleneck, regardless of the number or sizes of prior waves of clonal expansion. The rest of the lineages are dead ends. Therefore, mutations common to all tumor cells accumulate along the single lineage preceding this final founder cell whereas heterogeneous mutations may arise with clonal expansion (Fig. 1).
Figure 1.
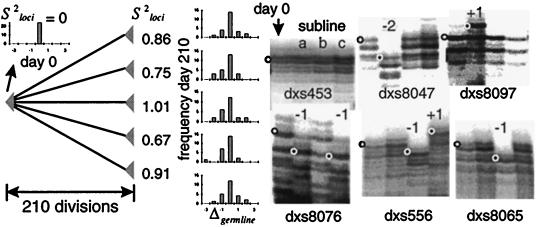
(a) A tumor history is “read” from the mutations present in its cells. All of these cells are related to a single founder cell that represents the final bottleneck along a progression pathway. The history of this founder cell can be traced back along a single lineage because of the bottleneck nature of progression. We define the start of this pathway as the somatic loss of MMR. MS loci can record this history. Consider a single MS locus. After loss of MMR, the locus will randomly become larger or smaller, with the difference from its germline length a function of the number of divisions since loss of MMR. Upon terminal clonal expansion, the locus will become polymorphic. The time since terminal clonal expansion is reflected in the width or variance (Salleles2) of the tumor allele frequency distribution. The MS allele in the founder cell can be inferred by the most common allele present in the tumor. The time since loss of MMR preceding the bottleneck is reflected in the difference (Δgermline) between the length of the germline allele and the founder allele. Although the stochastic nature of mutation makes a single MS locus relatively uninformative, the analysis repeated at 20–30 different loci is robust. (b) Simulations of MS mutation. Data represents the results of 1,000 trials with 20–30 MS loci, and a symmetric stepwise model with the chance of addition of one repeat of 0.0025, and loss of one repeat of 0.0025, with the total mutation rate of 0.005 per division. In each scenario, the final tumor size is 1.0 cm3 or one billion cells. Different patterns of MS mutations, summarized by the variances, Salleles2 and Sloci2, are obtained with identical numbers of divisions (2,000) but different tumor histories. Therefore, a history of a human tumor can be inferred by sampling its MS alleles and estimating Salleles2 and Sloci2. (c) Example of simulations with identical clonal expansion histories (20 terminal exponential divisions) but different times since loss of MMR. The average value of Sloci2 increases with the numbers of divisions since loss of MMR whereas Salleles2 is constant. The graph illustrates the mean and 95% confidence intervals (dotted lines) of the simulations. The triangles in the graph represent values obtained from 210- to 352-day-old tissue culture experiments that mirror these simulations (see Fig. 2).
We used a quantitative approach to infer MSI+ tumor histories based on the analysis of the common and unique somatic MS mutations that may accumulate along such a progression pathway. The start of this pathway is marked by the somatic loss of MMR, which increases mutation rates in noncoding CA-repeat MS loci ≈100-fold (12, 13) and allows them to function as molecular tumor clocks (14–16). Therefore the majority of MS mutations reflect divisions after loss of MMR. Note that changes in MS allele lengths are thought to arise with slippage during DNA replication (17, 18), so mutations are coupled with cell division. Differences between the final founder cell and its germline genotype reflect mitotic divisions that occur along a single lineage preceding the founder cell. The longer the intervals between loss of MMR and final tumor formation, the greater may be the differences between germline and tumor genotypes. The genotype of the final founder cell can be inferred by determining the most common allele present among the tumor cells. Polymorphic tumor alleles, which can only arise after clonal expansion, provide information on numbers of divisions after the final bottleneck.
Materials and Methods
Specimens.
DNA samples were extracted from formalin-fixed microscopic tissue sections (14) of 13 tumors from nine male patients. In most cases, tumors were subdivided into multiple regions based on phenotype or topography. Clinical information was obtain from medical records. Surveillance intervals were defined by times of colonoscopy or surgery. Patients with hereditary nonpolyposis colorectal cancer had germline mutations (Patients I, IV, and VI–IX, hMLH1; Patient III, hMSH2) confirmed by sequencing. The histories of Patients II and V did not meet criteria for hereditary nonpolyposis colorectal cancer. The MSI+ colorectal cell line HCT 116 [hMLH1-deficient (19)] was obtained from American Type Culture Collection and had a mitotic rate of approximately one division per day.
MS Analysis.
To simplify analysis, X chromosome CA-dinucleotide repeat MS loci and male patients were used. Tumor DNA was diluted before PCR such that ≈20–80% of assays produced products. Every measurement of a MS locus therefore essentially represents a single cell because MSI+ tumors characteristically lack aneuploidy (20). Products were labeled with 33P-dCTP (NEN) incorporated during 38–43 PCR cycles and were analyzed on 6% denaturing polyacrylamide sequencing gels and a PhosphorImager (Molecular Dynamics). Lengths different from germline were considered to be from the tumor. When distributions included the germline length, 40–50% of genomes were considered to originate from contaminating normal cells (estimated by visual inspection for each tumor), and frequencies at the germline size were reduced by this amount to approximate the tumor MS length distribution.
Defining an MSI+ tumor genotype can be problematic because their cells rapidly acquire further mutations. Multiple measurements at each locus are necessary to characterize a polymorphic tumor population. PCR after dilution of tumor DNA typically yields a variety of different alleles distributed around a single mode. Estimation of a mode from a tumor repeat length distribution is experimentally simpler than estimation of its mean because germline alleles from contaminating normal cells can be more easily eliminated. We summarize each MS locus by the difference (Δgermline) between the germline length and the mode of the tumor repeat length (Fig. 1b). At least 10 molecules were amplified at each locus until its mode became evident. We also estimate the sample variance (Salleles2) of the repeat length distribution at each locus. On average, ≈25 molecules were typed at each locus, and ≈500–1,000 molecules from 21–30 MS loci were typed from each tumor to characterize its genotype. For each tumor, we also summarize the variability among modal lengths (Δgermline) at different loci by their sample variance (Sloci2).
The human tumors were first screened with up to 37 MS loci (Research Genetics, Huntsville, AL; list available on request). The 21–30 MS loci used for analysis were chosen for their amplification quality and a germline CA-repeat length of 16 or greater. The criterion of a minimum length repeat was based on published (21, 22) and unpublished observations that shorter repeat lengths tend to have lower mutation rates, and to avoid a potential lower length boundary constraint (see below).
Modeling.
At the heart of our statistical approach is an algorithm for simulating the ancestry of a sample of cells (those used to estimate the genotype at each MS locus) taken from the tumor. We note that in our approach it is not necessary to simulate the history of the whole cell population that has been sampled. Once the ancestry of the sample has been simulated, mutations can be superimposed according to any mutation model [for example, a stepwise model (14, 23) in which loci accumulate random stepwise single repeat unit additions or deletions (reviewed in ref. 24)]. Thus, our method recreates both cellular division and mutation and the experimental process of sampling the final clonal expansion to ascertain the genotype (Δgermline) of each MS locus. The simulations start with the loss of MMR in a single cell with a germline genotype. A constant rate of 0.005 mutations per division [which is within the range of mutation rates observed in MMR-deficient cell lines (12, 13)] is assumed. Clonal expansion is defined as the time when this cell produces two daughter cells whose lineages persist. A tumor of approximately one billion cells is present at the end of each simulation.
To assess the sensitivity of the analysis to departures from the simple stepwise mutation model, we also used a model with range constraints (24) in which MS loci were not allowed to mutate to less than 12 repeats. For the range of estimated variances observed in Table 1 and starting (as in our experiments) with loci with lengths ≥16 repeats, the estimates of tumor age differ by at most 10% between the two scenarios (data not shown). Note that, if the mutation rate is halved, the estimated ages would be doubled. Simulation studies showed that age estimates obtained by sampling between 10 and 50 alleles or 20 and 30 loci varied little (data not shown).
Table 1.
Tumor ages
| Patient | Tumor | N* | Salleles2 | Expansion divisions | Sloci2 | Total divisions | Age, years | 95% CI, years | Clinical interval |
|---|---|---|---|---|---|---|---|---|---|
| I | Adenoma/Cancer-1 | ||||||||
| Adenoma 1.0 cm | 30 | 1.6 | 350 | 8.3 | 1,900 | 5.2 | 2.3–7.2 | — | |
| Cancer Dukes' C | 29 | 0.88 | 190 | 7.7 | 1,700 | 4.6 | 2.0–6.6 | — | |
| Cancer-2 Dukes' B | 30 | 1.6 | 350 | 9.2 | 2,100 | 5.7 | 2.5–7.9 | 0.5 yr | |
| II | Adenoma/Cancer | ||||||||
| Adenoma 1.0 cm | 28 | 1.3 | 280 | 15.8 | 3,400 | 9.2 | 3.6–13 | — | |
| Cancer Dukes' B | 28 | 1.2 | 260 | 9.8 | 2,100 | 5.9 | 2.4–8.3 | — | |
| III | Adenoma-1 0.5 cm | 25 | 0.96 | 210 | 7.5 | 1,700 | 4.6 | 1.9–6.6 | 2.0 yr |
| Cancer Dukes' B | 29 | 0.42 | 91 | 10.3 | 2,200 | 5.8 | 2.5–8.2 | — | |
| Adenoma-2 0.5 cm | 27 | 1.9 | 420 | 6.7 | 1,600 | 4.4 | 2.0–6.3 | 2.3 yr | |
| IV | Cancer Dukes' D | 21 | 3.0 | 660 | 9.0 | 2,200 | 6.1 | 2.2–8.6 | — |
| V | Cancer Dukes' D | 28 | 1.3 | 280 | 6.4 | 1,500 | 4.1 | 1.6–5.7 | — |
| VI | Adenoma 1.1 cm | 23 | 0.92 | 200 | 5.7 | 1,300 | 3.6 | 1.3–5.2 | 3.8 yr |
| Cancer Dukes' A | 23 | 0.46 | 100 | 9.0 | 1,900 | 5.1 | 2.0–7.6 | 3.8 yr | |
| VII | Cancer Dukes' B | 26 | 1.3 | 280 | 15.0 | 3,200 | 8.8 | 4.1–13 | — |
| VIII | Cancer Dukes' B | 24 | 0.87 | 190 | 10.7 | 2,300 | 6.3 | 2.7–9.2 | — |
| IX | Cancer Dukes' B | 24 | 1.6 | 350 | 22.3 | 4,700 | 12.9 | 5.3–19 | 1.0 yr |
| Average | 1.3 | 280 | 10.2 | 2,300 | 6.2 |
*Number of MS loci examined.
Estimating Tumor Histories.
Tumor histories were estimated by a computational inference method that involves matching the experimentally determined variances with the variances expected under a given model of cell division and MS mutation. A tumor history is defined by the time between loss of MMR and tumor removal and the time since clonal expansion (Fig. 1a). The simulations and theory indicate that, for a specific pattern of clonal expansion, the average value of Salleles2 is proportional to the time since initiation of the expansion. Therefore, the average of the experimental Salleles2 values obtained from all MS loci can be used to estimate the time since clonal expansion. Although expansion histories are likely to be variable for each tumor, simulations with a number of plausible scenarios (constant growth, immediate expansion followed by no growth, stepwise growth, and bottlenecks that reduce tumor populations up to 90%) indicate differences in average estimated ages of <20%. Therefore, we chose a model of constant growth to estimate the ages of the clonal expansions from Salleles2. For example, a cell is replaced by an average of 1.061 cells in each division during the last 350 divisions of the adenoma of Patient I.
To estimate the age of a tumor, defined as the number of divisions, g, between sampling and loss of MMR, we use a method of moments approach. First, the simulation algorithm is used repeatedly to estimate how the expected value E(Sloci2) of the variance Sloci2 varies as a function of g; say, E(Sloci2) = f(g). For a given tumor with an observed variance of s2, we estimate the age, g, by solving the equation s2 = f(g) to get the estimated age gest. Once more, f(g) is often an approximately linear function of g over the range of interest, so finding gest is simple. Confidence intervals for gest may be found by a parametric bootstrap approach (cf. ref. 25, Section 2.2). We simulate a number, B, of replicates of the cell division, mutation, and sampling process, each running for gest divisions. For each of them, we calculate the variance among the loci and use this to reestimate the age of the process. This results in B reestimates of the age: g1*,… ,gB*, say. Without loss of generality, we can assume they are listed in increasing order. The values of g1* − gest,… ,gB* − gest are used to approximate the distribution of gest − g. In particular, if α1 < α2 are such that l = Bα1 and m = Bα2 are integers, then a 100(1 − α1 − α2)% confidence interval is given by (2gest − g1*, 2gest − gm*).
Results
In multistep progression (1–3), the final tumor arises from a single founder cell (Fig. 1a). The numbers of divisions before and after this founder cell are potentially reconstructed by a quantitative analysis using MS loci as molecular tumor clocks. Initiation is usually defined by an onset of neoplasia (1–3). However in this analysis, the start of progression is defined as the somatic loss of MMR because this event triggers an ≈100-fold (12, 13) increase in MS mutations (estimated here at 0.005 mutations per division). Therefore, the majority of mutations reflect divisions after loss of MMR, which begin to accumulate after only a few hundred divisions in MSI+ tumors.
The quantitative analysis compares a tumor genotype with a starting genotype, which is likely identical to the germline genotype because normal tissues in our patients, as with most patients with MSI+ tumors, lack detectable mutations (3, 26–28). Theory and simulations (see Materials and Methods) illustrate that how tumors progress influences their final patterns of MS mutations (Fig. 1). Simplistically, MS loci become polymorphic after clonal expansion because each cell accumulates mutations independently. The distribution of the tumor alleles is characterized by its variance (Salleles2). The difference in length between a tumor allele and its germline allele (Δgermline) is largely a function of the time before terminal clonal expansion. This is because multiple alleles in a clonal expansion mutate randomly and tend to drift around the founder allele rather than to drift coherently. Drift from germline (summarized at all loci by Sloci2) predominately accumulates in the single cell lineage preceding the final clonal expansion.
The MS mutation patterns simulated under different progression scenarios are illustrated in Fig. 1b. If loss of MMR occurs at the time of terminal clonal expansion, drift from germline is small whereas tumor loci are polymorphic (Sloci2 low, Salleles2 high). If loss of MMR occurs early relative to expansion, drift from germline is greater. This is further illustrated in Fig. 1c when identical expansion histories but different numbers of divisions preceding the final founder are simulated. Sloci2 is a function of the number of divisions preceding terminal expansion. Therefore, we should be able to reconstruct the histories of individual MSI+ tumors by sampling their MS allelic distributions and estimating Sloci2 (proportional to the time since loss of MMR) and Salleles2 (proportional to the time since terminal clonal expansion). A rough simplification of our model is that an S2 of ≈1 is equivalent to ≈200 divisions.
Experimental Validation.
To test this model, we experimentally reconstructed progression pathways in which a single cell loses MMR and then subsequently undergoes clonal expansion at different times. A single clone of the MSI+ colorectal cancer cell line HCT 116 was isolated and grown for 190–332 days as different sublines (Fig. 2). Single clones from the sublines were again isolated and then clonally expanded for 20 more divisions. Therefore, each of the expanded subclones have similar histories of late terminal clonal expansion after various days since “initiation.” The MS genotypes of the final subline expansions were compared with the estimated genotype of the cell originally isolated at day zero. Consistent with the quantitative model, many of the loci had drifted from the “germline” lengths of the original clone (Fig. 2). The drift appeared random as each subline had alleles larger, smaller, or the same size as germline. For each subline, Salleles2 was essentially zero (data not shown) because clonal expansion was allowed for only 20 days. These control studies are consistent with our model as the subline Sloci2 values, ranging from 0.6 to 2.2, were within estimated 95% confidence intervals (Fig. 1c).
Figure 2.
A single HCT 116 cell was isolated, grown, and subsequently split into different sublines. After 190–332 days, single clones were isolated from each subclone, were expanded for 20 more divisions, and were typed at 24 MS loci (autoradiographs from three 210-day-old sublines and six loci are illustrated). Relative to germline (open circles or the allele size of the original clone), subline alleles are randomly the same, or larger or smaller (filled circles). The distribution of these changes (Δgermline) and their estimated variances (Sloci2) are small compared with human tumors (compare with Fig. 3). Variation between alleles at a single locus is minimal because only 20 divisions occurred during terminal expansions (Salleles2 ≈ 0). This experiment mimics five simulated trials with a history of 210 divisions since loss of MMR and a terminal clonal expansion of 20 exponential divisions (see Fig. 1c). The experimental Sloci2 values are consistent with the simulations (see triangles in the graph of Fig. 1c).
This experimental model also illustrates the bottleneck nature of progression. The sublines were passaged as conventional cultures rather than maintained as single cells. However, the analysis only depends on starting from a single MMR deficient cell and subsequently selecting a single founder cell for terminal clonal expansion. Only the numbers of divisions before and the expansion history of the founder cell and not the expansion history before the founder cell affect the MS mutations present in the final expansion.
Analysis of Human Tumors.
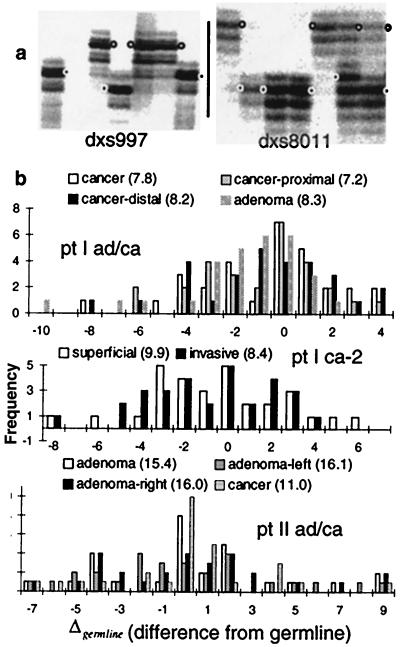
The MS alleles in the human MSI+ colorectal tumors were sampled by diluting tumor DNA before PCR (Fig. 3). Approximately 10–30 alleles from 21–30 MS loci were analyzed for 13 tumors from nine patients (Table 1). Compared with the cell line studies, the tumor MS distribution modes had drifted more (Fig. 3), with estimated Sloci2 values from 5.7 to 22.3 (Table 1). Estimated Salleles2 values were between 0.42 and 3.0. As a measure of consistency, Δgermline and Sloci2 from different parts of the same tumor should be similar and within simulated 95% confidence intervals because they presumably share identity by descent. This expectation was met (Fig. 3; data not shown).
Figure 3.
(a) Autoradiographs of human tumor MS alleles. After dilution and PCR, the germline alleles (open circles) and tumor specific alleles (filled circles) become evident. Although the tumor alleles are polymorphic, they exhibit a modal size [−2 for DXS997 in the invasive cancer of Patient V (left) and −5 for DXS8011 in the right adenoma region of Patient II (right)]. (b) The differences from germline (Δgermline) of the 21–30 different MS loci from Patients I or II. Broader distributions, summarized by their variances (Sloci2 in parentheses), indicate greater numbers of divisions since the loss of MMR (see Fig. 1 and compare with Fig. 2). Different regions from the same tumor have similar distributions and Sloci2 values.
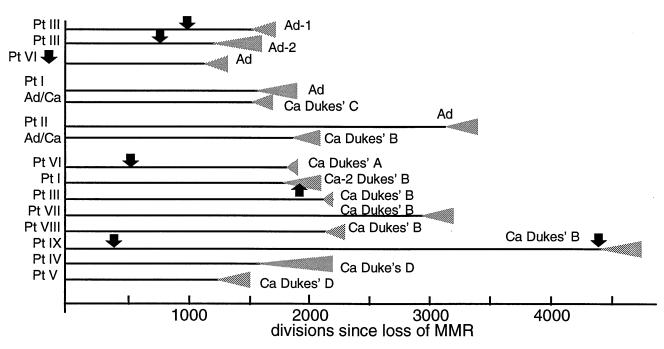
The tumor histories derived from their MS mutations are illustrated in Fig. 4. The histories were different between tumors with an average of 2,300 divisions since loss of MMR repair and 280 divisions since clonal expansion. Assuming one division per day [a value consistent with intestinal stem cell studies (29)], the average age of the tumors was 6.2 yr since loss of MMR. Adenoma clonal expansion ages were slightly greater than cancer expansion ages (290 versus 280 divisions), and adenoma ages since loss of MMR were slightly less than the cancer ages (2,000 versus 2,400 divisions). However, histologic stages did not correspond to tumor ages as there was considerable variation between tumor types (Fig. 4).
Figure 4.
Colorectal tumor histories inferred from their MS mutations. Each tumor history is unique, and most divisions occur before terminal clonal expansion (gray triangles). The average age is 2,300 divisions since loss of MMR. A trend toward increasing age with histologic progression is not apparent as the cancers and adenomas have similar ages. Negative clinical examinations (arrows, assuming one division per day) preceded six tumors, and for five tumors provided independent verification because they occurred before the postulated initiation of terminal clonal expansion. The negative examination in Patient I after the estimated initiation of the expansion of his Cancer-2 may indicate that it either was missed or was still too small to be detected. The data are consistent with genetic progression in the absence of visible progression, suggesting progenitor clone sizes are often below the threshold of clinical detection after loss of MMR.
Despite the variations in the tumor histories, the estimated ratios of divisions after terminal clonal expansion were relatively small (average 0.12) compared with the total divisions since loss of MMR. This finding suggests most divisions after loss of MMR occur before terminal clonal expansion. Note that our quantitative approach is limited to the division history of the final tumor and cannot recreate an entire progression history (such as an adenoma-cancer sequence) because it provides no information on clone sizes or phenotypes before the final founder cell. For example, the cancers may have arisen from and subsequently destroyed adenoma precursors or may have developed from occult precursors. Fortunately negative clinical examinations preceded six of our tumors, allowing independent verification of our model and knowledge of when a physically detectable precursor was probably absent.
The negative clinical examinations were consistent with the absence of clonal expansion inferred from five of our histories (Fig. 4). The history of the sole exception (Cancer-2 in Patient I) indicated terminal clonal initiated ≈170 days before the negative examination. However, this expansion may have been still too small to allow clinical detection. Under our scenario of constant growth, only ≈20,000 tumor cells would have been present at the time of the negative examination. In four of the tumors, the negative examinations occurred after loss of MMR but before terminal clonal expansion. Therefore, these tumors arising under clinical surveillance are more consistent with a scenario of occult progenitors rather than progression through a series of physically detectable clonal expansions.
Discussion
Quantitative sequence comparisons have provided a powerful method for reconstructing population or species histories. Here we translate this approach to the problem of multistep tumor progression and use it to estimate numbers of divisions since loss of MMR and since a last clonal expansion. Estimated times had large confidence intervals, and other models of MS mutation and progression could yield alternative tumor histories. Nevertheless, our model follows the basic principles of multistep tumor progression (1), provides histories consistent with clinical and experimental observations, and accounts for the complex patterns of MS mutations observed in MSI+ tumors with a simple mechanism of constant stepwise mutation.
Each of our tumors had a unique history, consistent with the hypothesis that tumor progression may follow different pathways (1, 2). The estimated intervals (Fig. 4) between initiation and removal (3.6–12.9 yr) were short, which may reflect the accelerated progression expected by a mutator phenotype (30). Despite this variability, one general feature was the early loss of MMR relative to terminal clonal expansion. The early loss of MMR is consistent with MS mutations distributed throughout MSI+ tumors (12) and frameshifts in short repetitive sequences frequently found in their tumor suppressor loci (31–33).
Our tumor histories suggest that most progression occurs before terminal clonal expansion. Although this finding is not surprising for cancers, adenomas also had long progression intervals before clonal expansion and were of similar ages (averages of 2,000 versus 2,400 divisions) as the cancers. This suggests that adenomas arise after long periods of occult progression rather than their classical roles at the start of colorectal cancer progression. Adenomas may still contain precursors to MSI+ cancers since the transition from adenoma to cancer may be rapid in the setting of MMR deficiency (7).
The negative surveillance examinations preceding some of our tumors also suggest progression occurs in the absence of visible manifestations. The emergence of a tumor shortly after a negative clinical examination may reflect a failure to detect a preexisting lesion, or rapid progression within the surveillance interval. Rapid progression seems unlikely because the interval tumors had similar numbers of MS mutations and estimated ages (6.0 versus 6.2 yr) as the tumors that arose in patients not under surveillance. Our analysis is more consistent with the absence of detectable tumors but the presence of occult progenitors at the times of the negative examinations. For example, although the physically detectable manifestations of the cancers of Patients I and IX (Fig. 4) arose within less than 1 yr, the cancer progenitors had already accumulated >90% of their MS mutations at the times of their last negative clinical examinations.
It is important to note that histories derived from our quantitative analysis may differ from a physical record of progression. A visible record of progression requires selection and detectable clonal expansion. Therefore, some phases of progression may be invisible if their consequences do not immediately result in detectable physical changes. In contrast, we measure progression through MS mutations that can accumulate in the absence of selection, changes in phenotype, or detectable clonal expansion.
A critical question is whether the genetic progression measured by the accumulation of MS mutations is relevant to tumor progression. The time of the last clonal expansion can be directly related to a physically important progression milestone, but our studies suggest the early loss of MMR is unlikely associated with a recognizable progenitor. MS mutations can accumulate in phenotypically normal cells (8–11) so at least some of the progression recorded by MS loci may occur in overtly normal appearing crypts. Therefore, many of the divisions recorded by MS tumor clocks may occur before visible cycles of selection and clonal dominance. However, if cancer is a genetic disease, the accumulation of pertinent somatic mutations regardless of immediate selection and clonal expansion is relevant to progression, especially because neoplasia may not occur until a human cell has acquired a number of alterations (34). The loss of MMR appears to influence subsequent progression because many tumor suppressor loci such as APC, TGF-β RII, Bax, and others have characteristic frameshift mutations in MSI+ tumors (31–33). Although we measure mutations in noncoding MS loci, these mutations likely reflect a similar accumulation of mutations in loci that ultimately confer visible selection.
The current quantitative studies are consistent with multistep progression (1, 2). After loss of MMR, mutations gradually accumulate over time until the combinations sufficient for visible clonal expansion are acquired. The major difference with the classical adenoma–carcinoma sequence (3) is that much of progression occurs in clone sizes or phenotypes below a threshold of clinical detection. Although clonal expansion effectively increases mutation rates (35), expansion to a visible precursor may not be required as a prerequisite for MSI+ progression because individual cells readily acquire mutations. At least in MSI+ colorectal tumors, it appears that this previously occult prologue before visible neoplasia is longer than generally appreciated. The relatively long lengths of these prologues suggest the final phenotypes are contingent on many of the mutations acquired during occult progression. Given the technical and ethical problems with direct observations, quantitative analysis may provide the only feasible and objective method to reconstruct human tumor progression and probe even beyond the earliest physically detectable precursors. Additional studies with more or different types of loci should help further characterize the unique histories of individual tumors.
Acknowledgments
We thank C. Fajado, A. Arakawa, N. Yum, K. Adzhyan, C. B. Blake, and A. Ghatan for their technical assistance and Drs. Jeremy R. Jass and Barbara Leggett for providing appropriate specimens. This work was supported by grants from the National Institutes of Health to D.S. (CA58704 and CA70858) and National Science Foundation to S.T. (BIR95-04393).
Abbreviations
- MS
microsatellite
- MMR
mismatch repair
References
- 1.Nowell PC. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Foulds L. Cancer Res. 1954;14:327–339. [PubMed] [Google Scholar]
- 3.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Otchy D P, Ransohoff D F, Wolff B G, Waver A, Ilstrup D, Carlson H, Rademacher D. Am J Gastroenterol. 1996;91:448–454. [PubMed] [Google Scholar]
- 5.Morson B C. Cancer. 1974;34:845–849. doi: 10.1002/1097-0142(197409)34:3+<845::aid-cncr2820340710>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 6.Winawer S J, Zauber A G, Ho M N, O'Brien M J, Gottlieb L S, Sternberg S S, Waye J D, Schapiro M, Bond J H, Panish J F, et al. N Engl J Med. 1993;329:1977–1981. doi: 10.1056/NEJM199312303292701. [DOI] [PubMed] [Google Scholar]
- 7.Jass J R, Stewart S M. Gut. 1992;33:783–786. doi: 10.1136/gut.33.6.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao X, Buermeyer A B, Narayanan L, Tran D, Baker S M, Prolla T A, Glazer P M, Liskay R M, Arnheim N. Proc Natl Acad Sci USA. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons R, Li G M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 10.Ricciardone M D, Ozcelik T, Cevher B, Ozdag H, Tuncer M, Gurgey A, Uzunalimoglu O, Cetinkaya H, Tanyeli A, Erken E, Ozturk M. Cancer Res. 1999;59:290–293. [PubMed] [Google Scholar]
- 11.Wang Q, Lasset C, Desseigne F, Frappaz D, Bergeron C, Navarro C, Ruano E, Puisieux A. Cancer Res. 1999;59:294–297. [PubMed] [Google Scholar]
- 12.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata D, Navidi W, Salovaara R, Li Z H, Aaltonen L A. Nat Med. 1996;2:676–681. doi: 10.1038/nm0696-676. [DOI] [PubMed] [Google Scholar]
- 15.Shibata D. Am J Pathol. 1997;151:643–646. [PMC free article] [PubMed] [Google Scholar]
- 16.Tsao J L, Tavaré S, Salovaara R, Jass J R, Aaltonen L A, Shibata D. Am J Pathol. 1999;154:815–824. doi: 10.1016/S0002-9440(10)65437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Strand M, Prolla T A, Liskay R M, Petes T D. Nature (London) 1993;365:274–277. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 19.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 20.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 21.Weber J L. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- 22.Hudson T J, Engelstein M, Lee M K, Ho E C, Rubenfield M J, Adams C P, Housman D E, Dracopoli N C. Genomics. 1992;13:622–629. doi: 10.1016/0888-7543(92)90133-d. [DOI] [PubMed] [Google Scholar]
- 23.Valdes A M, Slatkin M, Freimer N B. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein D B, Pollock D D. J Hered. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- 25.Davison A C, Hinkley D V. Bootstrap Methods and Their Application. Cambridge, U.K.: Cambridge Univ. Press; 1997. [Google Scholar]
- 26.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 27.Aaltonen L A, Peltomäki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Järvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 28.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 29.Potten C S, Loeffler M. Development (Cambridge, UK) 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 30.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 31.Huang J, Papadopoulos N, McKinley A J, Farrington S M, Curtis L J, Wyllie A H, Zheng S, Willson J K V, Markowitz S D, Morin P, et al. Proc Natl Acad Sci USA. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L Z, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 33.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 34.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 35.Tomlinson I P M, Novelli M R, Bodmer W F. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]