Abstract
The hydroxypyridone ciclopirox olamine belongs to the antimycotic drugs used for the treatment of superficial mycoses. In contrast to the azoles and other antimycotic drugs, its specific mode of action is only poorly understood. To investigate the mode of action of ciclopirox olamine on fungal viability, pathogenicity, and drug resistance, we examined the expression patterns of 47 Candida albicans genes in cells grown in the presence of a subinhibitory concentration (0.6 μg/ml) of ciclopirox olamine by reverse transcription-PCR. In addition, we used suppression-subtractive hybridization to further identify genes that are up-regulated in the presence of ciclopirox olamine. The expression of essential genes such as ACT1 was not significantly modified in cells exposed to ciclopirox olamine. Most putative and known virulence genes such as genes encoding secreted proteinases or lipases had no or only moderately reduced expression levels. In contrast, exposure of cells to ciclopirox olamine led to a distinct up- or down-regulation of genes encoding iron permeases or transporters (FTR1, FTR2, FTH1), a copper permease (CCC2), an iron reductase (CFL1), and a siderophore transporter (SIT1); these effects resembled those found under iron-limited conditions. Addition of FeCl3 to ciclopirox olamine-treated cells reversed the effect of the drug. Addition of the iron chelator bipyridine to the growth medium induced similar patterns of expression of distinct iron-regulated genes (FTR1, FTR2). While serum-induced yeast-to-hyphal phase transition of C. albicans was not affected in ciclopirox olamine-treated cells in the presence of subinhibitory conditions, a dramatic increase in sensitivity to oxidative stress was noted, which may indicate the reduced activities of iron-containing gene products responsible for detoxification. Although the Candida drug resistance genes CDR1 and CDR2 were up-regulated, no change in resistance or increased tolerance could be observed even after an incubation period of 6 months. This was in contrast to control experiments with fluconazole, in which the MICs for cells incubated with this drug had noticeably increased after 2 months. These data support the view that the antifungal activity of ciclopirox olamine may at least be partially caused by iron limitation. Furthermore, neither the expression of certain multiple-drug resistance genes nor other resistance mechanisms caused C. albicans resistance to this drug even after long-term exposure.
The dimorphic opportunistic fungus Candida albicans is a major cause of mucosal and invasive mycoses. Infections caused by C. albicans are frequently treated with fluconazole, other azole antimycotics, or nonazole antimycotics. The modes of action of the majority of these drugs are well known (24). The imidazoles and triazoles like clotrimazole, ketoconazole, itraconazole, miconazole, fluconazole, and a whole range of similar components act by inhibiting C14-demethylase, an enzyme which is involved in the conversion of lanosterol to ergosterol in the ergosterol biosynthetic pathway. Consequently, the amount of ergosterol, which is an essential component of the fungal cell membrane, is decreased, leading to destabilization of the membrane and leakage of cellular components (26).
Amphotericin B, nystatin, and natamycin belong to the class of polyene antimycotics. They act by complexing with the ergosterol in the cell membrane. This leads to destabilization and changes in the permeability of the membrane and, consequently, causes the loss of proteins, carbohydrates, and nucleotides (24). Amorolfine is a morpholine antimycotic that inhibits two other enzymes of the ergosterol synthesis. It interacts with Δ14-reductase and Δ8-Δ7-isomerase, thus inhibiting the transformation of 4,4-dimethylergosta-8,14,24,(28)-trien-3β-ol to 4,4-dimethylergosta-8,24,(28)-dien-3β-ol and fecosterol to episterol, respectively (44). The pyrimidine antimycotic flucytosine is a precursor of a cytosine antimetabolite. After uptake into the fungal cell by the fungus-specific enzyme cytosine permease, it is deamidized to the toxic substance 5-fluorouracil (14). In contrast to all of these drugs, for which the modes of action are well known, very little is known about the fungal target(s) of the hydroxypyridone antimycotic ciclopirox olamine, a component widely used for the topical treatment of mucocutaneous mycoses.
Ciclopirox olamine is the ethanolamine salt of ciclopirox, which is a 6-cyclohexyl-1-hydroxy-4-methyl-2(1H)-pyridone (40). Therefore, the therapeutic effect of 1% ciclopirox olamine is equivalent to that of 0.77% ciclopirox (25), as the ethanolamine part of this agent does not add to the antifungal effect. Ciclopirox olamine (originally termed HOE 296) was first reported as a possible antifungal agent in 1973 (17). It has a very broad spectrum of activity and inhibits nearly all clinically relevant dermatophytes, yeasts, and molds, including the frequently azole-resistant Candida species Candida glabrata, Candida krusei, and Candida guilliermondii. In addition, it acts against a wide range of bacteria including many gram-positive and gram-negative species pathogenic for humans (16). The MICs of ciclopirox olamine for dermatophytes and yeasts pathogenic for humans show great uniformity, ranging from 0.98 to 3.9 μg/ml. Another feature that influences the clinical potency of the drug is its steep dose-response curve (16). Most of the studies dealing with the mode of action of this drug were published approximately 20 years ago, at the time that ciclopirox olamine was synthesized and introduced into clinical therapy (16, 17, 23, 33, 40, 49). When the drug became more frequently used, the results of many investigations about its efficacy and comparisons of its clinical effectiveness with those of other antimicrobial agents were published (25); however, the primary mode of action was rarely investigated. Consequently, the knowledge about the fungal target(s) of this clinically established and very efficient antifungal drug, which has only a very few side effects, is poor.
Early data from Sakurai et al. (50) suggested that ciclopirox olamine is rapidly taken up by growing C. albicans cells in large amounts as a function of both the incubation time and the external concentration of the drug. Uptake of the drug seemed to be energy independent, and intracellular accumulation can lead to levels 200 times greater than those in the medium. More than 97% of the accumulated drug is bound largely irreversibly to different cell structures and organelles such as the cell membrane, cell wall, mitochondria, microsomes, and ribosomes, while only small amounts are found in the cytosolic fraction. The drug is neither metabolized nor degraded (49). Furthermore, it was demonstrated that ciclopirox olamine causes inhibition of protein, RNA, and DNA synthesis in growing fungal cells, possibly by blocking the uptake of precursors of the macromolecules or by blocking the uptake of essential ions such as potassium ions and phosphates (33). In addition, in the presence of higher drug concentrations, cellular leakage causes the loss of folin-positive substances and potassium ions from the cell. In contrast to the cell membrane, the fungal cell wall does not seem to be functionally affected.
Electron microscopy revealed modifications in particle distribution within the plasma membrane of dermatophytes and C. albicans under the influence of ciclopirox olamine treatment (23). Subinhibitory concentrations of ciclopirox olamine caused modifications of cellular processes, which in turn had an effect on the adherence of C. albicans to buccal and vaginal epithelial cells (7). Similar observations were made in adherence studies with subinhibitory concentrations of rilopirox, another hydroxypyridone antimycotic agent (6). It is thought that the mode of action of rilopirox resembles that of ciclopirox olamine to a high degree (36). Ultrastructural investigations with C. albicans cells exposed to rilopirox showed, in addition to morphological alterations similar to those detected by Gasparini et al. (23), that the plasma membrane exhibits elongated invaginations, that the numbers and sizes of lipid droplets increase, and that mitochondria are greatly enlarged, while the cell wall remains unaffected (48).
Early data from Dittmar and Lohaus (17) indicated that the MIC of ciclopirox olamine increases dramatically when thioglycolate-containing media were used for MIC determinations. They assumed that iron salts, which are present in these media, antagonized the inhibitory effect by forming complexes with the drug. Therefore, it was hypothesized that the chelation of metal ions and the inhibition of iron-dependent enzymes play a primary role in the action of ciclopirox olamine (1). For rilopirox, a high affinity for iron ions has been shown in complexometric studies (37). Furthermore, rilopirox inhibited the activity of a catalase from Aspergillus niger and the respiratory chain in mitochondria and submitochondrial particles in Saccharomyces cerevisiae (37). Since both catalases and enzymes of the respiratory chain are iron-dependent proteins, this further supported the view that iron binding may play a major role in the mode of action of hydroxypyridone antimycotics.
Another point of interest is the fact that for ciclopirox olamine, even though it was introduced into clinical therapy more than two decades ago and is frequently used to treat superficial mycoses or vaginal candidiasis, no single case of fungal resistance has been reported. Among drug-resistant fungi, mechanisms of drug resistance involving drug efflux pumps are among the most important for resistance to a wide variety of antifungal drugs and toxic molecules (54, 59). Two families of such efflux transporters are known. The gene products of the first multidrug resistance gene detected, MDR1 (5, 20), and the possible fluconazole resistance gene FLU1 (11) belong to the major facilitator family. In contrast, the putative Candida drug resistance genes CDR1 to CDR4 (2, 21, 46, 52) encode members of the ATP-binding-cassette (ABC) transporter family. In C. albicans the up-regulation of MDR1, CDR1, and CDR2 are implicated in the development of fluconazole resistance, while for CDR3 and CDR4, no correlation between mRNA levels and the degree of resistance was reported (12, 42, 53).
In order to investigate the modes of action of ciclopirox olamine on fungal viability, pathogenicity, and drug resistance, we examined the expression patterns of 47 essential genes, putative or known virulence genes, and genes involved in multiple-drug resistance of C. albicans in cells grown in the presence of subinhibitory concentrations of ciclopirox olamine by reverse transcription (RT)-PCR. To clarify whether ciclopirox olamine may act as a chelating agent, thus influencing iron-dependent cellular processes by chelating metal ions, we also studied the expression of genes encoding proteins of iron metabolism. In addition, we used a cDNA subtraction technology called suppression-subtractive hybridization (SSH) (15) to further identify genes that are up-regulated in the presence of ciclopirox olamine. Furthermore, the influences of iron ions and oxidative stress on ciclopirox olamine-treated C. albicans cells were examined to understand how this drug might act on fungi. Finally, a long-term resistance induction experiment with ciclopirox olamine and fluconazole was performed to further investigate whether resistance to this drug can be generated.
MATERIALS AND METHODS
Strains and growth cultures.
All experiments were carried out with C. albicans strain SC5314. Sabouraud glucose medium (2%) was used for cell culture growth, and RPMI 2% glucose medium and 2% Sabouraud glucose medium were used for MIC determinations. For cell culture growth curves, 220 ml of 2% Sabouraud glucose medium containing different concentrations of an antimycotic agent were inoculated with 105 cells/ml, and the mixture was shaken at 160 rpm and 37°C. Growth was measured photometrically at 630 nm. Iron(III) chloride hexahydrate (Merck, Darmstadt, Germany) or 2,2′-bipyridine (Sigma-Aldrich, Steinheim, Germany) was added to the medium at different concentrations for inhibition studies.
RNA extraction and RT-PCR.
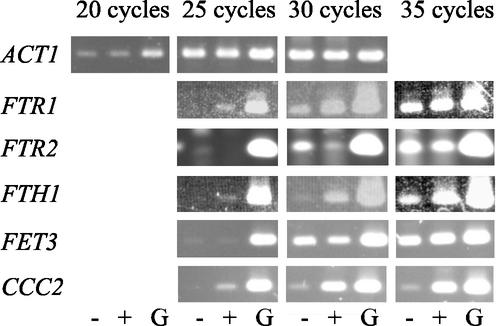
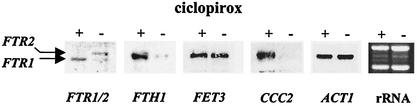
For RT-PCR expression analysis, cells were harvested and immediately frozen in liquid nitrogen. Frozen pellets of approximately 105 cells were homogenized and lysed by vortexing in 1 ml of peqGOLD RNApure (Peqlab, Erlangen, Germany) with hydrochloric acid-treated glass beads (diameters, 0.40 to 0.60 mm; Braun Biotech, Melsungen, Germany) for 10 min, and RNA extraction was carried out as described by the manufacturer. RNA quality was controlled by gel electrophoresis (51), and RNA concentrations were measured by determination of the absorbance at 260 nm (1 unit of the optical density [OD] at 260 nm is equal to 40 μg of RNA/ml of H2O, based on the extinction coefficient of RNA in H2O). One microgram of total RNA was treated with DNase, and cDNA was synthesized as described previously (19). The cDNA was diluted 1:3 with H2O, and 0.4 μl was used for each PCR. All PCRs were performed in 10-μl volumes containing 0.24 μl of 10 mM deoxynucleoside triphosphates, 0.3 μl of 50 mM MgCl2, 1.0 μl of 10× PCR buffer, 0.4 μl each of 10 μM forward and reverse primers, and 0.04 μl of 5 U of Taq polymerase (Gibco, Life Technologies, Karlsruhe, Germany) per μl. Primer sequences and annealing temperatures are shown in Table 1. cDNAs were amplified after initial denaturation at 94°C for 3 min, annealing for 3 min, and elongation at 72°C for 5 min, followed by 20, 25, 30, or 35 amplification cycles of denaturation at 94°C for 30 s, annealing for 30 s, and elongation at 72°C for 30 s and one final elongation step at 72°C for 10 min. The PCR products were analyzed on a 1% agarose gel. During PCR, the PCR product concentration is proportional to the starting cDNA concentration, as long as product accumulation remains exponential. The point at which exponential accumulation plateaus can be estimated by noting the point at which continued cycles do not produce significantly increased product yields. To ensure that samples from the exponential phase of PCR amplification were examined, we used 20, 25, 30, and 35 cycles for all genes of interest. All RT-PCR experiments were done in duplicate for ciclopirox olamine- and fluconazole-treated cells in samples from two independent biological experiments. The housekeeping gene ACT1 (41), which was expressed to a similar extent under all conditions investigated, was used as a standard control. The expressions of selected genes (ACT1, CCC2, FTR1, FTR2, FTH1, FET3) during the different cycles of the RT-PCR are shown in Fig. 1. For example, the expression of ACT1 with and without ciclopirox olamine remains unchanged, while the expression of CCC2 (a copper transporter gene) is strongly increased under the influence of this drug. To ensure the complete absence of genomic DNA contamination in the RT-PCR analysis, the RT-PCR was controlled by the use of primers whose sequences are specific for intron-containing genes EFB1 and IMH3 of C. albicans (35, 43). These genes were also found to be useful internal mRNA controls, as high levels of transcripts were detected at all time points tested. As an additional control, genomic DNA of C. albicans was used as a PCR template for every primer pair. After gel electrophoresis and ethidium bromide staining of the DNA bands, the relative level of expression of each gene of interest was semiquantified by measuring the DNA band fluorescence for DNA from cells grown with or without antifungal drug densitometrically by using video images (AlphaImager 2000 Documentation and Analysis system; Alpha Innotech, Hessisch Oldendorf, Germany) and AIDA image analyzer (version 2.0) software (Raytest Isotopenmeβgeräte GmbH, Straubenhardt, Germany). The background intensity was subtracted, and the relative intensities of the bands obtained from cells grown in the presence or absence of an antifungal drug were calculated. Data are presented as the averages (standard deviations) from the two independent experiments. Numbers are given as the fold increase or decrease in the level of expression relative to the level of expression of the housekeeping genes ACT1, IMH3, and EFB1, for which the maximum difference in the level of fluorescence between the two bands was 15%, possibly due to experimental differences. On this basis, gene expression levels were defined as unchanged at expression ratios between 0.78 and 1.29; when the fluorescence differences were between 0.77 and 0.51 or 1.3 and 1.99, the result was defined as moderately down-regulated or moderately up-regulated. Finally, differences of 0.5 or less and 2.0 or more relative to the control band were defined as strongly down-regulated or strongly up-regulated, respectively. If no expression of a gene could be detected, the result was defined as 0.
TABLE 1.
Primer sequences and annealing temperatures of primers used for RT-PCR
| Gene | Primer sequence
|
Annealing temp (°C) | |
|---|---|---|---|
| Forward primer | Reverse primer | ||
| ACT1 | 5′-ACCGAAGCTCCAATGAATCCA | 5′-GGATGGACCAGATTCGTCGTA | 59 |
| IMH3 | 5′-TCTCCTGAAGTCACTGTTGG | 5′-GGTACCAATGGCAGCACCAC | 59 |
| EFB1 | 5′-AGTCATTGAACGAATTCTTGGCTG | 5′-TTCTTCAACAGCAGCTTGTAAGTC | 59 |
| SAP1 | 5′-GATGTCATTAAAACTCCTGTTAATG | 5′-CCAGTTTCAATTCAGCTTGG | 60 |
| SAP2 | 5′-CTCCTAAAGCATTCCCAGTTAC | 5′-CATCATCACCTTGTAAAGAAGC | 60 |
| SAP3 | 5′-CATGTCAAGCTGGTCAAGGAC | 5′-ATAGGCTGATCTCAAGAAATTATC | 60 |
| SAP4 | 5′-GTTCCAGATTCAAATGCCG | 5′-CTTGAGCCATGGAGATCTTTC | 60 |
| SAP5 | 5′-TGAGACTGGTAGAGATGGTG | 5′-GGTTTACCACTAGTGTAATATGT | 60 |
| SAP6 | 5′-AAACCAACGAAGCTACCAGAAC | 5′-TAACTTGAGCCATGGAGATTTTC | 60 |
| SAP7 | 5′-GATAAGGCATCAGGTACTATGG | 5′-AGGAACAACGGCATGGTTATC | 60 |
| SAP8 | 5′-CTGTTATTGTTGACACAGGTTC | 5′-GTAGAAATACTTGAAGAAGTAGTG | 60 |
| SAP9 | 5′-CACCATAAGCAACGTGACTG | 5′-GCGAAAGCAACAACCCATAC | 60 |
| SAP10 | 5′-ACGTCAGAAGACTTTTCCATTG | 5′-ATATGGCGATCCATGAACGTG | 60 |
| LIP1 | 5′-ACAAATTCAATGGGATCAAGAG | 5′-ATAAGTGACATGGACGTTACTG | 56 |
| LIP2 | 5′-TTTCCGACTTTGCTGTTCCAG | 5′-ATAATACTGCTTACAAGACCAAG | 56 |
| LIP3 | 5′-AAATGCCAGGCAAAATGAGC | 5′-TGTTGTAAAGCTCCTTCATATC | 56 |
| LIP4 | 5′-TGATCAATTATATTGGTAAGCAC | 5′-TCCTTTTTGGATGAGTATATTC | 56 |
| LIP5 | 5′-AATCGTCCCTATTGTCGATACC | 5′-AAGTCCGAGATGGAGAACAAC | 56 |
| LIP6 | 5′-TTAAACCTGGTGCCAAAGCTG | 5′-TCGATGCCCTGGTGGTGAAC | 56 |
| LIP7 | 5′-TTCCATCATTCGAGACATTTCAG | 5′-ACGGAAGTACTGACTGAGAAATG | 56 |
| LIP8 | 5′-AGAGTGATACAGACAAAAAATCAG | 5′-AAGACCATTCAGCATGGTG | 56 |
| LIP9 | 5′-TTTATAAAGTATGTGGGAGCTAG | 5′-TAGGACCAAGCCCTTGTTGTG | 56 |
| LIP10 | 5′-TTGGGTTTGCAACTGCTAAGC | 5′-ATAGTAGATCTAGCACTGAGC | 56 |
| RBT1 | 5′-CTGAAACTGAAACCACTCCAACTGCTCAC | 5′-CAGATGTAACAACAACAACACCAGTGG | 59 |
| HYR1 | 5′-GCTGGTAACGGATCAAACGAAG | 5′-GGAAACAATTAAACCAAACAAAATG | 59 |
| HWP1 | 5′-CACAACAACAACAGCAGGAGGAAC | 5′-TGGAGTAGTAGCTGGAGTTGTTGG | 59 |
| TUP1 | 5′-CAAAATCTTGAGGGGCCACGAAC | 5′-CTAAGGACCCGGAAGCGATTTGTT | 59 |
| PLB1 | 5′-TGCCGTTTCCAATATTGATC | 5′-CCAAGGATCCACTTGATCTA | 54 |
| PLB2 | 5′-CCAGAACTTGACATCATTAACTGA | 5′-TATAAAGTTATCTACGAGAATGG | 54 |
| PLB3 | 5′-CTTTCTAATGATGATAATGATATT | 5′-TTATCCAAGGATAAATACACTTA | 54 |
| PLC1 | 5′-TTTGGGATTCATTAGCTCAAGG | 5′-TAACATTAGCAAAGCTTATTGG | 54 |
| PLD1 | 5′-CAGCTTGTTTCATCGACGGT | 5′-AGCTGCCATATATGGCTTACC | 54 |
| MDR1 | 5′-AGAGCCATCACCGGTAACGACAG | 5′-CCAACCAAAAATGAAAAGACCTGAAG | 59 |
| FLU1 | 5′-CACTGCCTTGGCTGGTAAC | 5′-ACATCGTGCAAAAGGAAGAAC | 56 |
| CDR1 | 5′-TTTCTGGTGCCATGACTCCTGCTAC | 5′-CAATATAAATGGCCAAAAAGAATACG | 56 |
| CDR2 | 5′-GGGTATTGGCTGGTCCTAATGTGATTC | 5′-CTAGCCAACCAGTAAAAGAAAATAGTAA | 56 |
| CDR3 | 5′-ATTACCGGGCTGTACAAAATG | 5′-GTGGCTGCTAAAACCTGATAAG | 54 |
| CDR4 | 5′-TACGGTGCTCCAAAATGTCC | 5′-TGGGCAGCAATAAATGTAATCC | 54 |
| CAT1 | 5′-TTACGTTCAAGTTCATTTCATCAG | 5′-CAATCTGTGCGGTCTGTGAG | 54 |
| FTR1 | 5′-CCATCGGTGCGTCCTAC | 5′-GAGAACAAACCAGCAGCAATC | 54 |
| FTR2 | 5′-GGTCCAGAGTTGAAGAAAAAGTTG | 5′-GGGAAAGATGAAGCAGGAC | 54 |
| FTH1 | 5′-CCCAACTGAATCTGATGACCTTAC | 5′-GCCAACACAGCAGCATTTAC | 54 |
| FTH2 | 5′-GATGGGAATAACAATGGTGAGG | 5′-CCGATAATGGAATAGAATGAGGTG | 54 |
| FET3 | 5′-GGCGAAGCTGCAACAAACG | 5′-AACGCAGCAACACATGAAAACAC | 54 |
| CCC2 | 5′-TGGAGGCAACCAAAGTAGTAGTG | 5′-ATTAACGGCGCTTTTTCTTTTC | 54 |
| CFL1 | 5′-TCATTTGATTGCCGACAGG | 5′-TGAAATCGCCATATCCTTGAC | 56 |
| SIT1 | 5′-GCCGTTGGTACTAGATGGAAATG | 5′-AACCCGTGACAACAGATACAAATG | 56 |
| PCK1 | 5′-AAGGGGTTGAAAAAGGTGATGTC | 5′-ATGCGGAGAATGTAGCGTGTG | 56 |
| WHI1 | 5′-TGTCCGACTTAGGTAGAAAAGATA | 5′-TGGAATCACCAAAAATAGCAT | 56 |
| NAM7 | 5′-TTCATATGCTACCCACGACACC | 5′-CTCATCAAAATTGGCAAAACTTCT | 56 |
| IPF12201 | 5′-GGGGTATGATTATGGTGAGATTG | 5′-AAGGTAACACGGTGAAAAAGAAC | 56 |
| IPF10138 | 5′-AAGGGTTGTTTTATGGTGGTG | 5′-TTCTTGGGCGACTTTATTAGC | 56 |
FIG. 1.
Analysis of RT-PCR products amplified from RNA samples collected from cultures without ciclopirox olamine (−), cultures with 0.6 μg of ciclopirox olamine/ml (+), and control PCRs with genomic DNA (G) of C. albicans SC5314. Results are shown for primer pairs amplifying cDNA from the housekeeping actin gene ACT1, the high-affinity iron permease gene FTR1, the low-affinity iron permease gene FTR2, the putative iron transporter FTH1, the multicopper oxidase gene FET3, and the copper transporter gene CCC2 after 20, 25, 30, and 35 cycles of PCR. The levels of expression of ACT1 and FET3 remained unchanged in the drug-treated culture, while the levels of expression of FTR1, FTH1, and CCC2 were strongly increased in the presence of a subinhibitory concentration of ciclopirox olamine. In contrast, low-affinity iron permease gene FTR2 was strongly down-regulated in the presence of the drug. Similar data were observed by Northern analysis (Fig. 5).
Northern blot analysis.
To confirm the expression patterns of selected genes observed by RT-PCR, frozen cell pellets were homogenized and lysed by vortexing in 2 ml solution of GTC buffer (4 M guanidine thiocyanate, 2% lauroylsarconisate, 10 mM EDTA, 25 mM Tris HCl [pH 7.5]) and phenol (Roti-Phenol/Chloroform; Roth, Karlsruhe, Germany) 1:1 with hydrochloric acid-treated glass beads for 10 min. After centrifugation, the aqueous phase was treated twice with phenol, and then the nucleic acids were precipitated with isopropanol.
Twenty micrograms of total RNA from each sample was loaded onto a formaldehyde gel as described previously (31a). mRNA levels were measured relative to the rRNA levels by loading approximately equal amounts of total RNA in each lane of the Northern blots. Blots were hybridized with digoxigenin (DIG)-labeled probes (the primers used are listed in Table 1) at 42°C in DIG easy Hyb solution (Roche, Mannheim, Germany) for 12 h. After hybridization, the blots were washed twice at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 5 min, twice at 60°C in 0.1× SSC-0.1% sodium dodecyl sulfate for 15 min, and once in Tris-buffered saline (0.3 M NaCl, 0.1 M Tris-HCl [pH 7.5]) plus 0.3% Tween. DIG-labeled probes were detected after incubation with an anti-DIG antibody (Roche) (diluted 1:10,000 in blocking solution [Roche] at room temperature for 0.5 h) by using the CDP star reagent (Roche) and exposed to an X-ray film (Roche). The ACT1 gene was used as an additional internal loading control.
SSH.
To identify genes that were up- or down-regulated due to treatment with ciclopirox olamine, we used the cDNA SSH protocol previously described by Diatchenko et al. (15). This protocol is based on comparison of two populations of mRNA to obtain clones of genes that are expressed in one population but not in the other. Briefly, both mRNA populations are converted into cDNA, and the two cDNA populations are hybridized. The hybrids are removed, leaving unhybridized cDNAs that are expressed in one population but that are absent from the other. Two cultures of SC5314 cells were grown in 2% Sabouraud glucose medium for 5 h. At this time point 1.0 μg of ciclopirox olamine per ml was added to one of the cultures. One hour later, the cells were harvested and immediately deep frozen in liquid nitrogen. For the subtraction, mRNA was isolated from total RNA by using the Dynabeads mRNA Purification kit (Dynal, Hamburg, Germany) according to the instructions of the manufacturer and transcribed into cDNA as described above. The cDNA subtraction was performed by using the PCR-Select cDNA Subtraction kit (CLONTECH Laboratories, Heidelberg, Germany) according to the instructions of the manufacturer. The subtracted PCR products were cloned and sequenced as described below.
Cloning, sequencing, and sequence alignments.
The subtracted PCR products obtained by the SSH protocol were ligated into the pCR2.1-TOPO plasmid vector and transformed into competent TOP10F′One Shot Escherichia coli cells (Invitrogen, Karlsruhe, Germany) with the TOPO TA Cloning kit (Invitrogen). Ninety-six of the transformed colonies were picked and cultured overnight in two 48-well microtiter plates in which each well contained 400 μl of Luria-Bertani medium containing ampicillin at 100 mg/liter. A PCR screen was performed for single transformants by using 1.5 μl of each 400 μl of overnight cell suspension per 20 μl of PCR mixture, and primer pair 1 and 2R, which had been used in the subtraction protocol, was used to amplify the plasmids containing gene inserts of plasmids. Gene inserts were sequenced in the sequencing service unit of the Robert Koch-Institut (Berlin, Germany) by using a T7 primer and the ABI BigDye kit (Applied Biosystems, Weiterstadt, Germany). The sequences were compared with those in the Candida databases (Candida DB) of the Institut Pasteur, Paris, France, and the National Center for Biotechnology Information by using the BLASTn mode of the BLAST search program (http://genomeweb.pasteur.fr/lfrangeu/ca/IPFblast.html; http://genolist.pasteur.fr/CandidaDB; http://ncbi.nlm.nih.gov/blast/). Sequence data from these websites are based on data from the Stanford DNA Sequencing and Technology Center website (http://www-sequence.stanford.edu/group/candida). Sequencing of C. albicans at the Stanford DNA Sequencing and Technology Center was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. To avoid the analyses of clones carrying identical cDNA fragments, the PCR products of the screened clones were blotted onto a positively charged nylon membrane (Roche) and hybridized to DIG-labeled (Roche) probes (51) containing sequences which turned out to be abundant after several rounds of sequencing. Only PCR products that did not hybridize with these probes were selected for further sequencing.
Hyphal induction.
To induce the yeast-to-hyphal phase transition of C. albicans, a preculture of strain SC5314 was grown overnight in 2% Sabouraud glucose medium at 25°C and shaken at 160 rpm. An inoculum of 106 cells/ml was taken for further incubation in 5% fetal bovine serum (Sigma-Aldrich) containing 0 and 0.6 μg of ciclopirox olamine/ml at 37°C and 160 rpm. Samples were taken every 30 min and briefly subjected to ultrasonication, and the percentages of yeast and hyphal cells were counted by light microscopy in a counting chamber.
Hydrogen peroxide resistance.
A 24-h preculture of strain SC5314 grown at 37°C and 160 rpm in 2% Sabouraud glucose medium was taken to measure the susceptibilities of those cells to H2O2, as described previously (32). Cells from the preculture were subcultured and grown in 2% Sabouraud glucose medium with ciclopirox olamine (0.6 μg/ml) and without ciclopirox olamine to a concentration of 5 × 106 cells/ml, harvested, and resuspended in 0.1 M potassium phosphate buffer (pH 7.0) to give an OD at 600 nm of 0.1. Various concentrations of H2O2 (30% [wt/wt] hydrogen peroxide solution; Sigma-Aldrich) were added to the cell suspension to obtain H2O2 concentrations of 0, 0.5, 1, 1.5, 2, and 2.5 mM. After 1 h of incubation at 30°C, aliquots were taken from the cell suspensions, diluted 1:100 in the same buffer, plated on solid yeast peptone dextrose agar plates, and incubated for 3 days at 28°C. The number of growing colonies was representative of the number of surviving cells.
Long-term resistance induction.
In order to investigate whether long-term treatment with ciclopirox olamine or fluconazole may induce antifungal resistance in C. albicans SC5314, cells were treated as follows. All cultures were grown in 1,000-ml Erlenmeyer flasks containing 220 ml of 2% Sabouraud glucose medium at 37°C and shaken at 160 rpm. This large culture size was chosen to allow a large number of cell divisions and, therefore, a greater possibility of genetic alteration. Initially, concentrations of both ciclopirox olamine (0.6 μg/ml) and fluconazole (8 μg/ml) which still allowed growth of the cells were chosen. Two milliliters of these cell cultures was routinely subcultured three times a week into fresh medium containing the same concentration of antifungal agent in the original culture. When increasing cell growth under the given conditions was detected, the concentrations of the antifungal agents were doubled. Additionally, at regular intervals susceptibility tests were carried out to determine the ciclopirox olamine and fluconazole MICs for the cells by the NCCLS M27-A (45) broth microdilution method with RPMI medium containing 2% glucose.
Electron microscopy.
Electron microscopic pictures of SC5314 cells were taken to investigate the effect of the subinhibitory concentration of 0.6 μg of ciclopirox olamine/ml. Cells were grown with and without 0.6 μg of ciclopirox olamine/ml in 2% Sabouraud glucose medium as described above and were harvested after 15 h. Cells were fixed in Karnovsky solution (34) at room temperature. After centrifugation at 800 × g for 15 min, the pellets were embedded in glycide ether. The blocks containing cells were cut with an ultramicrotome. Semithin sections were studied with a light microscope after staining with 1% toluidine blue and 1% pyronin G. Ultrathin sections were stained with 0.5% uranyl acetate for 10 min at 30°C and 2.7% lead citrate for 5 min at 20°C. Ultrathin sections were examined with an EM 902 transmission electron microscope (Zeiss) operating at 80 kV at magnifications between ×3,000 and ×85,000.
RESULTS
Inhibition of C. albicans growth by ciclopirox olamine, fluconazole, and bipyridine.
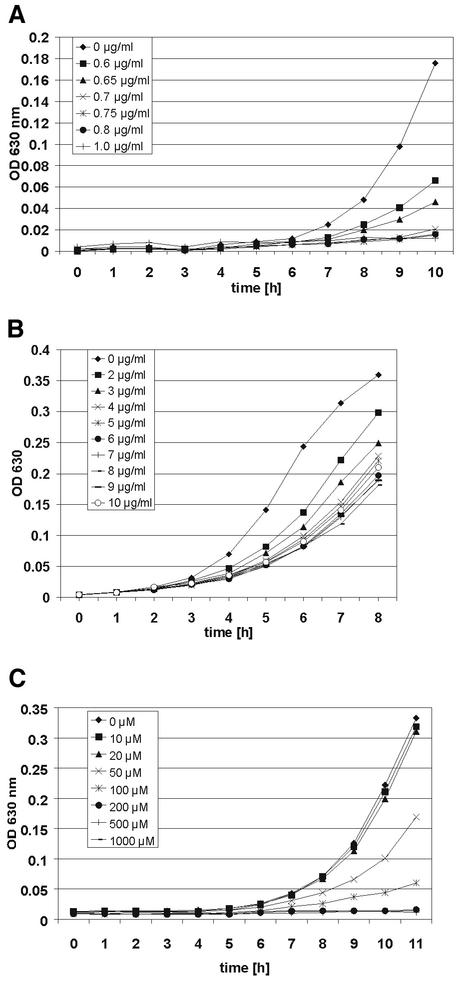
In order to investigate the influence of the antifungal agent ciclopirox olamine on C. albicans, cells of strain SC5314 were grown in the presence of several concentrations of this drug and growth curves were determined. For further investigations, the subinhibitory concentrations which reduced cell culture growth without being fungicidal were determined. A concentration of 0.6 μg of ciclopirox olamine/ml proved to be the optimal subinhibitory concentration that significantly inhibited but that did not completely block the cell culture growth (Fig. 2A). Growth decreased dramatically in the presence of concentrations greater than 0.6 μg/ml. A concentration of 0.7 μg/ml almost completely inhibited growth. In order to determine that the gene expression patterns of ciclopirox olamine-treated cells observed (see below) were due to distinct and drug-specific effects on certain genes rather than a general inhibition phenomenon, the experiments were repeated with fluconazole. Growth curves with fluconazole showed that 2.0 μg of fluconazole/ml was the optimal subinhibitory concentration that caused similarly reduced cell growth (Fig. 2B). Growth curves in the presence of fluconazole showed a much wider range of concentrations with intermediate inhibition. Growth could be detected even in the presence of 10 μg of fluconazole/ml of medium. To compare these results with standard MICs, the ciclopirox olamine and fluconazole MICs at which 80% of isolates were inhibited (MIC80s) for SC5314 were determined by the NCCLS M27-A standard method (45) by a microdilution test in RPMI 2% glucose or 2% Sabouraud glucose medium. The MIC80s of ciclopirox olamine in RPMI 2% glucose medium and 2% Sabouraud glucose medium were 2.0 and 1.0 μg/ml, respectively; and the MIC80s of fluconazole in RPMI 2% glucose and 2% Sabouraud glucose medium were 0.12 and 2.0 μg/ml, respectively.
FIG. 2.
Growth curves for C. albicans SC5314 in 2% Sabouraud glucose medium measured at an OD of 630 nm containing different concentrations of ciclopirox olamine (A), fluconazole (B), and bipyridine (C). In these experiments 0.6 μg of ciclopirox olamine/ml, 2.0 μg of fluconazole/ml, and 100 μM bipyridine turned out to be the optimal subinhibitory concentrations which notably inhibited the cells but still allowed growth.
One possible effect of ciclopirox olamine may be the binding of iron ions, which in turn causes iron-limiting conditions and therefore reduced cell growth. In order to study the effect of iron-limiting conditions on cell growth, we added the well-known iron chelator bipyridine. The addition of 100 μM bipyridine caused reduced cell growth, similar to the effect of 0.6 μg of ciclopirox olamine/ml (Fig. 2C).
Reversal of the effect of ciclopirox olamine by addition of iron(III) chloride hexahydrate.
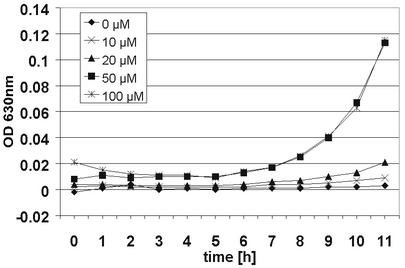
Since a number of experimental observations suggested that ciclopirox olamine may act by binding to iron ions, we assumed that such an effect might be reversed by the addition of excessive iron ions. Growth curve experiments for C. albicans SC5314 grown in the presence and the absence of an inhibitory concentration of ciclopirox olamine (1.0 μg/ml) were repeated with media containing different concentrations of iron(III) chloride hexahydrate (Fig. 3). No growth of ciclopirox olamine-treated cells could be detected without iron(III) chloride supplementation. However, supplementation with increasing concentrations of iron(III) chloride increased the levels of growth of the Candida cultures. In a control experiment without ciclopirox olamine, an inverse effect of iron(III) chloride supplementation was observed (data not shown). In that case, increasing concentrations of iron(III) chloride ions decreased the level of growth of the Candida culture, indicating that excess iron ions have toxic effects on the fungal cells.
FIG. 3.
Growth curves for C. albicans SC5314 in 2% Sabouraud glucose medium containing 1.0 μg of ciclopirox olamine/ml supplemented with different concentrations of iron(III) chloride measured at an OD of 630 nm. Iron(III) chloride at concentrations of 20 μM and greater increased the levels of growth of the Candida cultures.
Effect of ciclopirox olamine on the cellular morphology of C. albicans.
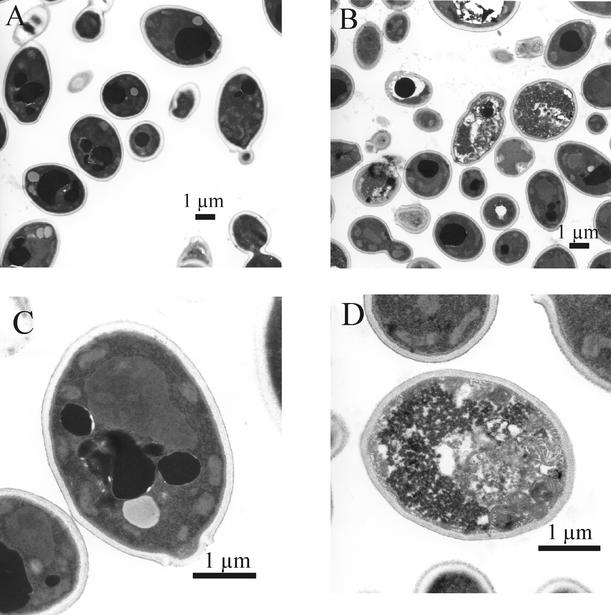
Subinhibitory concentrations of ciclopirox olamine still allowed reduced levels of growth of C. albicans cells. We used electron microscopy to investigate whether such drug concentrations have effects on cell morphology (Fig. 4A to D). Interestingly, not all cells grown in the presence of 0.6 μg of ciclopirox olamine/ml were damaged to the same extent. Compared with untreated cells (Fig. 4A and C), some cells appeared normal, while others had severe morphological defects (Fig. 4B and D). These defects included enlargements of the vacuole, which looked homogenous in untreated cells. The cell membranes of damaged cells showed numerous invaginations, and in many areas the cytoplasmic margin was separated from the cell wall. In addition, mitochondria appeared bloated and fissures in the inner membrane system were largely widened. With the exception of the cell wall, which looked unaffected, most parts and organelles of the damaged cells were severely affected by ciclopirox olamine at a concentration of 0.6 μg/ml. Therefore, it seemed that the reduced growth was caused by a fungicidal effect on a number of cells, even though other cells remained unaffected.
FIG. 4.
Electron microscopy of C. albicans SC5314 cells grown in 2% Sabouraud glucose medium without (A) and with (B) 0.6 μg of ciclopirox olamine/ml for 15 h. Not all cells were damaged to the same extent. Some cells appear normal, while others have severe morphological defects. (C) Detail of a single cell grown without ciclopirox olamine. (D) Detail of a cell grown under the influence of 0.6 μg of ciclopirox olamine/ml with enlargements of the vacuole and numerous invaginations of the cell membrane. In some areas the cytoplasmic margin was separated from the cell wall. In addition, mitochondria appeared bloated and fissures in the inner membrane system were largely widened. With the exception of the cell wall, which looked unaffected, most parts and organelles of the damaged cells were severely affected by the drug.
Gene expression in C. albicans treated with subinhibitory concentrations of ciclopirox olamine, fluconazole, and bipyridine.
We assumed that growth in the presence of antifungal drugs may directly or indirectly influence the gene expression patterns of treated cells. After 15 h of growth in media containing the subinhibitory concentrations of ciclopirox olamine (0.6 μg/ml) and fluconazole (2.0 μg/ml), C. albicans cells were harvested, total RNA was extracted, and cDNA was synthesized. This cDNA was used as a template for a series of RT-PCRs with 47 selected genes of interest. The expression data observed by RT-PCR from two independent experiments are summarized in Tables 2 to 4. First, the levels of expression of the housekeeping genes ACT1 (41) (Fig. 1), IMH3 (35) (data not shown), and EFB1 (43) (data not shown) were determined. All three genes were constitutively expressed in the presence of ciclopirox olamine, suggesting that basic cell functions still take place in growing cells and that the changes in expression patterns of other genes could be validly compared to the expression patterns of these housekeeping genes. Antifungal drugs not only may cause inhibition of cell viability but may also influence the expression of genes required for pathogenicity. Therefore, we next investigated the effects of ciclopirox olamine and fluconazole on the expression of putative or known virulence genes (Table 2). The levels of expression of most genes that encode potential virulence factors, such as members of the SAP gene family (SAP1 to SAP10) (30) and the LIP gene family (LIP1 to LIP10) (22, 31), were either not affected or moderately decreased. Conversely, no cases of increased gene expression could be detected. Furthermore, some genes involved in the yeast-to-hyphal phase transition were investigated (Table 2). One gene known as a central repressor of filamentous growth, TUP1 (9), showed a moderate up-regulation of expression. Similarly, the levels of expression of one gene known to be repressed by TUP1 (8, 10), RBT1, which encodes a cell wall protein, and gene HYR1, which encodes a hyphal wall protein, were moderately increased. Since the treatment of C. albicans cells with azole drugs is known to cause up-regulation of a number of drug resistance genes (12, 53), we also investigated the expression patterns of these resistance genes in ciclopirox olamine-treated cells (Table 3). The expression of Candida drug resistance gene CDR1 was strongly up-regulated, while the levels of expression of CDR2 and FLU1 were moderately increased. The level of expression of the major facilitator gene MDR1 showed no change.
TABLE 2.
Effects of ciclopirox olamine and fluconazole on levels of expression of putative or known virulence genes
| Gene | Gene function (reference) | Expression levela
|
|
|---|---|---|---|
| CIC | FLC | ||
| SAP1 | Secreted protease (30) | =, 0.89 (0.095) | ↑, 2.28 (0.81) |
| SAP2 | Secreted protease (30) | (↓), 0.76 (0.11) | =, 0.79 (0.145) |
| SAP3 | Secreted protease (30) | =, 0.95 (0.275) | =, 1.1 (0.105) |
| SAP4 | Secreted protease (30) | ↓, 0.50 (0.195) | 0 |
| SAP5 | Secreted protease (30) | ↓, 0.5 (0.09) | =, 0.85 (0.13) |
| SAP6 | Secreted protease (30) | =, 0.83 (0.04) | =, 1.05 (0.13) |
| SAP7 | Secreted protease (30) | =, 0.79 (0.165) | 0 |
| SAP8 | Secreted protease (30) | =, 0.81 (0.155) | (↓), 0.75 (0.005) |
| SAP9 | Secreted protease (30) | =, 0.97 (0.03) | =, 0.84 (0.15) |
| SAP10 | Secreted protease (30) | (↓), 0.74 (0.03) | =, 0.8 (0.13) |
| LIP1 | Secreted lipase (22) | ↓, 0.35 (0.125) | =, 0.8 (0.17) |
| LIP2 | Secreted lipase (31) | (↓), 0.53 (0.215) | =, 0.99 (0.135) |
| LIP3 | Secreted lipase (31) | 0 | 0 |
| LIP4 | Secreted lipase (31) | =, 0.91 (0.27) | (↓), 0.76 (0.11) |
| LIP5 | Secreted lipase (31) | (↓), 0.64 (0.175) | =, 1.06 (0.275) |
| LIP6 | Secreted lipase (31) | =, 1.04 (0.045) | =, 0.97 (0.015) |
| LIP7 | Secreted lipase (31) | =, 0.91 (0.085) | 0 |
| LIP8 | Secreted lipase (31) | =, 0.88 (0.07) | ↑b |
| LIP9 | Secreted lipase (31) | 0 | 0 |
| LIP10 | Secreted lipase (31) | 0 | 0 |
| RBT1 | Hyphal wall protein (8) | (↑), 1.32 (0.2) | =, 0.91 (0.075) |
| HYR1 | Hyphal wall protein (10) | (↑), 1.48 (0.065) | =, 0.87 (0.01) |
| HWP1 | Hyphal wall protein (10) | 0 | ↓, 0.44 (0.25) |
| TUP1 | Transcriptional repressor (9) | (↑), 1.32 (0.045) | =, 0.91 (0) |
| PLB1 | Secreted phospholipase (28) | 0 | 0 |
| PLB2 | Secreted phospholipase (56) | 0 | 0 |
| PLB3 | Secreted phospholipase (47% homology to PLB3 of S. cerevisiae) | ↑, 2.1 (0.04) | ↓, 0.27 (0.035) |
| PLC1 | Secreted phospholipase (4) | 0 | (↑), 1.63 (0.41) |
| PLD1 | Secreted phospholipase (29) | 0 | 0 |
The numbers are the averages (standard deviations) of the relative expression levels in cell culture with and without antifungal agents (see Materials and Methods). Expression levels are defined as follows: ↓, strongly down-regulated (0.5 and less); (↓), moderately down-regulated (0.77 and less); =, unchanged (0.78 to 1.29); (↑), moderately up-regulated (1.3 and greater); and ↑, strongly up-regulated 2.0 and greater; CIC, ciclopirox olamine; FLC, fluconazole.
No gene expression was detectable without fluconazole.
TABLE 4.
Effects of ciclopirox olamine, fluconazole, and bipyridine on levels of expression of selected genes coding for iron-dependent proteins or proteins of iron metabolism
| Gene | Gene function (reference) | Expression levela
|
||
|---|---|---|---|---|
| CIC | FLC | BIP | ||
| CAT1 | Catalase (60) | ↓, 0.4 (0.065) | (↑), 1.52 (0.005) | 0 |
| FTR1 | High-affinity iron permease (47) | ↑, 2.5 (0.49) | =, 0.93 (0.055) | ↑, 2.23 |
| FTR2 | Low-affinity iron permease (47) | ↓, 0.26 (0.025) | =, 0.89 (0.04) | ↓, 0.27 |
| FTH1 | Putative iron transporter [63% homology to FTH1 of S. cerevisiae] | ↑, 8.64 (0.045) | =, 0.85 (0.02) | ↑, 2.4 |
| FTH2 | Putative iron transporter [30% homology to FTH1 of S. cerevisiae] | =, 1.26 (0.15) | =, 0.99 (0.06) | =, 1.0 |
| FET3 | Multicopper oxidase (18) | =, 1.04 (0.325) | =, 0.9 (0.03) | ↓, 0.16 |
| CCC2 | Copper transporter (58) | ↑, 4.82 (2.03) | =, 0.91 (0.08) | ↓, 0.29 |
| CFL1 | Ferric reductase (27) | ↑, 3.31 (2.0) | =, 0.8 (0.02) | =, 1.08 |
| SIT1 | Siderophore transporter (38) | ↑, 3.54 (0.21) | ND | ND |
The numbers are the averages (standard deviations) of the relative expression levels in cell culture with and without antifungal agents (see Materials and Methods). Expression levels are defined as follows: ↓, strongly down-regulated (0.5 and less); (↓), moderately down-regulated (0.77 and less); =, unchanged (0.78 to 1.29); (↑), moderately up-regulated (1.3 and greater); and ↑, strongly up-regulated (2.0 and greater). ND, not determined; CIC, ciclopirox olamine; FLC, fluconazole; BIP, bipyridine.
TABLE 3.
Levels of expression of selected housekeeping genes, resistance genes, and genes identified after SSH in the presence of ciclopirox olamine and fluconazole
| Gene | Gene function (reference) | Expression levela
|
|
|---|---|---|---|
| CIC | FLC | ||
| ACT1 | Actin (cytoskeleton) (41) | =, 1.08 (0.03) | =, 1.06 (0.065) |
| IMH3 | IMP dehydrogenase (nucleotide synthesis) (35) | =, 1.1 (0.055) | =, 0.81 (0.035) |
| EFB1 | Elongation factor (43) | =, 1.15 (0) | =, 1.02 (0.01) |
| MDR1 | Multidrug resistance transporter (20) | =, 1.1 (0.03) | ↑, 2.08 (0.81) |
| FLU1 | Fluconazole resistance transporter (11) | (↑), 1.83 (0.165) | (↑), 1.78 (0.36) |
| CDR1 | Candida drug resistance transporter (46) | ↑, 2.78 (0.835) | ↑, 2.16 (0.505) |
| CDR2 | Candida drug resistance transporter (52) | (↑), 1.66 (0.01) | =, 1.01 (0.005) |
| CDR3 | Candida drug resistance transporter (2) | =, 1.09 (0.32) | =, 0.91 (0.015) |
| CDR4 | Candida drug resistance transporter (21) | =, 0.99 (0.205) | =, 0.88 (0.045) |
| PCK1 | Phosphoenolpyruvate carboxykinase (39) | (↑), 1.31 (0.235) | ND |
| WHI1 | White colony protein (55) | (↑), 1.48 (0.345) | ND |
| NAM7 | Putative nonsense-mediated mRNA decay protein (73% homology to NAM7 of S. cerevisiae) | (↑), 1.3 (0.01) | ND |
| IPF12201 | Putative Na+ nucleoside cotransporter (41% homology to a Na+ nucleoside cotransporter of M. musculus) | (↑), 1.61 (0.185) | ND |
| IPF10138 | Putative membrane protein (58% homology to YNR018W of S. cerevisiae) | ↑, 2.46 (1.13) | ND |
Numbers are the averages (standard deviations) of the relative expression levels in cell culture with and without antifungal agents. (see Materials and Methods). Expression levels are defined as follows: ↓, strongly down-regulated (0.5 and less); (↓), moderately down-regulated (0.77 and less); =, unchanged (0.78 to 1.29); (↑), moderately up-regulated (1.3 and greater); ↑, strongly up-regulated (2.0 and greater). ND, not determined; CIC, ciclopirox olamine; FLC, fluconazole.
Ciclopirox olamine may act as a chelator of ions. This in turn may influence iron metabolism and thus the levels of expression of genes involved in iron metabolism or genes encoding iron-dependent proteins (Table 4). CAT1, which encodes an iron-containing catalase (60), was strongly down-regulated. Among the genes coding for proteins involved in iron metabolism, the high-affinity iron permease gene FTR1 (47), the siderophore transporter gene SIT1 (38), and FTH1, a homologue of vacuolar membrane iron transporter gene ScFTH1 (57) of Saccharomyces cerevisiae, were strongly up-regulated. In addition, the transcript levels of the ferric reductase gene CFL1 (27) and the copper transporter gene CCC2 (58) (Fig. 1) were strongly increased. In contrast, the low-affinity iron permease gene FTR2 (47) was strongly down-regulated. The transcript levels of the multicopper oxidase gene FET3 (18) and another gene encoding a putative iron transporter as identified by homology, FTH2, showed no changes in levels of expression.
The control experiments with 2 μg of fluconazole/ml showed that the expression patterns of the same genes differed depending on the drug used (Tables 2 to 4). Among the selected set of known or putative virulence genes, we observed both moderately or strongly up-regulated levels of transcription and moderately or strongly down-regulated levels of transcription, while the levels of transcription of the other genes remained unchanged during drug treatment. Up-regulation of the known resistance genes MDR1 and CDR1 was detected in fluconazole-treated cells, as reported previously (12, 42). CDR2 showed no changes in expression levels. The level of expression of the catalase gene CAT1 was moderately increased, while the levels of expression of other genes involved in iron metabolism were unchanged.
As the expression of a number of genes involved in iron metabolism was clearly influenced by treatment with ciclopirox olamine, we investigated whether this was due to an iron-chelating effect of this drug. Therefore, we determined the levels of expression of these genes in cells treated with the known iron chelator bipyridine (Table 4). Similar to the expression patterns observed for iron-starved cells (47), the high-affinity iron permease gene FTR1 was strongly up-regulated, while the low-affinity iron permease gene FTR2 was strongly down-regulated. In addition, high transcript levels of the iron transporter gene FTH1 were detected. The levels of expression of all other genes investigated were either decreased or remained unchanged in the presence of bipyridine.
To further confirm the influence of ciclopirox olamine on the expression of genes involved in iron metabolism, we also investigated the transcript levels of selected genes by Northern analysis (Fig. 5). Similar to the data observed by RT-PCR analysis, we observed high transcript levels of FTR1, FTH1, and CCC2 in ciclopirox olamine-treated cells. Since the gene sequence of low-affinity iron permease gene FTR1 is highly similar to that of FTR2, the same hybridization probe detected the mRNAs of both genes. However, since the same probe detected two different mRNAs with different lengths and different intensities in samples from nontreated and ciclopirox olamine-treated cells, we concluded that the transcript of FTR1 is smaller than that of FTR2.
FIG. 5.
Northern analysis of selected genes involved in iron metabolism. Similar to the data observed by RT-PCR analysis (Fig. 1; Table 4), high transcript levels were detected for FTR1 (lane 1), FTH1 (lane 3), and CCC2 (lane 7) but not FET1 (lane 5) in ciclopirox olamine-treated cells compared with the levels for untreated cells (lanes 2, 4, 6, and 8, respectively). In contrast, FTR2 was up-regulated in untreated cells (lane 2) compared with the regulation in treated cells. mRNA levels were measured relative to the rRNA levels by loading approximately equal amounts of total RNA in each lane of the Northern blots (examples of ethidium bromide-stained membranes for the two samples with the ACT1 gene are shown in lanes 11 and 12). The ACT1 gene was hybridized as an additional internal loading control (lanes 9 and 10). Due to the high degree of similarity of FTR1 and FTR2, transcripts of both genes were detected with the same hybridization probe (lanes 1 and 2). Since this FTR1-FTR2 probe detected two different mRNAs with different lengths and different intensities in samples from nontreated (lane 2) and ciclopirox olamine-treated (lane 1) cells, we concluded that the transcript of FTR1 is smaller than that of FTR2. The lanes are unnumbered but represent lanes 1 to 12, from left to right, respectively.
C. albicans genes up-regulated in ciclopirox olamine-treated cells identified by SSH.
In addition to assessing the levels of expression of a selection of known genes, we also aimed to identify other novel genes whose expression levels may be altered by treatment with ciclopirox olamine. To achieve this, we used an SSH protocol to identify up-regulated genes in ciclopirox olamine-treated cells. Since we assumed that the levels of expression of a number of genes might be affected shortly after exposure to ciclopirox olamine, we used cells that were grown for 5 h without drug and treated them for 1 h with 1.0 μg of ciclopirox olamine/ml. Growth in the presence of the drug was clearly decreased within 1 hour compared to the level of growth in the culture without the drug (data not shown). After subtraction, cloning, and PCR screening, 63 clones containing plasmids were sequenced. Eight different genes, PCK1 (phosphoenolpyruvate carboxykinase), WH11 (white colony protein), NAM7 (a putative nonsense-mediated mRNA decay protein with 73% homology to NAM7 of S. cerevisiae), IPF12201 (putative Na+ nucleoside cotransporter with 41% homology to an Na+ nucleoside cotransporter of Mus musculus), IPF10138 (a putative membrane protein with 58% homology to YNR018W of S. cerevisiae), BPT1 (a putative bile pigment transporter with a high degree of homology to BPT1 of S. cerevisiae), RPS8A (a putative ribosomal protein with 87% homology to RPS8B of S. cerevisiae), and RPL3 (a putative ribosomal protein with 86% homology to RPL3 of S. cerevisiae), were identified (Table 3). Among the 63 clones, PCK1 was found in 8 clones, WH11 and NAM7 were each found in 2 clones, IPF12201 was found in 3 clones, IPF10138 was found in 11 clones, BPT1 was found in 7 clones, and RPS8A and RPL3 were each found in 1 clone. Other clones contained plasmid sequences or sequences that could not be identified within the C. albicans genome.
To prove whether the genes identified by SSH were indeed up-regulated in the presence of subinhibitory concentrations of ciclopirox olamine, RT-PCR analysis was performed with PCK1, WH11, NAM7, IPF12201, and IPF10138. The expression of IPF10138 was strongly up-regulated, while the levels of expression of the other genes were moderately increased (Table 3), thus confirming the SSH data.
Hyphal induction of C. albicans in the presence of subinhibitory concentrations of ciclopirox olamine.
In order to investigate whether ciclopirox olamine influenced the yeast-to-hyphal phase transition, we performed a hyphal phase induction assay. C. albicans cells treated with 0.6 μg of ciclopirox olamine/ml showed the same ability as untreated cells to produce hyphal cells by induction with serum (data not shown).
Hydrogen peroxide susceptibilities of ciclopirox olamine-treated cells.
Our RT-PCR expression studies showed that the level of transcription of the catalase gene CAT1 was strongly down-regulated. This may consequently cause a decrease in catalase activity, which would lead to an insufficient detoxification of H2O2. Therefore, we studied the sensitivities of ciclopirox olamine-treated cells to H2O2 in comparison with those of untreated cells. The numbers of untreated cells surviving the treatment with H2O2 decreased with rising concentrations of H2O2. Addition of ciclopirox olamine further increased the susceptibility to oxidative stress caused by H2O2 (Table 5). Only 1.7% of the cells survived in the ciclopirox olamine-containing cultures with 1 mM H2O2, and no growth was detected with 1.5 mM H2O2. This was in clear contrast to the results for cultures not treated with ciclopirox olamine. In that case, 16.6% of the cells survived in the presence of 1 mM H2O2 and 3.4% of the cells were still viable in the presence of 2.5 mM H2O2.
TABLE 5.
H2O2 resistance of C. albicans SC5314 grown in cultures with and without 0.6 μg of ciclopirox olamine/mla
| H2O2 concn (mM) | No. (%) of surviving cells
|
|
|---|---|---|
| Without ciclopirox olamine | With 0.6 μg of ciclopirox olamine/ml | |
| 0.0 | 670 (100) | 59 (100) |
| 0.5 | 492 (73.4) | 5 (8.5) |
| 1.0 | 111 (16.6) | 1 (1.7) |
| 1.5 | 31 (4.6) | 0 |
| 2.0 | 33 (4.9) | 0 |
| 2.5 | 23 (3.4) | 0 |
The numbers (percentages) of surviving cells after treatment with various concentrations of H2O2 (see Materials and Methods) are shown.
Induction of resistance by long-term treatment with ciclopirox olamine.
In order to investigate whether long-term treatment with ciclopirox olamine may induce antifungal resistance in C. albicans, cells were cultured over a period of 6 months in the presence of a subinhibitory concentration of ciclopirox olamine (0.6 μg/ml). During this long period, no changes in the susceptibilities of the SC5314 cells to the drug could be detected. Since the cells remained susceptible to ciclopirox olamine at 0.6 μg/ml, it was not possible to investigate the effect of increasing the drug concentration. Increasing the ciclopirox olamine concentration to 1.0 μg/ml completely inhibited growth at all time points. In contrast, the concentration of fluconazole could be increased from 8 to 128 μg/ml within 2 months (61 days). While 8 μg/ml was the highest concentration of fluconazole that still allowed growth under the given starting conditions, fluconazole concentrations could be increased to 16, 32, 64, and 128 μg/ml after different periods of incubation.
DISCUSSION
Although ciclopirox olamine is a well-known antifungal drug and has been in use for more than two decades, its mode of action is poorly understood. One way of elucidating the mode of action of a particular drug is to examine the gene expression profiles of cells treated with the drug (3, 13). Microorganisms must adapt to changing environmental conditions by expressing genes necessary for optimal growth or viability. Therefore, the gene expression pattern may reflect a certain environmental condition or the effect of a particular antimicrobial drug. Antimicrobial drugs may act by disturbing essential biological functions of the target cell. However, they may also act directly against virulence attributes which are not important for in vitro cell culture growth. Such activity may at least enhance the antimicrobial potential of a drug in vivo. In order to elucidate the mode of action of ciclopirox olamine, we investigated the expression profiles of selected genes essential for growth and viability and genes encoding known or putative virulence factors in cells exposed to this drug using gene-specific primers in a series of RT-PCRs with different cycle numbers and cDNA templates from two independent experiments. Northern analysis of selected genes involved in iron metabolism confirmed the data obtained by RT-PCR (Fig. 1 and 5). Furthermore, we used SSH to identify known or novel genes up-regulated during treatment with this drug.
The levels of expression of selected housekeeping genes such as those encoding actin (ACT1), inosine 5′-monophosphate dehydrogenase (IMH3), or translation elongation factor 1 (EFB1) did not change when cells were treated with subinhibitory concentrations of ciclopirox olamine. This indicated that essential functions such as the cytoskeleton, protein translation, and nucleotide synthesis were not directly affected by treatment with this drug under the chosen conditions. However, the fact that genes encoding a phosphoenolpyruvate carboxykinase (PCK1), a membrane transporter (BPT1), or ribosomal proteins (RPS8A, RPL3) were up-regulated in the presence of ciclopirox olamine (Table 3) showed that this drug has multiple direct or indirect effects on the cells. It was interesting that within a population of cells treated with ciclopirox olamine, a number of cells seemed to be heavily damaged, while others appeared to be unharmed (Fig. 4A to D). It is therefore likely that the gene expression pattern within unharmed cells may differ from the expression pattern within damaged cells. In most cases, the levels of expression of all genes encoding secretory aspartic proteinases (SAP genes) or lipases (LIP genes) were either unaltered or only moderately down-regulated when cells were treated with ciclopirox olamine. It seems unlikely that the drug treatment directly influenced the expression of these genes; rather, it may reflect a general reduction in vitality, which in turn caused lower levels of expression of these putative virulence genes. Nevertheless, these reduced levels of expression of certain virulence genes may contribute to the antifungal activity of ciclopirox olamine in vivo. Interestingly, the transcript level of PLB3, a putative secretory phospholipase gene, was clearly increased, indicating that at least certain genes are expressed at higher levels in drug-treated cells.
Transcript levels of genes associated with the yeast-to-hyphal phase transition, another virulence attribute of C. albicans, such as TUP1, RBT1, and HYR1, were either unaltered or moderately up-regulated. However, this up-regulation had no influence on cellular morphology since subinhibitory concentrations of ciclopirox olamine did not inhibit germ tube formation induced by serum. However, the possibilities that serum components such as proteins reduced the inhibitory effect of ciclopirox olamine by binding to the drug and that ciclopirox olamine does have an inhibitory effect on germ tube formation in media which do not contain serum cannot be excluded.
Because a number of observations suggested that ciclopirox olamine might act as a chelator of iron ions (1, 36, 37), we studied the expression patterns of several genes involved in iron metabolism. Two genes encoding iron permeases, FTR1 and FTR2, were differentially regulated compared with the regulation in untreated cells (Table 4). While FTR1, which encodes a permease with a high affinity for Fe2+ (47), was strongly up-regulated, FTR2, the gene which codes for a low-affinity Fe2+ permease (47), was strongly down-regulated. This inverse regulation of these permease-encoding genes has been shown to reflect iron-limiting conditions (47), suggesting that cells treated with ciclopirox olamine grow in an environment with insufficient iron concentrations. This view is supported by the fact that another gene encoding a putative iron transporter, FTH1, was also strongly up-regulated. Fth1 is a homologue of the iron transporter ScFth1 from S. cerevisiae, which is located in the vacuolar membrane and which may be responsible for mobilization of intravacuolar stored iron (57). Reduction of Fe3+ to Fe2+ is triggered by ferric reductases and is an essential step for the provision of cells with biologically active iron ions. In C. albicans, the CFL1 gene encodes a cell surface ferric reductase which is associated with iron uptake. Under iron-replete conditions this gene is expressed at basal levels, while under low-iron conditions transcription is up-regulated (27). The observation that CFL1 is strongly up-regulated in ciclopirox olamine-treated cultures is in agreement with the view that this drug causes iron-limiting conditions. In addition to extracellular Fe3+ reduction, C. albicans is also able to utilize various siderophores, which are transported into the cell by siderophore transporters after binding to iron ions (38). The siderophore transporter gene Sit1 is induced during iron deprivation (38), and in our experiments we found that this siderophore transporter gene was strongly up-regulated, which again supports the view that ciclopirox olamine causes iron limitation. If ciclopirox olamine acts as an iron chelator, this drug may also be actively taken up by a transporter of the major facilitator family, like SIT1. This is supported by the observation that ciclopirox olamine can be found intracellularly at 200-fold higher levels (50). Finally, it is known that efficient iron import also depends on proteins involved in copper metabolism, and thus, it was not surprising to discover that the copper transporter gene CCC2 was strongly up-regulated in ciclopirox olamine-treated cells.
If ciclopirox olamine acts as an iron chelator, one would expect that the addition of a known iron chelator to C. albicans cultures would cause a similar effect on growth and gene expression. The addition of bipyridine indeed (Fig. 2C; Table 4) reduced the level of culture growth in a manner to similar to that in which ciclopirox olamine did and led to the strong up-regulation of FTR1 and FTH1 but the down-regulation of FTR2, as predicted for iron-limiting conditions (47). Finally, the inhibitory effect of ciclopirox olamine could be reversed in a dose-dependent manner by the supplementation of media containing 1.0 μg of ciclopirox olamine/ml with iron(III) chloride, adding further evidence that the antifungal affect of ciclopirox olamine is principally caused by iron-chelating activity.
Iron limitation may have multiple effects on the fungal cells. One possible consequence should be the loss of activity of iron-dependent enzymes. In fact, we found that the expression of the catalase gene CAT1 is strongly down-regulated, suggesting that CAT1 regulation depends directly on available iron ions. Furthermore, the lack of active catalases was indicative of the high degree of susceptibility of ciclopirox olamine-treated cells to oxidative stress induced by H2O2. However, we cannot exclude the possibility that the higher degrees of susceptibility of ciclopirox olamine-treated cells may also be due to a synergistic effect of both components.
In addition to the possible lack of detoxification activities, iron limitation may lead to a general loss of energy since a number of enzymes of the respiratory chain in the mitochondria depend on iron ions. This in turn would cause multiple effects on most cellular functions and would explain the variety of effects of ciclopirox olamine.
One may argue that the observed changes in the gene expression pattern may be a general consequence of cell growth inhibition. However, treatment of C. albicans with another antifungal drug, fluconazole, caused expression profiles that clearly differed from those for ciclopirox olamine-treated cells (Tables 2 to 4). For example, genes involved in iron metabolism, such as CCC2, CFL1, and FTH1, were up-regulated in ciclopirox olamine-treated cells but were down-regulated or remained unchanged in fluconazole-treated cells.
Although ciclopirox olamine is one of the most commonly used topical antimycotic drugs, a ciclopirox olamine-resistant strain has not been described to date. This is in sharp contrast to the case for azole drugs, to which increasing numbers of strains are resistant. For azoles such as fluconazole, the mechanisms for the development of resistance are well known and include a number of drug resistance genes (54, 59). In this study, we questioned how ciclopirox olamine may act on these resistance genes and whether resistance can be induced by long-term treatment with this drug. While the levels of expression of MDR1, which encodes a member of the major facilitator family, and ABC transporter genes CDR3 and CDR4 showed no change in the presence of ciclopirox olamine compared to the levels of expression of untreated cells, CDR1, and CDR2 were up-regulated. In addition, the transcript level of FLU1, another member of the major facilitator gene family, was clearly increased. Even though FLU1 encodes a transporter for fluconazole, the expression of this gene in C. albicans clinical isolates could not be related to the degree of azole resistance. Only when FLU1 was expressed in S. cerevisiae did it mediate specific resistance to fluconazole (11). However, it is well known that CDR1 and CDR2 play an important part in resistance to fluconazole (12, 42). Even though the expression of these drug resistance genes was induced by ciclopirox olamine, the susceptibility of C. albicans to ciclopirox olamine did not change, even after 6 months of continual treatment. In contrast, the MIC of fluconazole could be increased up to 16-fold for cells incubated with this drug, starting from 8 μg/ml (the highest concentration which permitted reduced growth) to 128 μg/ml after a 2-month period. One possible reason for the inability of C. albicans to generate tolerance of or resistance to ciclopirox olamine may be its fungicidal mode of action and its steep dose-response curve. Cells still grew in the presence of 0.6 μg of ciclopirox olamine/ml, but an increase in this concentration (to 1.0 μg/ml) completely inhibited growth. On the contrary, fluconazole acts in a fungistatic manner and has a flat dose-response curve (26). The MIC80 of fluconazole was 2 μg/ml in 2% Sabouraud glucose medium, but cells were still able to grow in the presence of a concentration of 8 μg/ml. Another possible reason for the inability of C. albicans to generate resistance to ciclopirox olamine could be due to an irreversible binding of the drug to intracellular structures (50), which would make it an unlikely substrate for efflux pumps such as the MDR or CDR gene products.
In summary, our data strongly indicate that the principal mode of antifungal activity of ciclopirox olamine is due to iron chelation, which restricts the availability of iron to the fungal cell and consequently inhibits growth. Furthermore, up-regulation of certain multiple-drug resistance genes, other resistance mechanisms, or 6 months of exposure to the drug could not generate ciclopirox olamine resistance in C. albicans. Finally, we propose that determination of the gene expression profile induced by an external stimulus, whether the stimulus is a drug or some other stimulus, may provide valuable information regarding the mode of action of this stimulus.
Acknowledgments
This work was supported by the Manfred Plempel-Stipendium to M.N. and by Aventis Pharma, Germany.
We thank Julian Naglik, Guy's Hospital, London, United Kingdom, and Hans-Christian Sigle, Traunstein-Trostberg, Germany, for critical reading of the manuscript. We thank Horst Emmel, Siegfried Pociuli, and Hans-Günter Bredow, Robert Koch-Institut, Germany, for technical help.
REFERENCES
- 1.Abrams, B. B., H. Hänel, and T. Hoehler. 1992. Ciclopirox olamine: a hydroxypyridone antifungal agent. Clin. Dermatol. 9:471-477. [DOI] [PubMed] [Google Scholar]
- 2.Balan, I., A. M. Alarco, and M. Raymond. 1997. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J. Bacteriol. 179:7210-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, D. E., C. E. McCreary, and D. C. Coleman. 1998. Genetic characterization of a phospholipase C gene from Candida albicans: presence of homologous sequences in Candida species other than Candida albicans. Microbiology 144:55-72. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Yaacov, R., S. Knoller, G. A. Caldwell, J. M. Becker, and Y. Koltin. 1994. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob. Agents Chemother. 38:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braga, P. C., S. Maci, M. DalSasso, and M. Bohn. 1996. Experimental evidences for a role of subinhibitory concentrations of rilopirox, nystatin and fluconazole on adherence of Candida spp. to vaginal epithelial cells. Chemotherapy (Basel) 42:259-265. [DOI] [PubMed] [Google Scholar]
- 7.Braga, P. C., G. Piatti, E. Conti, and F. Vignali. 1992. Effects of subinhibitory concentrations of ciclopirox on the adherence of Candida albicans to human buccal and vaginal epithelial cells. Drug Res. 42:1368-1371. [PubMed] [Google Scholar]
- 8.Braun, B. B., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, B. B., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 10.Braun, B. B., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146:2743-2754. [DOI] [PubMed] [Google Scholar]
- 12.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. M. Luyten, and H. Vanden Bosche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diasio, R. B., J. E. Bennett, and C. E. Myers. 1978. Mode of action of 5-fluorocytosine. Biochem. Pharmacol. 27:703.. [DOI] [PubMed] [Google Scholar]
- 15.Diatchenko, L., Y. F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmar, W., W. Grau, W. Raether, E. Schrinner, and W. H. Wagner. 1981. Microbiological laboratory studies with ciclopiroxolamine. Drug Res. 31:1317-1322. [PubMed] [Google Scholar]
- 17.Dittmar, W., and G. Lohaus. 1973. HOE 296, a new antimycotic compound with a broad antimicrobial spectrum. Drug Res. 23:670-674. [PubMed] [Google Scholar]
- 18.Eck, R., S. Hundt, A. Härtl, E. Roemer, and W. Künkel. 1999. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 145:2415-2422. [DOI] [PubMed] [Google Scholar]
- 19.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schäfer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of the parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fling, M. E., J. Kopf, A. Tamarkin, J. A. Gorman, H. A. Smith, and Y. Koltin. 1991. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol. Gen. Genet. 227:318-329. [DOI] [PubMed] [Google Scholar]
- 21.Franz, R., S. Michel, and J. Morschhäuser. 1998. A fourth gene from the Candida albicans CDR family of ABC transporters. Gene 220:91-98. [DOI] [PubMed] [Google Scholar]
- 22.Fu, Y., A. S. Ibrahim, W. Fonzi, X. Zhou, C. F. Ramos, and M. A. Ghannoum. 1997. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology 143:331-340. [DOI] [PubMed] [Google Scholar]
- 23.Gasparini, G., D. Contini, A. Torti, C. Guidarelli, A. Lasagni, and R. Caputo. 1986. The effect of ciclopiroxolamine investigated by means of the freeze-fracture technique. Mykosen 29:539-544. [DOI] [PubMed] [Google Scholar]
- 24.Groll, A. H., S. C. Piscitelli, and T. J. Walsh. 1998. Clinical pharmacology of systemic antifungal agents: a comprehensive review of agents in clinical use, current investigational compounds, and putative targets for antifungal drug development. Adv. Pharmacol. 44:343-500. [DOI] [PubMed] [Google Scholar]
- 25.Gupta, A. K. 2001. Ciclopirox: an overview. Int. J. Dermatol. 40:305-310. [DOI] [PubMed] [Google Scholar]
- 26.Gupta, A. K., D. N. Sauder, and N. H. Shear. 1994. Antifungal agents: an overview. Part II. J. Am. Acad. Dermatol. 30:911-933. [DOI] [PubMed] [Google Scholar]
- 27.Hammacott, J. E., P. H. Williams, and A. Cashmore. 2000. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology 146:869-876. [DOI] [PubMed] [Google Scholar]
- 28.Hoover, C. I., M. J. Jantapour, G. Newport, N. Agabian, and S. J. Fisher. 1998. Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol. Lett. 167:163-169. [DOI] [PubMed] [Google Scholar]
- 29.Hube, B., D. Hess, C. A. Baker, M. Schaller, W. Schäfer, and J. W. Dolan. 2001. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147:879-889. [DOI] [PubMed] [Google Scholar]
- 30.Hube, B., and J. Naglik. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997-2005. [DOI] [PubMed] [Google Scholar]
- 31.Hube, B., F. Stehr, M. Bossenz, A. Mazur, M. Kretschmar, and W. Schäfer. 2000. Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 174:362-374. [DOI] [PubMed] [Google Scholar]
- 31a.Hube, B., M. Monod, D. A. Schofield, A. J. P. Brown, and N. A. R. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 32.Huh, W. K., S. T. Kim, H. Kim, G. Jeon, and S. O. Kang. 2001. Deficiency of d-erythroascorbic acid attenuates hyphal growth and virulence of Candida albicans. Infect. Immun. 69:3939-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata, K., and H. Yamaguchi. 1981. Studies on the mechanism of antifungal action of ciclopiroxolamine. Drug Res. 31:1323-1327. [PubMed] [Google Scholar]
- 34.Karnovsky, M. J. 1965. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27:137A-138A.
- 35.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korting, H. C., and M. Grundmann-Kollmann. 1997. The hydroxypyridones: a class of antimycotics of its own. Mycoses 40:243-247. [DOI] [PubMed] [Google Scholar]
- 37.Kruse, R., W. Hengstenberg, H. Hänel, and W. Raether. 1991. Studies for the elucidation of the mode of action of the antimycotic hydroxypyridone compound, rilopirox. Pharmacology 43:247-255. [DOI] [PubMed] [Google Scholar]
- 38.Lesuisse, E., S. A. B. Knight, J. M. Camadro, and A. Dancis. 2002. Siderophore uptake by Candida albicans: effect of serum treatment and comparison with Saccharomyces cerevisiae. Yeast 19:329-340. [DOI] [PubMed] [Google Scholar]
- 39.Leuker, C. E., A. Sonneborn, S. Delbrück, and J. F. Ernst. 1997. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192:235-240. [DOI] [PubMed] [Google Scholar]
- 40.Lohaus, G., and W. Dittmar. 1981. The chemistry of antimicrobially active 1-hydroxy-2-pyridones. Drug Res. 31:1311-1316. [PubMed] [Google Scholar]
- 41.Losberger, C., and J. F. Ernst. 1989. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 17:9488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maebashi, K., M. Niimi, M. Kudoh, F. J. Fischer, K. Makimura, K. Niimi, R. J. Piper, K. Uchida, M. Arisawa, R. D. Cannon, and H. Yamaguchi. 2001. Mechanisms of fluconazole resistance in Candida albicans isolates from Japanese AIDS patients. J. Antimicrob. Chemother. 47:527-536. [DOI] [PubMed] [Google Scholar]
- 43.Maneu, V., A. M. Cervera, J. P. Martinez, and D. Gozalbo. 1996. Molecular cloning and characterization of a Candida albicans gene (EFB1) coding for the elongation factor EF-1β. FEMS Microbiol. Lett. 145:157-162. [DOI] [PubMed] [Google Scholar]
- 44.Mercer, E. I. 1991. Morpholine antifungals and their mode of action. Biochem. Soc. London Trans. 19:788-793. [DOI] [PubMed] [Google Scholar]
- 45.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 46.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 47.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 288:1062-1064. [DOI] [PubMed] [Google Scholar]
- 48.Reitze, H. K., D. R. Dannhorn, H. Hänel, and K. A. Seitz. 1993. Ultrastructural investigations of Candida albicans after in vitro treatment with the new fungicidal agent rilopirox. Mycoses 36:385-395. [DOI] [PubMed] [Google Scholar]
- 49.Sakurai, K., T. Sakaguchi, H. Yamaguchi, and K. Iwata. 1978. Mode of action of 6-cyclohexyl-4-methyl-2(1H)-pyridone ethanolamine salt (HOE 296). Chemotherapy (Tokyo) 24:68-76. [DOI] [PubMed] [Google Scholar]
- 50.Sakurai, K., T. Sakaguchi, H. Yamaguchi, and K. Iwata. 1978. Studies on uptake of 6-cyclohexyl-1-hydroxy-4-methyl-2(1H)-pyridone ethanolamine salt (HOE 296) by Candida albicans. Chemotherapy (Tokyo) 24:146-153. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 53.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 55.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]
- 56.Sugiyama, Y., S. Nakashima, F. Mirbod, H. Kanoh, Y. Kitajima, M. A. Ghannoum, and Y. Nozawa. 1999. Molecular cloning of a second phospholipase B gene, CaPLB2 from Candida albicans. Med. Mycol. 37:61-67. [PubMed] [Google Scholar]
- 57.Urbanowski, J. L., and R. C. Piper. 1999. The iron transporter Fth1p forms complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274:38061-38070. [DOI] [PubMed] [Google Scholar]
- 58.Weissman, Z., Shemer, R., and D. Kornitzer. 2002. Deletion of the copper transporter CaCCC2 reveals two distinct pathways for iron acquisition in Candida albicans. Mol. Microbiol. 44:1551-1560. [DOI] [PubMed] [Google Scholar]
- 59.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wysong, D. R., L. Christin, A. M. Sugar, P. W. Robbins, and R. D. Diamond. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]