Abstract
From May 1997 to December 2001, a serotype O:6 multidrug-resistant strain of Pseudomonas aeruginosa colonized or infected 201 patients in the University Hospital of Besançon (France). The susceptibility profile of this epidemic clone to fluoroquinolones and aminoglycosides was relatively stable during the outbreak but showed important isolate-to-isolate variations (up to 64-fold) in the MICs of β-lactams. Analysis of 18 genotypically related isolates selected on a quaterly basis demonstrated alterations in the two DNA topoisomerases II and IV (Thr83→Ile in GyrA and Ser87→Leu in ParC) and production of an ANT(2")-I enzyme. Although constitutively overproduced in these bacteria, the MexXY efflux system did not appear to contribute significantly to aminoglycoside resistance. β-Lactam resistance was associated with derepression of intrinsic AmpC β-lactamase (with isolate-to-isolate variations of up to 58-fold) and sporadic deficiency in a 46-kDa protein identified as the carbapenem-selective porin OprD. Of the 18 isolates, 14 were also found to overproduce the efflux system MexAB-OprM as a result of alteration of the repressor protein MexR (His107→Pro). However, complementation experiments with the cloned mexR gene demonstrated that MexAB-OprM contributed only marginally to β-lactam and fluoroquinolone resistance. Of the four isolates exhibiting wild-type MexAB-OprM expression despite the MexR alteration, two appeared to harbor secondary mutations in the mexA-mexR intergenic region and one harbored secondary mutations in the putative ribosome binding site located upstream of the mexAB oprM operon. In conclusion, this study shows that many mechanisms were involved in the multiresistance phenotype of this highly epidemic strain of P. aeruginosa. Our results also demonstrate that the clone sporadically underwent substantial genetic and phenotypic variations during the course of the outbreak, perhaps in relation to local or individual selective drug pressures.
Pseudomonas aeruginosa is a well-known opportunistic pathogen. As such, it has repeatedly been associated with sporadic or clustered cases of infection generally confined to single hospitalization units. In contrast, it has much more rarely been involved in large hospital outbreaks or interhospital spreads (11, 15, 16, 33, 34). The reason why some strains of P. aeruginosa demonstrate higher epidemic potency remains unclear. Propagation of such clones proceeds through complex routes that may involve, for example, the hands of health care personnel, environmental reservoirs, medical equipment, or reagents (reviewed in reference 6). Whether these epidemic clones possess intrinsic features enabling them to spread more efficiently in patient populations than sporadic isolates is unresolved to date. Interestingly, most of them appear to be multidrug resistant, a property which may account for their maintenance in clinical environments under high antibiotic pressure (11, 32, 33). On the other hand, wild-type or moderately resistant epidemic clones may be underestimated simply because they are more difficult to distinguish from all the sporadic strains.
We recently witnessed the propagation of a multiresistant P. aeruginosa epidemic clone in the surgical intensive care unit (ICU) of the Besançon public hospital, France (8). The outbreak was controlled in the ward after implementation of strict hygiene measures but has disseminated inside and outside the hospital since then, affecting more than 200 patients. Examination of the resistance profile of this clone to β-lactam antibiotics showed significant isolate-to-isolate variations, suggesting an adaptation of the strain to local or individual selective conditions. The present study reports some of the genetic and phenotypic variations associated with drug resistance that have been observed in this clone during the 4.5-year outbreak.
MATERIALS AND METHODS
Hospital setting.
The hospital of Besançon (eastern France) is a university-affiliated public institution located at two distinct geographic sites that are 7 km apart. It contains 1,228 acute-care beds distributed among 59 different medical and surgical units, including two adult ICUs of 15 beds each, one surgical and one medical. About 50,000 patients are admitted each year, for a total of 350,000 patient-days of hospitalization.
Data collection.
A surveillance program was instituted from May 1997 to December 2001 in the two adult ICUs of the hospital. During the investigation period, routine clinical specimens and systematic screening samples (rectal and nasal swabs and tracheal aspirates) were collected from all the patients hospitalized in the ICUs and tested for P. aeruginosa by conventional methods of identification. Screening cultures were performed on samples each patient from on the day of admission and then once a week throughout the stay in the ICU. Recognition of an outbreak due to a specific clone of multiresistant P. aeruginosa O:6 in May 1999 (8) led us to extend the survey to all the wards of the hospital. Every serotype O:6 strain isolated during routine diagnosis was then stored for possible further analysis.
Identification of the epidemic clone.
Resistance patterns of all the P. aeruginosa strains isolated in the laboratory were determined by the Kirby-Bauer disk method on Mueller-Hinton agar (Bio-Rad, Ivry sur Seine, France), as recommended by the Antibiogram Committee of the French Society for Microbiology (3). A Sirscan automated image analyzer (I2A, Perols, France) was used to precisely measure inhibition diameters and to compile resistance data (26). In addition to its O:6 serotype determined by slide agglutination with specific antisera (Bio-Rad), the epidemic clone displayed a stable, easily recognizable resistance profile to amikacin (disk load, 30 μg; inhibition zone, 22 ± 2.3 mm [mean ± standard deviation]), gentamicin (15 μg; 6 mm), tobramycin (10 μg; 9 ± 2.0 mm), and ciprofloxacin (5 μg; 8.2 ± 2.5 mm). The accuracy (run-to-run variation) with which the Sirscan apparatus measures inhibition zones is in the millimeter range (26). The genetic relatedness of isolates exhibiting the clone resistance phenotype was systematically investigated by pulsed-field gel electrophoresis (PFGE) (CHEF DRIII apparatus; Bio-Rad) of DraI-macrorestricted genomic DNA (43). According to consensual guidelines (44), an isolate was considered to belong to the epidemic clone when its PFGE profile was identical or closely related (fewer than three different bands) to the profile of the first isolate recovered in May 1997. Eighteen isolates belonging to the epidemic clone were randomly selected on a quaterly basis for further analysis. All these bacteria displayed strictly identical PFGE macrorestriction patterns.
Antibiotic susceptibility tests.
MICs of antimicrobial agents were determined by the macrodilution method in agar (Muller-Hinton medium adjusted to 20 mg of Ca2+ per liter and 10 mg of Mg2+ per liter [BBL, Cockeysville, Md.]) with inocula of 104 bacteria per spot (5). Inoculated plates were incubated for 18 h at 37°C before bacterial growth was evaluated. Resistance mechanisms to aminoglycosides were tentatively deduced from the resistance profiles of the isolates to test antibiotics by using the aminoglycoside resistance test kit provided by the Schering Plough Research Institute. The kit is composed of disks of apramycin, fortimicin, kanamycin, tobramycin, gentamicin, amikacin, isepamicin, netilmicin, neomycin, 5-epi-netilmicin, 2′-netilmicin, and 6′-netilmicin. Inoculation and incubation of Muller-Hinton agar plates were performed as recommended by the manufacturer, and the inhibition diameters were measured accurately with the Sirscan zone reader.
Characterization of β-lactamases.
Bacteria were cultured in 200 ml of Muller-Hinton broth to mid-log phase, collected by centrifugation, resuspended in 20 ml of 10 mM HEPES buffer (pH 7.2), and lysed twice through a French pressure cell (1,500 lb/in2) (Spectronics Instruments, Rochester, N.Y.). The crude lysate was subjected to electric focusing on pH 3.5 to 9.5 Ampholine PAG plate gel (Pharmacia, Piscataway, N.J.) using an LKB Multiphor 2117 apparatus (Pharmacia) and nitrocefin (1 mM in 66 mM Sörensen buffer [pH 7.0] for β-lactamase detection. The TEM-1 (pI 5.4), TEM-2 (pI 5.6), TEM-3 (pI 6.3), TEM-24 (pI 6.5), SHV-1 (pI 7.6), and SHV-4 (pI 7.8) enzymes served as reference markers for gel calibration. In an attempt to identify the secondary β-lactamase (pI 7.7) produced by isolate 97-4, we tried to amplify the structural gene coding for this enzyme by using various sets of consensus primers designed for the characterization of OXA-, SHV- or PSE/CARB-type β-lactamases (4, 7). In contrast to reference strains, isolate 97-4 yielded no amplification (data not shown). β-Lactamase activities were determined spectrophotometrically in the 18 selected isolates as previously described (9). Briefly, the enzymatic reaction was initiated by the addition of an appropriate volume of bacterial lysate to 1 ml of a 50-μg/ml solution of nitrocefin in 0.05 M phosphate buffer (pH 7.0). The shift in the absorbance due to nitrocefin hydrolysis was then recorded for 5 min at 482 nm. Measurements were done in duplicate for each strain. The protein concentration in the lysates was determined by the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.).
PCR amplification and DNA sequencing.
Chromosomal DNA was prepared by the procedure of Chen and Kuo (10). The primers used for amplification or sequencing are listed in Table 1. The gyrA, gyrB, and parE quinolone-determining resistance regions (QRDR) of P. aeruginosa were amplified by PCR as described by Kureishi et al. (20), Mouneimné et al. (28), and Akasaka et al. (2), respectively. Two 21-mer primers, PARC1 and PARC2, designed on the basis of the parC sequence of P. aeruginosa (1), were used to amplify a 209-bp fragment encompassing the QRDR of parC. The mexR coding sequence and the mexA-mexR intergenic region were amplified with primers MexR20 and MexRINT (Table 1). A second pair of primers (P860 and P793) was necessary to sequence mexR. A genomic fragment encompassing the open reading frame of mexZ and the mexX-mexZ intergenic region was amplified with primers MexZ1026 and MexZ2060 and then sequenced with two additional primers, MexZ2 and MexZ2P (Table 1). Amplifications were carried out in 50-μl volumes containing 0.4 U of RedTaq DNA polymerase (Sigma, Saint-Quentin Fallavier, France). The reactions were performed in a DNA thermal cycler (Perkin-Elmer, Norwalk, Conn.) for 35 cycles, with each cycle consisting of 30 s at 94°C for denaturation; 30 s at 57°C (parC), 1 min at 53°C (mexR), or 50 s at 70°C (mexZ) for annealing; and 1 min at 72°C for polymerization. Sequencing of the amplicons was done on both strands with an Applied Biosystems 373A sequencer (Genome Express, Meylan, France).
TABLE 1.
Primers used for DNA amplification
| Primer | Nucleotide sequence (5′ → 3′) | Position (nt)a | Reference |
|---|---|---|---|
| GyrA1 | TTATGCCATGAGCGAGCTGGGCAACGACT | 147-176 in gyrA | 20 |
| GyrA2 | AACCGTTGACCAGCAGGTTGGGAATCTT | 484-512 in gyrA | 20 |
| GyrB1 | GCGCGTGAGATGACCCGCCGT | 1162-1182 in gyrB | 28 |
| GyrB2 | CTGGCGGTAGAAGAAGGTCAG | 1531-1551 in gyrB | 28 |
| PARC1 | ATGAGCGAACTGGGGCTGGAT | 166-187 in parC | This study |
| PARC2 | ATGGCGGCGAAGGACTTGGGA | 354-375 in parC | This study |
| ParE1 | CGGCGTTCGTCTCGGGCGTGGTGAAGGA | 1223-1250 in parE | 2 |
| ParE2 | TCGAGGGCGTAGTAGATGTCCTTGCCGA | 1787-1814 in parE | 2 |
| MexR20 | CCAGTAAGCGGATAC | Downstream of mexR | This study |
| MexRINT | GGATGATGCCGTTCACCTG | 205-223 in mexA | This study |
| P860b | CACATGCTGGAAGACCG | 320-336 in mexR | This study |
| P793b | CATTAGGTTTACTCGGC | 253-269 in mexR | This study |
| MexZ2060 | CCAGCAGGAATAGGCCGACCAGGGC | 43-47 in mexX | This study |
| MexZ1026 | CAGCGTGGAGATCGAAGGCAGCCGG | Upstream of mexZ | This study |
| MexZ2c | GGCTCGTTCTCGTCGCTG | 345-362 in mexZ | This study |
| MexZ2Pc | CAGCGACGAGAACGAGCC | 345-362 in mexZ | This study |
Numbering of nucleotides (nt) is according to deposited sequences of gyrA (L29417 [20]), gyrB (Y16286 [28]), parC (AB003428 [1]), parE (AB003429 [1]), mexA (L11616 [37]), mexR (U23763 [38]), and mexZ and mexX (AF073776 [39]).
Primers used to facilitate mexR and mexR-mexA intergenic region sequencing.
Primers used to facilitate mexZ and mexZ-mexX intergenic region sequencing.
Immunodetection of OprM and MexY.
Membrane proteins were extracted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electrotransferred to a nitrocellulose membrane, and labeled with a MexY antibody (total membrane fraction) or an OprM antibody (outer membrane fraction) as described by Hocquet et al. (18) and Ziha-Zarifi et al. (48), respectively. To ensure that equal amounts of membrane proteins were subjected to SDS-PAGE, a prerequisite before lane-to-lane comparisons, an additional gel was run in parallel under the same conditions and stained with Coomassie blue.
Complementation with the mexR gene.
The mexR gene has previously been cloned on broad-host-range plasmid vector pAK1900, which codes for amoxicillin-ticarcillin resistance, yielding plasmid pAKR4 (48). Plasmids pAK1900 and pAKR4 were purified with the Qiagen Midi kit and transferred by electroporation into selected isolates of P. aeruginosa (41). The resulting transformants were selected on Mueller-Hinton agar containing 250 mg of ticarcillin per liter.
Quantitative RealTime PCR.
A detailed description of the quantitative RealTime PCR procedure will be described elsewhere (J.-L. Delmas, C. Van Delden, and T. Köhler, submitted for publication). Briefly, total RNA was isolated from cultures at an absorbance at 600 nm of 1 using the Qiagen RNeasy protocol. The RNA samples were further treated with DNase (RQ1 DNase; Promega) and purified by phenol-chloroform extraction and ethanol precipitation. Total RNA (1 μg) was reverse transcribed with SuperscriptII reverse transcriptase (Invitrogen) as specified by the supplier. The amount of cDNA was determined in a Rotor Gene RG3000 RealTime PCR machine (Corbett Research, Sydney, Australia), using a SybrGreen Quantitect kit (Qiagen) and rpsL transcripts as an internal control. The results (see Table 2) represent relative expression levels (fold change) for a given target gene in the various clinical isolates compared to the PAO1 wild-type strain.
TABLE 2.
Phenotypic variations of the epidemic clone
| Strain | Date of recovery | MIC (μg/ml) ofa
|
β-Lactamase activityb | Expression of production of
|
Generation timef (min) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | TOB | TIC | PIP | FEP | CAZ | ATM | IMP | CIP | mexAc | oprMc | OprMd | mexXc | mexYc | MexYd | 46-kDa proteine | ||||
| PAO1 | 4 | 2 | 1 | 16 | 32 | 2 | 2 | 4 | 1 | 0.5 | 16 | 1.00 | 1.00 | ± | 1.00 | 1.00 | − | + | 27 | |
| Epidemic clone | ||||||||||||||||||||
| 97-2 | May 1997 | 2 | >512 | 32 | 64 | 128 | 8 | 8 | 16 | 4 | 32 | 10,400 | 1.85 | 1.45 | ++ | 3.70 | 1.95 | + | + | 34 |
| 97-3 | Aug. 1997 | 4 | 256 | 16 | 64 | 128 | 4 | 4 | 8 | 2 | 32 | 3,600 | 1.85 | 3.40 | +++ | 3.70 | 4.00 | + | + | 42.5 |
| 97-4 | Dec. 1997 | 2 | 128 | 16 | >512 | 128 | 64 | 16 | 32 | 8 | 32 | 7,400 | 3.45 | 4.60 | +++ | 4.25 | 5.95 | ++ | + | 37 |
| 98-1 | Feb. 1998 | 4 | 256 | 16 | 128 | 128 | 16 | 8 | 32 | 16 | 32 | 3,000 | 2.60 | 4.35 | +++ | 2.50 | 3.95 | + | − | 35 |
| 98-2 | June 1998 | 4 | 256 | 16 | 128 | 128 | 16 | 8 | 32 | 16 | 32 | 3,000 | 3.35 | 3.30 | +++ | 3.25 | 4.40 | + | − | 42 |
| 98-3 | July 1998 | 4 | 128 | 16 | 128 | 128 | 16 | 16 | 32 | 16 | 32 | 1,900 | 2.85 | 3.00 | +++ | 2.85 | 4.40 | + | − | 40 |
| 98-4 | Nov. 1998 | 4 | 128 | 16 | 64 | 128 | 16 | 8 | 32 | 2 | 32 | 2,000 | 3.35 | 4.40 | +++ | 1.30 | 2.70 | + | + | 36 |
| 99-1 | Mar. 1999 | 4 | 128 | 32 | 64 | 128 | 16 | 8 | 32 | 2 | 32 | 4,400 | 1.95 | 2.30 | +++ | 1.95 | 2.75 | + | + | 37 |
| 99-2 | May 1999 | 4 | 256 | 16 | 64 | 128 | 8 | 4 | 16 | 4 | 32 | 5,600 | 1.65 | 3.50 | ++ | 1.65 | 1.60 | + | + | 39.5 |
| 99-3 | July 1999 | 4 | 256 | 16 | 64 | 128 | 4 | 4 | 16 | 2 | 32 | 6,100 | 1.70 | 1.85 | +++ | 4.10 | 1.95 | + | + | 45 |
| 99-4 | Oct. 1999 | 4 | 256 | 16 | 64 | 128 | 4 | 4 | 16 | 4 | 32 | 5,500 | 2.90 | 9.65 | +++ | 3.35 | 3.60 | ++ | + | 36 |
| 00-1 | Jan. 2000 | 4 | 128 | 16 | 64 | 128 | 8 | 8 | 16 | 2 | 32 | 1,800 | 1.60 | 2.15 | + | 3.10 | 2.75 | + | + | 32 |
| 00-2 | Apr. 2000 | 4 | 256 | 32 | <8 | 64 | 1 | 1 | 4 | 2 | 16 | 530 | 0.50 | 0.60 | ++ | 3.45 | 3.65 | + | + | 34 |
| 00-3 | Aug. 2000 | 4 | 512 | 32 | 16 | 32 | 2 | 1 | 2 | 4 | 16 | 180 | 0.75 | 0.95 | ± | 3.80 | 4.05 | + | + | 32.5 |
| 00-4 | Nov. 2000 | 4 | 512 | 32 | 32 | 256 | 8 | 4 | 16 | 4 | 16 | 4,100 | 0.50 | 0.70 | + | 0.85 | 1.00 | ++ | + | 35 |
| 01-1 | Feb. 2000 | 8 | 256 | 16 | 16 | 128 | 4 | 4 | 8 | 4 | 8 | 1,500 | 0.80 | 1.15 | + | 2.90 | 2.80 | ++ | + | 25 |
| 01-2 | June 2001 | 4 | 512 | 32 | 64 | 128 | 16 | 8 | 32 | 8 | 32 | 6,200 | 2.15 | 3.10 | + | 4.70 | 2.45 | + | ± | 28 |
| 01-3 | Aug. 2001 | 2 | 256 | 16 | 64 | 128 | 16 | 8 | 16 | 16 | 32 | 4,000 | 3.10 | 7.75 | +++ | 3.70 | 5.00 | + | − | 39 |
| Mode | 4 | 256 | 16 | 64 | 128 | 16 | 8 | 16 | 4 | 32 | ||||||||||
AMK, amikacin; GEN, gentamicin; TOB, tobramycin; TIC, ticarcillin; PIP, piperacillin; FEP, cefepime; CAZ, ceftazidime; ATM, aztreonam; IMP, imipenem; CIP, ciprofloxacin.
Cephalosporinase activities are expressed in nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. Values are means of two determinations.
mRNA quantification by reverse transcriptase PCR (see Materials and Methods). Values are means of two determinations. Numerical values are expressed relative to PAO1, which is set at 1.00.
As determined by Western blot analysis with an OprM or MexY antiserum. Semiquantitative values are expressed as a range fro − (negative) or ± (weakly positive) to +++ (strongly positive).
putative OprD, as determined by SDS-PAGE of outer membrane preparations stained with Coomassie blue.
Values are means of four determinations calculated at the logarithmic phase by regression analysis from different cultures (see Materials and Methods).
Growth rate parameters.
A bacterial suspension adjusted to 2 × 108 CFU/ml in Muller-Hinton broth was used in 100-μl volumes to inoculate the wells (four per isolate) of a Nunclon Microwell plate (Nunc, Naperville, Ill.). The microplate was incubated at 37°C for 5 h with constant agitation (200 rpm) and scanned at 30-min intervals in a Titertek Multiscan MCC-340 apparatus (Flow Laboratories SA, Puteaux, France) set at 540 nm. The growth rate at the logarithmic phase was estimated with the nonlinear regression model X = X0 · eμt, where t is the time (in hours), X is the biomass at t, X0 is the biomass at t = 0, and μ is the growth rate (h−1). The generation time (θ, in minutes) was calculated from the formula θ = (ln 2 × 60)/μ.
Statistical analysis.
Using nonparametric analysis-of-variance statistics, we tested the null hypothesis that there was no difference in growth time among the 18 isolates. Then we examined the temporal evolution of the clone (Spearman's nonparametric correlation test), using the average growth time of each isolate (x -axis) and the time elapsed since the isolation of the first isolate, 97-2 (y -axis).
RESULTS
Hospital dissemination of the epidemic clone.
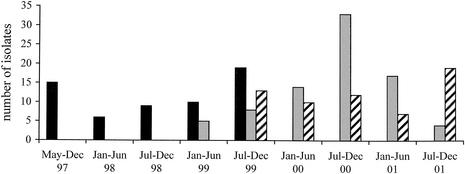
During the survey period, 201 patients admitted to the hospital in Besançon were colonized and/or infected by an epidemic clone of P. aeruginosa serotype O:6. The outbreak was initially detected in the surgical ICU in May 1999 (8). However, a retrospective study showed that the epidemic strain was already present in the institution in May 1997. The surgical ICU (from May 1997 to November 1999) and the medical ICU (from May 1999 to June 2001) were the successive epicenters of the outbreak, which finally reached 27 other wards. Figure 1 represents the distribution of patients colonized or infected by the strain according to the type of hospitalization unit. PFGE analysis of the isolates (one per patient) confirmed the clonal origin of the strain (data not shown).
FIG. 1.
Distribution of the epidemic clone during the study period according to the type of hospitalization unit. One isolate was counted per infected or colonized patient. Solid bars represent the isolates recovered from the surgical ICU, shaded bars represent those recovered from the medical ICU, and hatched bars represent those recovered from other wards.
Susceptibility to antibiotics and β-lactamase production.
A study of 18 isolates selected on a quarterly basis demonstrated that the clone had a rather stable susceptibility profile to aminoglycosides and fluoroquinolones during the 4.5-year outbreak but underwent significant variations in its resistance to β-lactams (Table 2). No trend toward higher resistance was observed with time. While remaining rather susceptible to amikacin, most of the isolates exhibited moderate to high resistance to gentamicin, tobramycin, ticarcillin, piperacillin, cefepime, ceftazidime, aztreonam, imipenem, and ciprofloxacin. As shown in Table 2, the greatest isolate-to-isolate MIC variations were those of ticarcillin and cefepime (up to 64-fold). Comparison of all 201 isolates by the diffusion method in agar also highlighted notable variations in the inhibition zones around disks of β-lactams (Fig. 2).
FIG. 2.
Variations in the drug susceptibility of the epidemic clone. The histogram represents mean inhibition diameters (in millimeters) determined by the disk diffusion method for 201 isolates (one per patient). Error bars indicate standard deviations. Abbreviations of antibiotics are the same as in Table 2.
A single β-lactamase (pI ≥ 8) corresponding to chromosomally encoded enzyme AmpC was detected in all 18 selected isolates (Table 2). Its activity differed greatly from one bacterium to another (from 11 to 650 times that of wild-type strain PAO1), without having any apparent correlation with the MICs of broad-spectrum cephalosporins such as ceftazidime and cefepime. Interestingly, a secondary β-lactamase with pI 7.7 was also found in one isolate (97-4) that was highly resistant to ticarcillin and cefepime (see Materials and Methods). Although no plasmid could be detected in isolate 97-4, the presence of this additional β-lactamase demonstrated sporadic acquisition of exogenous resistance determinants by the clone.
Porin OprD.
Outer membrane proteins of the 18 isolates were extracted and analyzed by SDS-PAGE. Comparison of the resulting electrophoregrams with the banding profile of susceptible reference strain PAO1 revealed the absence of a 46-kDa protein, consistent with porin OprD (formerly D2), in four clinical isolates resistant to imipenem (98-1, 98-2, 98-3, and 01-3; MIC = 16 μg/ml) (Table 2). In addition, the intensity of the putative OprD band was notably diminished in another isolate (01-2) with intermediate susceptibility to imipenem.
Overproduction of MexAB-OprM.
The contribution of the MexAB-OprM active efflux system to the resistance of epidemic P. aeruginosa was investigated by comparing the amounts of OprM, the outer membrane component of the export system, in the isolates and in the reference strain PAO1. Western blot analysis showed that most of the selected isolates overproduced OprM, except for isolate 00-3, which was comparable to wild-type PAO1 (Table 2). Of note, some epidemic isolates such as 00-1, 00-4, 01-1, and 01-2 appeared to produce less OprM than did others. Although the oprM gene belongs to the mexAB oprM operon, its expression appears to be influenced specifically by the activity of a second promoter located within mexB (47). To confirm the overexpression of the whole efflux operon mexAB oprM in the epidemic clone, we determined the transcription levels of mexA and oprM by quantitative real-time PCR. As shown in Table 2, production of OprM was not strictly correlated with transcription of mexA and oprM. With the exception of isolate 00-2, all the bacteria displaying relatively large amounts of OprM in their outer membranes (++ or +++ in Table 2) appeared to also overexpress mexA (1.65 to 3.45 times more) and oprM (1.45 to 9.65 times more) compared with strain PAO1. On the other hand, bacteria (except 00-1 and 01-2) producing wild-type or moderate amounts of OprM (± or +, respectively, in Table 2) were found to express mexA and oprM to a similar or lesser degree with respect to PAO1. Taken together, these data suggested that while most of the epidemic isolates were MexAB-OprM-overproducing mutants (12 of 18), reverse mutants had emerged during the outbreak (at least 3 of 18).
Mutations in the repressor gene mexR.
To identify the mutational events responsible for MexAB-OprM upregulation, we amplified and sequenced mexR, the repressor gene of the mexAB oprM operon (38), as well as the intergenic region between mexA and mexR, in four OprM-overproducing isolates (97-2, 98-4, 00-1, and 01-3) and in four other isolates (00-2, 00-3, 00-4, and 01-1). All the bacteria exhibited the same mexR nucleotide sequence, which differed from that of PAO1 (38) by one silent mutation at position 171 (C→T) and by a base substitution at position 323 (A→C), converting a histidine to a proline at position 107 in the MexR protein. To further determine whether this latter mutation contributed to multidrug resistance in the epidemic clone, isolates 97-2, 98-4, 00-1, and 01-3 were complemented with an intact mexR gene cloned on broad-host-range vector pAK1900 (construct pAKR4). The overexpression of mexR from plasmid pAKR4 resulted in a twofold decrease in the MICs of aztreonam and ciprofloxacin (data not shown), two antibiotics exported by the MexAB-OprM efflux system (25). Conversely, plasmid pAKR4 did not alter the resistance of wild-type strain PAO1, a result that supports the notion that the four clinical isolates tested were nalB mutants defective in MexR (His107→Pro).
Interestingly, isolates 00-2 and 00-3, which expressed mexA and oprM weakly, both harbored a 3-bp deletion (- - - AA instead of TAAAA) in the mexA-mexR intergenic region at a site where the repressor MexR binds to the mexR and mexA overlapping promoters (12). Another isolate with low mexA and oprM transcription, 00-4, displayed an A→C base substitution in the putative Shine-Dalgarno sequence (CGGA instead of AGGA) located 8 nucleotides upstream of the mexA start codon. No mutation could be detected in the mexA-mexR intergenic region of isolate 01-1.
Resistance to aminoglycosides.
Susceptibility profiles of the isolates to a panel of 10 test aminoglycosides were consistent with the presence of an aminoglycoside-2"-O-nucleotidyltransferase [ANT(2")-I] enzyme in the epidemic clone. In agreement with these results, bacteria were susceptible to apramycin and fortimicin, two antibiotics unaffected by ANT(2")-I. However, because of the role of the MexXY-OprM efflux pump in aminoglycoside resistance (39, 46), we investigated the expression of this pump in the 18 isolates. Immunoblot experiments clearly revealed overexpression of protein MexY in all the clinical isolates compared to PAO1 (like MexX [24], MexY was barely detectable in the reference strain) (Table 2). These data were concordant with the transcription levels of mexX and mexY determined by quantitative real-time PCR, except for isolate 00-4, which expressed mexX and mexY weakly and overproduced protein MexY quite strongly (Table 2). To assess whether the overproduction of MexXY resulted from a mutation in the putative repressor gene mexZ (39), the open reading frames of mexZ and the mexZ-mexX intergenic region were sequenced in the four above-mentioned isolates 97-2, 98-4, 00-1, and 01-3. All the mexZ sequences were identical and differed from that reported by Ramos Aires et al. (39) by two silent mutations at positions 236 (C→T) and 341 (A→G) and by an 8-bp deletion from nucleotide 255 to 362, introducing a frameshift in the coding sequence of the gene. No mutation was detected in the mexZ-mexX intergenic region of any of the sequenced isolates.
Resistance to quinolones.
The QRDR of the gyrA, gyrB, parC, and parE genes were amplified and sequenced in isolates 97-2, 98-4, 00-1, and 01-3 to identify the mechanisms responsible for the high resistance of the clone to fluoroquinolones (Table 2). Compared to the gyrA sequence of strain PAO1 (20), all bacteria contained an ACC-to-ATC mutation in gyrA codon 83, leading to a Thr→Ile substitution in the A subunit of DNA gyrase. In addition, the four isolates exhibited a mutation in codon 87 (TCG→TTG) of the QRDR of parC, resulting in a Ser→Leu substitution in the C subunit of topoisomerase IV. gyrB and parE did not display sequence changes responsible for amino acid changes in the QRDR of the GyrB and ParE subunits, respectively (28).
Given that when overexpressed efflux systems MexCD-OprJ and MexEF-OprN may also cause significant quinolone resistance in P. aeruginosa (19, 36), we assessed the transcription of mexC and mexE by quantitative real-time PCR. The 18 epidemic isolates actually expressed both genes very poorly, at levels comparable to that of wild-type PAO1 and far below those of nfxB and nfxC efflux mutants (data not shown).
Bacterial growth.
To assess the energetic cost of drug resistance in the epidemic clone, we compared the generation times of the 18 selected isolates. As shown in Table 2, important isolate-to-isolate variations (ranging from 25 to 45 min, with a mean of 36.1 min) were observed. Overall, the clone did not increase its growth rate during the 4.5-year outbreak (P = 0.09). No particular impact of active efflux or β-lactamase production on bacterial growth rate was noted (compare isolates 99-4, 00-3, and 00-4, which exhibited similar generation times [Table 2]).
DISCUSSION
This study highlights some of the phenotypic and genetic variations undergone by a P. aeruginosa serotype O:6 epidemic clone during a large outbreak which affected 201 patients from May 1997 to December 2001. This clone was initially recognized because of its particular susceptibility profile to aminoglycosides [conferred by an ANT (2")-I enzyme] and fluoroquinolones (caused by mutations in the QRDR of gyrA and parC) and because of its elevated resistance to many β-lactams. Regardless of the emergence of sporadic resistance to imipenem (correlated with decreased expression of an OprD-like protein), notable differences were observed in the β-lactam susceptibilities of epidemic bacteria.
Of the 18 isolates studied, 14 (78%) exhibited the initial multidrug resistance phenotype. These isolates, which were recovered throughout the outbreak (97-2, 97-3, 98-1, 98-2, 98-3, 98-4, 99-1, 99-2, 99-3, 99-4, 00-1, 00-4, 01-2, and 01-3 [Table 2]), displayed all the clone attributes in terms of resistance, except for variations in susceptibility to imipenem. It is well known that mutants selectively resistant to carbapenems emerge frequently in P. aeruginosa during antibiotic chemotherapy (13). Pai et al. (31) found that the resistance of clinical strains to imipenem was determined mostly by the expression levels of the porin OprD, the specific uptake pathway for carbapenems in P. aeruginosa (45). In this study, the MICs of imipenem were also correlated with the production of a 46-kDa outer membrane protein (probably OprD), regardless of AmpC or MexAB-OprM expression.
The contribution of the MexAB-OprM efflux system to β-lactam resistance in the epidemic bacteria appeared marginal compared to the effects of overexpressed β-lactamase. As indicated in Table 2, MICs of β-lactam antibiotics were not overridingly determined by the expression levels of MexAB-OprM. For example, resistance to ticarcillin and aztreonam, two antibiotics which are rather stable to hydrolysis by the AmpC enzyme (22) but are efficiently extruded by MexAB-OprM (21, 25), was not reduced (isolates 00-1 and 01-2) or was only slightly reduced (isolate 00-4) in bacteria expressing lower levels of the pump than in strong MexAB-OprM producers (e.g., isolates 98-2 and 98-4). In agreement with this observation, repression of the mexAB-oprM operon in several isolates with a plasmid-borne mexR gene had only a modest effect on the MICs of aztreonam (a twofold decrease). These results confirm other findings suggesting that P. aeruginosa mutants expressing high, constitutive levels of β-lactamase AmpC are not significantly more resistant to β-lactams when overproducing MexAB-OprM (29).
Regarding fluoroquinolone resistance, the contribution of MexAB-OprM again appeared limited in the epidemic clone since variations in the expression of the efflux system had little influence on the elevated MICs of ciprofloxacin determined by mutations in gyrA and parC (Table 2). Indeed, complementation of selected isolates with the cloned mexR gene lowered resistance to ciprofloxacin only twofold. Our results thus tend to contradict recent data suggesting synergistic interplays between efflux mechanisms and drug target alterations in P. aeruginosa (23). Also, the QRDR mutations (GyrA, Thr83→ Ile; ParC, Ser87→Leu) identified in the epidemic clone are rather common among fluoroquinolone-resistant clinical strains (2, 28, 30).
A new mutation leading to increased expression of mexAB oprM was characterized in this study. As reported previously, inactivation of the MexR repressor may result from frameshift mutations or single-base substitutions in the mexR gene (31, 40, 42, 48) or outside mexR (42). The mutation found here (His107→Pro) is expected to compromise MexR repressor activity since it disrupts a predicted α-helix of 18 amino acid residues located in the C-terminal half of the protein (14). The results of complementation experiments with the wild-type mexR allele support this assumption (see above). For the first time, to our knowledge, it was observed that MexAB-OprM expression may revert to wild-type levels in efflux mutants with altered mexR as a result of mutations in the promoter sequences of mexA and mexR (isolates 00-2 and 00-3) (12) or in the putative ribosome binding site of operon mexAB oprM (isolate 00-4). These compensatory mutations might help bacteria to diminish the energetic burden associated with increased efflux when resistance to antibiotics is not crucial for survival. As indicated in Table 2, the epidemic isolates tended to grow more slowly than did reference strain PAO1.
Another aspect of isolate-to-isolate variations concerned the production of AmpC β-lactamase, which in some cases was considerably reduced compared with that of the majority of bacteria, probably as a result of secondary mutations (isolate 00-3). In addition, some bacteria (isolate 00-2) turned out to be much more susceptible to β-lactams (ticarcillin, ceftazidime, and cefepime) despite their overproduction of both AmpC and MexAB-OprM. A similar phenotype was observed in an Enterobacter aerogenes clinical strain derepressed for AmpC and was attributed to increased permeability of the porin uptake pathway (27). However, the use of the hydrophilic probe tetramethyl-p-phenylenediamine (35) failed to demonstrate stronger outer membrane permeability in isolate 00-2 than in the 17 other isolates and PAO1 (data not shown). In contrast to isolates becoming more susceptible, our study also identified epidemic bacteria acquiring new resistance determinants (β-lactamase with pI of 7.7) by horizontal transfer.
Amikacin is not a substrate for ANT(2")-I enzymes but is a recognized substrate for the multidrug transporter MexXY-OprM (25, 39). It is noteworthy that despite overexpression of MexXY, the epidemic bacteria exhibited wild-type levels of susceptibility to this antibiotic. This tends to support the previous proposal that when upregulated following inactivation of the putative repressor MexZ, this efflux system by itself is insufficient to promote the development of significant aminoglycoside resistance in P. aeruginosa (24, 46).
In the absence of a recognized source of contamination (8), the reasons for the wide dissemination of the clone in our hospital still remain unclear. Recently, it was shown that the MexAB-OprM efflux system plays an important role in the invasiveness of P. aeruginosa (17). The authors suggested the pump might deliver virulence factors to host cells. If this hypothesis is correct, overexpression of MexAB-OprM might be essential for epidemic bacteria to colonize and invade hospitalized patients. MexAB-OprM would contribute to the epidemic success of the clone more as a virulence mechanism than as a resistance mechanism. In connection with this, it is interesting that Dubois et al. (11) described an OprM-overproducing strain of P. aeruginosa involved in a large hospital outbreak.
In conclusion, our study shows that epidemic P. aeruginosa strains may sporadically acquire or lose resistance determinants during the course of an outbreak. We are currently investigating whether such genetic and phenotypic variations result from local or individual antibiotic selective pressures.
Acknowledgments
We thank Bruno Challier from the Departement d'Information Médicale for help in statistical analysis and Christelle Vogne and Catherine Llanes-Barakat for providing primers for DNA amplification. We are also grateful to Isabelle Patry and Sandra Rochey for their excellent technical assistance.
REFERENCES
- 1.Akasaka, T., Y. Onodera, M. Tanaka, and K. Sato. 1999. Cloning, expression, and enzymatic characterization of Pseudomonas aeruginosa topoisomerase IV. Antimicrob. Agents Chemother. 43:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akasaka, T., M. Tanaka, A. Yamaguchi, and K. Sato. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antibiogram Committee of the French Society for Microbiology. 1996. Technical recommendations for in vitro susceptibility testing. Clin. Microbiol. Infect. 2(Suppl. 1):S11-S25. [PubMed] [Google Scholar]
- 4.Babini, G. S., and D. M. Livermore. 2000. Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob. Agents Chemother. 44:2230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balows, A., W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. 1991. Manual of clinical microbiology, 5th ed. ASM Press, Washington, D.C.
- 6.Beck-Sagué, C. M., S. N. Banerjee, and W. R. Jarvis. 1994. Epidemiology and control of Pseudomonas aeruginosa in U.S. hospitals, p. 51-71. In A. L. Baltch and R. P. Smith (ed.), Pseudomonas aeruginosa: infections and treatment. Marcel Dekker, Inc., New York, N.Y.
- 7.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand, X., P. Bailly, G. Blasco, P. Balvay, A. Boillot, and D. Talon. 2000. Large outbreak in a surgical intensive care unit of colonization or infection with Pseudomonas aeruginosa that overexpressed an active efflux pump. Clin. Infect. Dis. 31:E9-E14. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, L. E., and A. J. Godfrey. 1991. β-Lactam antibiotics: mode of action and bacterial resistance, p. 599-664. In V. Lorian (ed.), Antibiotics in laboratory medicine, 3rd ed. The Williams & Wilkins Co., Baltimore, Md.
- 10.Chen, W. P., and T. T. Kuo. 1993. A simple and rapid method for the preparation of Gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, V., C. Arpin, M. Melon, B. Melon, C. André, C. Frigo, and C. Quentin. 2001. Nosocomial outbreak due to a multiresistant strain of Pseudomonas aeruginosa P12: efficacy of cefepime-amikacin therapy and analysis of β-lactam resistance. J. Clin. Microbiol. 39:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish, D. N., S. C. Piscitelli, and L. H. Danziger. 1995. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy 15:279-291. [PubMed] [Google Scholar]
- 14.Frishman, D., and P. Argos. 1997. 75% accuracy in protein secondary structure prediction. Proteins 27:329-335. [DOI] [PubMed] [Google Scholar]
- 15.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M.-F. I. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaiIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grattard, F., O. G. Gaudin, B. Pozzetto, A. Ros, and A. D. Mbida. 1993. Genotypic homogeneity of nosocomial Pseudomonas aeruginosa O12 strains demonstrated by analysis of protein profiles, DNA fingerprints and rRNA gene restriction patterns. Eur. J. Clin. Microbiol. Infect. Dis. 12:57-61. [DOI] [PubMed] [Google Scholar]
- 17.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. W. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plésiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler, T., M. Michéa Hamzehpour, U. Henze, N. Gotoh, L. Kocjancic Curty, and J. C. Pechère. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 20.Kureishi, A., J. M. Diver, B. Beckthold, T. Schollaardt, and L. E. Bryan. 1994. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 38:1944-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X. Z., D. Ma, D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob. Agents Chemother. 38:1742-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livermore, D. M., and Y. Y. Yang. 1987. β-Lactamase lability and inducer power of newer β-lactam antibiotics in relation to their activity against β-lactamase-inducibility mutants of Pseudomonas aeruginosa. J. Infect. Dis. 155:775-782. [DOI] [PubMed] [Google Scholar]
- 23.Lomovskaya, O., A. Lee, K. Hoshino, H. Ishida, A. Mistry, M. S. Warren, E. Boyer, S. Chamberland, and V. J. Lee. 1999. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1340-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medeiros, A. A., and J. Crellin. 2000. Evaluation of the Sirscan automated zone reader in a clinical microbiology laboratory. J. Clin. Microbiol. 38:1688-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellencamp, M. A., J. S. Roccaforte, L. C. Preheim, C. C. Sanders, C. A. Anene, and M. J. Bittner. 1990. Isolation of Enterobacter aerogenes susceptible to beta-lactam antibiotics despite high level beta-lactamase production. Eur. J. Clin. Microbiol. Infect. Dis. 9:827-830. [DOI] [PubMed] [Google Scholar]
- 28.Mouneimné, H., J. Robert, V. Jarlier, and E. Cambau. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakae, T., A. Nakajima, T. Ono, K. Saito, and H. Yoneyama. 1999. Resistance to β-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob. Agents Chemother. 43:1301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano, M., T. Deguchi, T. Kawamura, M. Yasuda, M. Kimura, Y. Okano, and Y. Kawada. 1997. Mutations in the gyrA and parC genes in fluoroquinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2289-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai, H., J. W. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panzig, B., G. Schröder, F.-A. Pitten, and M. Gründling. 1999. A large outbreak of multiresistant Pseudomonas aeruginosa strains in north-eastern Germany. J. Antimicrob. Chemother. 43:415-418. [DOI] [PubMed] [Google Scholar]
- 33.Pellegrino, F. L., L. M. Teixeira, M. G. Siqueira Carvalho, S. Aranha Nouér, M. Pinto de Oliveira, J. L. Mello Sampaio, A. D'Avila Freitas, A. L. P. Ferreira, E. L. T. Amorim, L. W. Riley, and B. M. Moreira. 2002. Occurrence of a multidrug-resistant Pseudomonas aeruginosa clone in different hospitals in Rio de Janeira, Brazil. J. Clin. Microbiol. 40:2420-2424. [DOI] [PMC free article] [PubMed]
- 34.Pitt, T. L., D. M. Livermore, D. Pitcher, A. C. Vatopoulos, and N. J. Legakis. 1989. Multiresistant serotype O12 Pseudomonas aeruginosa: evidence for a common strain in Europe. Epidemiol. Infect. 103:565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plésiat, P., J. Ramos Aires, C. Godard, and T. Köhler. 1997. Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 179:7004-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, K., N. Gotoh, H. Tsujimoto, Q. X. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. I. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 37.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K., K. Tetro, Q. X. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 41.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talon, D., V. Cailleaux, M. Thouverez, and Y. Michel-Briand. 1996. Discriminatory power and usefullness of pulsed-field gel electrophoresis in epidemiological studies of Pseudomonas aeruginosa. J. Hosp. Infect. 32:135-145. [DOI] [PubMed] [Google Scholar]
- 44.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrobiol. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, Q., X. Z. Li, R. Srikumar, and K. Poole. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob. Agents Chemother. 42:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziha-Zarifi, I., C. Llanes, T. Köhler, J. C. Pechère, and P. Plésiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]