Abstract
Brassinosteroids (BRs) are steroidal plant hormones essential for normal plant growth and development. Mutants in the biosynthesis or perception of BRs are usually dwarf. The tomato Dwarf gene (D), which was predicted to encode a cytochrome P450 enzyme (P450) with homology to other P450s involved in BR biosynthesis, was cloned previously. Here, we show that DWARF catalyses the C-6 oxidation of 6-deoxocastasterone (6-deoxoCS) to castasterone (CS), the immediate precursor of brassinolide. To do this, we first confirmed that the D cDNA complemented the mutant light- and dark-grown phenotypes of the extreme dwarf (dx) allele of tomato. To identify a substrate for the DWARF enzyme, exogenous application of BR intermediates to dx plants was carried out. C-6 oxoBR intermediates enhanced hypocotyl elongation whereas the C-6 deoxoBR, 6-deoxoCS, had little effect. Quantitative analysis of endogenous BR levels in tomato showed mainly the presence of 6-deoxoBRs. Furthermore, dx plants were found to lack CS and had a high level of 6-deoxoCS in comparison to D plants that had CS and a lower level of 6-deoxoCS. Confirmation that DWARF catalyzed the C-6 oxidation of 6-deoxoCS to CS was obtained by functional expression of DWARF in yeast. In these experiments, the intermediate 6α-hydroxycastasterone was identified, indicating that DWARF catalyzes two steps in BR biosynthesis. These data show that DWARF is involved in the C-6 oxidation in BR biosynthesis.
Plant dwarfism is associated with many genetic defects, and those that are involved in the biosynthesis and perception of plant hormones represent one of the largest groups. Dwarf mutants that have defective hormone biosynthesis usually can be restored to a normal phenotype by the exogenous application of a hormone intermediate downstream of the blocked enzyme reaction. The prime example of this has been the study of the maize dwarf mutants in the biosynthesis of gibberellins (1) and, more recently, intermediates of the biosynthetic pathway leading to the production of brassinolide (BL), the most bioactive brassinosteroid (BR), have been used to rescue the phenotype of many dwarf mutants (2–5).
BL originally was isolated from rape pollen on the basis of its growth promoting activity (6) and was shown to be a steroidal lactone (7). The biosynthetic pathways for BL production have been established by feeding labeled compounds of possible BR intermediates to cultured cells of Catharanthus roseus and analyzing the metabolites by gas chromatography–mass spectrometry (GC-MS) (reviewed in refs. 8 and 9). These experiments revealed that both early and late oxidation of the C-6 carbon atom occurs, allowing two parallel pathways for the production of BL, namely the early and late C-6 oxidation pathways. Analysis of endogenous BR content of Arabidopsis suggests that both late and early C-6 oxidation pathways are occurring (10) whereas, in tomato, the late C-6 oxidation pathway may predominate, as shown by the detection of the relatively high level of 6-deoxocastasterone (6-deoxoCS) (11).
Arabidopsis BR biosynthesis mutants have a distinctive dwarf phenotype with dark green rugose leaves. When grown in the dark, they have reduced hypocotyl elongation and have a deetiolated phenotype (2–4), suggesting that BL plays an important role in both skotomorphogenesis and photomorphogenesis, the respective dark and light developmental pathways. DEETIOLATED2 was found to encode a steroid 5α-reductase catalyzing a conversion in the early stages of BR biosynthesis (2, 12, 13). CONSTITUTIVE PHOTOMORPHOGENESIS and DWARFISM (3) and DWARF4 (4) are the only known BR biosynthesis genes that are predicted to encode cytochrome P450 enzymes (P450s or CYPs). Through BR intermediate feeding experiments, DWARF4 is thought to encode a C-22α-hydroxylase, and CONSTITUTIVE PHOTOMORPHOGENESIS and DWARFISM is thought to encode a C-23α-hydroxylase (CYP90B and CYP90A, respectively), although functional expression of these genes has not been shown.
The tomato Dwarf gene (D), isolated by transposon tagging, was found to have homology to P450s, especially CYP90A (14) and CYP90B (4), and was classified as CYP85 (14). Mutant alleles of D, especially the strong extreme dwarf (dx) allele, share phenotypic similarities with the Arabidopsis BR mutants. Furthermore, dx plants are not recovered to a normal phenotype by the exogenous application of gibberellins (15). These data suggested that DWARF/CYP85 could be an enzyme in BR biosynthesis (14). To show the function of DWARF, like any plant P450, is difficult (recently reviewed in ref. 16). This difficulty arises because functional expression of P450s in heterologous hosts is not easy and matching a substrate to the enzyme can require detailed chemical analysis of material from lines with defective P450 activity.
To show the role of DWARF in BR biosynthesis, we first initiated rescue experiments of dx plants by using intermediates from the BR biosynthetic pathway. This suggested that dx plants lacked C-6 oxidation of 6-deoxoCS. The biochemical lesion in dx plants was determined by quantitative analysis of endogenous BRs of material from D, dx, and a complementing line overexpressing D (35S∷D). The dx plants lacked castasterone (CS) but had increased levels of 6-deoxoCS, suggesting that DWARF acts at this C-6 oxidation step. This was confirmed by functional expression of DWARF in yeast. This enzyme, therefore, provides the crucial link between 6-deoxoCS and CS in the latter part of the BR biosynthetic pathway.
MATERIALS AND METHODS
Plant Material and Growth.
Tomato stocks used were near isogenic lines of Ailsa Craig carrying either D (GCR 358) or dx (GCR 567) obtained from J. Maxon Smith (PPG, Practical Plant Genetics, Littlehampton, United Kingdom). 35S∷D plants were generated by transforming GCR 567 with T-DNA construct pGB1421 by using an Agrobacterium-mediated transformation protocol as described (17, 18). For BR analysis, plants were grown in cabinets (25°C, 16 h/day with 50 μmol⋅m−2⋅s−1 light intensity at soil level).
Plasmids.
A detailed description of the plasmid construction is available on request. The strategy used to make 35S∷D T-DNA plasmid pGB1421 included PCR amplification of D cDNA with primers to introduce two restriction sites, an NcoI site at the initiation codon (DW22, TGAGGTGCATCCATGGCCTTCTTC) and a BglII site at the 3′ end (DW23, TGCATCCTACAGATCTCTTCTC). PCR fragments were cut with NcoI and BglII and were ligated to NcoI- and BamHI-cut plasmid pSLJ4D4 (19) to generate pGB140-7. pGB140-7 was cut with SstI and XbaI, the gene fusion was cloned into pSLJ6562, a nos∷NPT binary vector derived from pSLJ3922 (19), cut with SstI and XbaI, and the resulting plasmid was designated pGB1421. The GAL∷D plasmid pGB233A1 was made by amplifying D cDNA by PCR using primers that introduced a BamHI site at the initiation codon (DW43, TGAGGTGGATCCATGGCCTTCTTCTTA) and an EcoRI site at the 3′ end (DW44, CATTCATGAATGAATTCTTAGTGAGCTGAAAC). PCR products were cut with BamHI and EcoRI and were ligated to BamHI- and EcoRI-cut pYeDP60 plasmid (20, 21) to generate pGB233A1. For both plasmids, D sequences were checked to ensure that no PCR errors were introduced.
BR Rescue Experiments.
On agar media: Seed was surface sterilized in 10% domestic bleach for 30 min, was rinsed and plated on MS medium with 1% glucose, and was supplemented with appropriate concentrations of BR dissolved in ethanol. Growth was maintained at 25°C, 16 h/day with 20 μmol⋅m−2⋅s−1 light intensity or in continuous dark, and seedlings were photographed after 8 days.
In liquid culture: One-week-old seedlings grown in 16-h days on MS medium with 1% glucose were removed and were placed in 9-cm Petri dishes containing 20 ml of liquid MS medium with appropriate concentrations of BRs. Gentle rotary shaking maintained continuous contact of the seedlings with the BR solution, at a constant 22°C with low level light. Hypocotyl length was measured after 4 days and was compared with control hypocotyl length.
BR Analysis of Plant Material.
BR analysis of tomato was carried out as described (11), but with the following modifications. Plants were harvested after 2 months’ growth. All plant material from 60 dx (401 g), 30 D (535 g), and 22 35S∷D (350 g) plants (except for fruit and flowers) down to the eighth leaf below the apex was harvested in small pieces in methanol. Methanol extracts were spiked with 100 ng each of 2H6-labeled internal standards [2H6]BL, [2H6]CS, [2H6]typhasterol, [2H6]3-dehydroteasterone, [2H6]teasterone, [2H6]cathasterone, [2H6]6-deoxotyphasterol, [2H6]3-dehydro-6-deoxoteasterone, and [2H6]6-deoxoteasterone [2H6]6-deoxoCT and 500 ng of [2H6]6-deoxoCS before reduction to an aqueous residue. The aqueous residue was partitioned between ethyl acetate and 0.5 M dipotassium phosphate. Biologically active fractions from silica gel purification columns were subjected to Sephadex LH-20 column chromatography using methanol:chloroform (4:1) as a mobile phase. Fractions 34–38 were combined and subjected to reversed phase HPLC. Detailed description of fractionation and GC–selected ion monitoring quantification is available on request but is essentially the same as described (11).
RNA Gel Blot Analysis.
Young leaf and stem material from plants at the same stage of development as used in BR analysis was frozen in liquid nitrogen. Extraction of RNA by using Trizol (GIBCO/BRL) was carried out following the manufacturer’s recommendations. Approximately 20 μg of total RNA was electrophoresed and transferred to Hybond N+ (Amersham Pharmacia) filter. Hybridization of the membrane with an antisense riboprobe of D sequences was carried out by using the method described in ref. 22. The final wash of the membrane was in an aqueous solution containing 30 mM sodium chloride, 3 mM sodium citrate, and 1% sodium dodecyl sulfate at 65°C. Hybridization signals were visualized after a 48-h exposure to an imaging plate and were analyzed by using a Fuji Bas station. Control hybridization of pea 18S ribosomal RNA was carried out in a similar manner by using a radiolabeled DNA probe, and detection was after a 15-min exposure to an imaging plate.
Yeast Functional Assay.
pGB233A1 and pYeDP6o were transformed into yeast strains WAT11 and WAT21 (23) by the lithium acetate procedure described by Gietz et al. (24). Newly streaked transformed colonies were used to inoculate 35 ml of YPG media (20 g/liter glucose/10 g/liter yeast extract/10 g/liter bactopeptone) and were grown at 28°C for 2 days. Cells were pelleted and resuspended in 200 ml of inducing media, YPL (20 g/l galactose/10 g/l yeast extract/10 g/l bactopeptone) for 12 h at 28°C. Induced cells were diluted in YPL to obtain an OD550 of 0.5 and 20 ml and were incubated for 6 h at 28°C with 5 μg each of [2H6]6-deoxoCS, [2H6]6α-hydroxycastasterone (6-OHCS), and [2H6]campestanol (CN). For the incubations with [2H6]6-deoxoCS and [2H6]6-OHCS, the mixtures were extracted with ethyl acetate and then were purified with a Sep-Pak Vac silica column (Waters), which was eluted with chloroform, 2% methanol in chloroform, and 10% methanol in chloroform. The last fraction was purified with a Sep-Pak PLUS C18 column (Waters). The eluate with methanol was subjected to HPLC on a 150- × 4.6-mm Senshu Pak ODS-1151-D column (Senshu Scientific, Tokyo) by using 45% acetonitrile at flow rate of 1.0 ml/min. CS and 6-OHCS were collected after retention times of 10–11 min and 8–9 min, respectively, before GC-MS analysis, as described in ref. 12. For analysis of the incubation with [2H6]CN, the mixture was spiked with 3 μg each of unlabeled CN, 6-OHCN, and 6-oxoCN before partitioning. Neutral ethyl acetate fraction obtained was chromatographed on a short C-300 silica gel column (Wako Biochemicals, Osaka). Successive elution with alcohol-free chloroform and 3% methanol in chloroform gave CN and 6-OHCN/6-oxoCN fractions, respectively. These fractions were trimethylsilylated and analyzed by GC-MS under the same conditions as in ref. 11.
RESULTS
Genetic Complementation of dx.
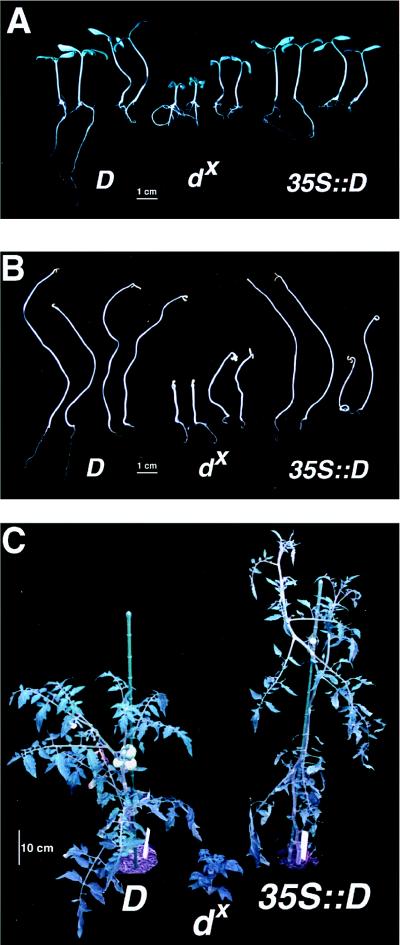
Initially, we confirmed that the D cDNA could complement the dx phenotype. This was done by Agrobacterium-mediated transformation of dx material by using a T-DNA construct pGB1421 that has D cDNA under the control of the cauliflower mosaic virus 35S promoter, i.e., 35S∷D. Five independent transformed lines showed cosegregation of the complemented phenotype with kanamycin resistance, indicating the presence of the D-containing T-DNA (data not shown). For this study, we focused on line GB1421JJ, a single locus 35S∷D complementing line. Fig. 1 A and B shows complementation of the light- and dark-grown seedling phenotypes, respectively, and Fig. 1C shows the complementation after 2 months’ growth.
Figure 1.
Growth responses of the wild type (D), the dx mutant, and the 35S∷D line (a dx-complementing line) under light/dark conditions in the presence or absence of BL. (A) Group of four light-grown seedlings (16 h/day) of D, dx, and 35S∷D. In each group, the two on the left were grown on media only and the two on the right were grown on media containing 10−6 M brassinolide. (B) Same as A, but dark-grown. (C) Representative phenotypes of 2-month-old light-grown (16 h/day) plants of each line before BR analysis.
The dark-grown phenotype of dx seedlings is similar to that reported for the Arabidopsis mutants involved in BL biosynthesis, i.e., short hypocotyl, lack of apical hook, and expansion of cotyledons (3, 25, 26). When BL was added to the media, both light- and dark-grown dx plants showed partial restoration to a normal phenotype (Fig. 1 A and B). The genetic complementation of dx, however, restored the normal light- and dark-grown phenotypes fully. Of interest, BL caused root inhibition of all lines tested and did not increase the elongation of dark-grown D plants. Unlike Arabidopsis mutants in BR biosynthesis (20, 25), dx plants do not have reduced apical dominance.
The average height of six representative individuals from dx, D, and 35S∷D lines (Fig. 2A) indicates the effective complementation of the dx phenotype using the 35S∷D construct. Although the 35S∷D line is larger than the D line, more independent 35S∷D transformants need to be analyzed in detail to see whether this is a general phenomenon. Gel blot analysis (Fig. 2B) carried out on RNA extracted from dx, D, and 35S∷D plant material showed that the D transcript was not detected from dx plants and was overexpressed in 35S∷D plants. This is consistent with dx having a null phenotype for D, which aids the interpretation of the BR analysis and complementation experiments.
Figure 2.
(A) Average height of plants (in millimeters) before harvesting for chemical analysis. (B) RNA gel blot analysis. Shown are D transcript levels in D, dx, and 35S∷D plant material. No hybridization of D sequences to a D transcript was detected in dx mRNA, suggesting this as a null allele. Low and high transcript accumulation was seen in the D and 35S∷D lines, respectively. 18S RNA hybridization is shown as RNA loading control.
Effect of BR Intermediates on the dx Phenotype.
To identify the metabolic block in BR biosynthesis, phenotypic restoration of the dwarf phenotype was sought by placing dwarf seedlings in continuous contact with solutions containing various BRs. Under these conditions, dose–response curves were made for the various intermediates in BR biosynthesis (Fig. 3). Increased responsiveness of dx hypocotyls compared with D hypocotyls to BL is shown in Fig. 3A as a greater percentage elongation of dx hypocotyls compared with D hypocotyls. This increased responsiveness was used to identify BRs that had a strong effect on dx hypocotyls. This assay showed that most C-6 oxidized BRs tested elicited a strong response (Fig. 3A) and that the C-6 oxidized intermediates earlier in the biosynthetic pathway produced a less dramatic response because of their lower biological activities (9). Fig. 3B shows that 6-deoxoCS elicited little response on the dx hypocotyl length whereas CS and BL elicited a greater response. This suggested that dx plants lack C-6 oxidation converting 6-deoxoCS to CS.
Figure 3.
(A) Dose–response curves showing percentage increase in hypocotyl length of dx (solid line) and D plants (dashed line) for various BRs. Each point is mean ± SEM; n = 6. CT, cathasterone; TE, teasterone; 3DT, 3-dehydroteasterone; TY, typhasterol. (B) Dose–response curves showing increase of hypocotyl length of dx and D plants for 6-deoxoCS (solid line), CS (small dashed line) and BL (long dashed line). Each point is mean ± SEM; n = 4–7.
Quantitative Analysis of Tomato BRs.
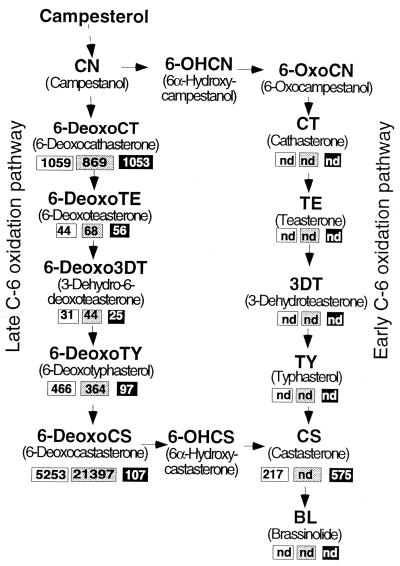
To further characterize the step in BR biosynthesis blocked in the dx mutants, chemical analysis by using GC-MS was performed on dx, D, and 35S∷D plant material from 2-month-old plants, as shown in Fig. 1C. Analysis of the content of steroid intermediates from steps preceding the committed conversion of campesterol into the BR biosynthetic pathway did not show any large differences between the three samples (data not shown). The first striking observation of the endogenous BR levels in the lines analyzed is that tomato lacks intermediates of the early C-6 oxidation pathway (Fig. 4). More importantly, this analysis showed that dx plants lacked CS and had increased amounts of a precursor 6-deoxoCS when compared with D plants, suggesting a role for D in the C-6 oxidation of 6-deoxoCS to CS (Fig. 4). The 35S∷D plants had reduced amounts of 6-deoxoCS and increased amounts of CS, further supporting a role for D in the C-6 oxidation of 6-deoxoCS to CS. 28-norcastasterone, a C-6-oxidized BR lacking C-28, was found to be abundant in D and 35S∷D lines (234 and 558 ng/kg fresh weight, respectively) but present at a very low level in dx (1 ng/kg fresh weight). The reduced level of this C-6-oxidized BR in dx plants provides further evidence suggesting that D is involved in C-6 oxidation of BRs.
Figure 4.
Brassinolide biosynthetic pathway and brassinosteroid content of tomato lines. Brassinosteroid amounts, in nanograms per kilogram fresh weight, of D, dx, and 35S∷D plants are shown in white, gray, and black boxes, respectively. nd, not detected, i.e., the BR concentration was below the detection limit.
Functional Expression of DWARF in Yeast.
To confirm that DWARF catalyzes C-6 oxidation of 6-deoxoCS, we carried out functional expression in yeast. D cDNA was cloned into plasmid pYeDP6o, which allows galactose-inducible expression of plant P450s in yeast (20). This plasmid, designated pGB233A1, was transformed into yeast strains WAT11 and WAT21. These yeast strains are engineered to overexpress the Arabidopsis NADPH P450 reductases in the presence of galactose (20, 23) and therefore provide an optimal environment for plant P450 activity. Induced cultures of the yeast transformants were used to metabolize ≈5 μg of deuterated [2H6]BRs. Products from this incubation were analyzed by GC-MS.
Incubation of the putative substrates for DWARF, including [2H6]CN, [2H6]6-deoxoCS, and [2H6]6-OHCS, was carried out with induced pGB233A1 transformants. Table 1 indicates the results of BR analysis from a pGB233A1 transformant incubated with [2H6]6-deoxoCS. [2H6]CS (major product) and [2H6]6-OHCS (minor product) were identified as metabolites of [2H6]6-deoxoCS by GC-MS. When the pGB233A1 yeast strains were incubated with [2H6]6-OHCS, only limited conversion to [2H6]CS was detected. No conversion of [2H6]CN was detected in both pGB233A1 transformants and vector-transformed yeast. Similarly, no conversion of [2H6]6-deoxoCS to [2H6]6-OHCS or [2H6]CS in transformants with the vector only was seen.
Table 1.
GC-MS identification of [2H6]6-OHCS and [2H6]CS converted from [2H6]6-deoxocastasterone in transformed yeast cultures
| Compound | Retention time on GC, min | Characteristic ions, m/z (relative intensity percentage) |
|---|---|---|
| [2H6]6-OHCS* | ||
| Standard | 11.43 | 592 [M+] (1), 577 (15), 502 (50), 443 (19), 271 (75), 161 (100) |
| Metabolite | 11.42 | 592 [M+] (1), 577 (14), 502 (47), 443 (19), 271 (71), 161 (100) |
| [2H6]CS† | ||
| Standard | 11.82 | 518 [M+] (26), 441 (2), 399 (8), 358 (10), 287 (27), 161 (100) |
| Metabolite | 11.82 | 518 [M+] (28), 441 (2), 399 (7), 358 (10), 287 (25), 161 (100) |
M+, molecular ion; m/z, mass–charge ratio.
Bismethaneboronate-trimethylsilyl derivative.
Bismethaneboronate derivative.
DISCUSSION
Here, we have shown that the tomato dx mutant is defective in BR biosynthesis because of a block in C-6 oxidation of BRs. Similar to the Arabidopsis BR biosynthesis mutants, dx plants have reduced hypocotyl elongation in both light and dark conditions. Genetic complementation experiments using the 35S∷D construct effectively restored both light and dark grown dx phenotypes (Fig. 1). Partial restoration of dx phenotype to the wild type was seen by exogenous application of BRs to tomato roots or in shaking culture. The lack of complete phenotypic recovery may be attributable to poor BR transport in tomato. This was suggested previously to account for the formation of dwarf and wild-type sectors in transposon-tagged mutants of D in which somatic excision of the transposon led to sectors of restored D function (14). Conceivably, the poor restoration of dx material by BRs also could be attributable to the increased levels of 6-deoxoCS in the dx plants acting as competitive inhibitors to the BR receptor and thus could prevent or reduce the signaling for the downstream responses. The dark-grown 35s∷D seedlings in the presence of BL had a reduced etiolation response compared with wild-type seedlings grown on the same concentration of BL. This can be attributed to an inhibitory effect of high concentrations of active hormone, and it suggests that the 35S∷D plants have exceeded a threshold level of active hormone concentration that results in inhibition of growth.
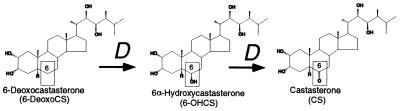
The endogenous levels of BR intermediates determined in this study contribute substantially to the limited information of BR levels in tomato. This comprehensive analysis suggests that, in tomato, the late C-6 oxidation pathway may be the major pathway leading to active BR production. The combination of this information and the fact that, in rice lamina inclination assays, the 6-deoxoBR-to-6-oxoBR conversion confers the largest increase in BR biological activity implies that DWARF plays a critical role in the latter stages of active BR biosynthesis. In fact, the 6-deoxoCS to CS conversion catalyzed by DWARF (Fig. 5) increases the bioactivity of the BR by 200× (27), and the further conversion to BL only would increase this activity by 7-fold. This is the largest increase in bioactivity known to be catalyzed by a single enzyme in the BR biosynthetic pathway, which could make it a key step in regulation of active BR production. Furthermore, CS has been suggested to have biological activity (discussed in ref. 9). If CS is an active BR and BL is absent in tomato, then DWARF may act in the final activation step in BR biosynthesis. Whether CS has activity could be clarified if a mutant in the CS to BL conversion is found.
Figure 5.
Two-step oxidation of 6-deoxocastasterone catalyzed by DWARF (D).
Overexpression of D does not increase the amount of all 6-oxoBRs in tomato, suggesting that DWARF does not catalyze the C-6 oxidation of all BR intermediates and has higher specificity for 6-deoxoCS. Of interest, the 50-fold decrease in 6-deoxoCS in the 35S∷D line compared with D translates to only a 2.5-fold increase in CS. This hints that, in the D overexpresser, the deactivation pathway of BRs could be induced to reduce the overproduction of active hormone, which in tomato cell cultures has been reported to occur via both cytochrome P450-dependent and -independent routes (28).
Unlike tomato, Arabidopsis has both early and late C-6 oxidation, as both 6-oxoBR and 6-deoxoBR intermediates have been detected (10). This suggests that, in Arabidopsis, either the homologue of DWARF will be able to catalyze the C-6 oxidation of both CN and 6-deoxoCS or that an additional enzyme exists that can catalyze the early C-6 oxidation. It has been suggested that the early C-6 oxidation is predominant in dark-grown Arabidopsis seedlings and that the late C-6 oxidation pathway occurs mainly in light-grown plants (4, 12). If this is true, it would suggest that the expression of D and the Arabidopsis homologue could be light-regulated and could provide a crucial link between light-regulated development and BR production.
Well documented techniques are available for heterologous expression of P450s in many systems, including yeast (23, 29), vaccinia virus (30), baculovirus (31), bacteria (32), and COS cells (21). The first example of functional expression of a P450 involved in mammalian sex hormone production was 17α-hydroxylase of steroids in COS cells (33). The enzymatic activity of plant P450s involved in sterol production has been shown in yeast (34) and bacteria (35). However, until now, none of the activities of the plant P450s involved in gibberellins or BR hormone biosynthesis has been confirmed by a functional assay. Here, we have shown a successful enzyme assay of DWARF by using yeast, which confirmed the role of DWARF in catalyzing the conversion of 6-deoxoCS to CS (Fig. 5 and Table 1). The lack of metabolism of [2H6]CN to [2H6]6-oxoCN in yeast provides further evidence that DWARF does not oxidize the C-6 position of all BRs. However, such a lack of [2H6]CN metabolism and the reduced oxidation of the [2H6]6-OHCS intermediate to [2H6]CS could be attributable to problems of transport of these substrates into yeast cells or access to the active site.
DWARF can be considered as a multifunctional P450 catalyzing two reactions (Fig. 5). However, 6-OHCS may not be a genuine free intermediate leaving the active site but a transient, yet stable, intermediate. Such consecutive reactions forming stable intermediates catalyzed by a single P450 in the biosynthesis of steroids is not uncommon (36). Examples include the the 14α-demethylation of lanosterol, which occurs by sequential oxidation of the methyl group via stable alcohol and aldehyde moieties (37); the cholesterol side chain cleavage P450 (P450scc), which catalyzes C-20-C-22 bond cleavage and which occurs in three steps, with the successive intermediates having increased binding to the P450 (38); and aromatase, which catalyzes two carbon hydroxylations and a carbon–carbon bond cleavage similar to P450scc (39).
As discussed in ref. 16, assigning function to a given P450 sequence is difficult. A mutant phenotype of the disrupted P450 gene may suggest a functional role for the gene in question; however, detailed chemical analysis is required to confirm the enzyme activity of the P450. Here, we have shown the function of DWARF by synergistic use of genetics, molecular biology, and chemistry. Now that we know that mutants of D are defective in BR biosynthesis, we should be able to compare BR action in Arabidopsis and tomato in such a way that manipulation of BR levels can be used for crop improvement.
Acknowledgments
We thank M. Schalk, F. Durst, and D. Werck-Reichhart for showing us how to carry out experiments and for helpful comments on P450 expression. We thank P. Urban and D. Pompon for pYEPD60 and yeast strains WAT11 and WAT21, T. Kaneta for photography, and D. Jones for critical comments. We also thank P. Bovil, D. Baker, and Y. Tachiyama for sequencing work and J. Maxon-Smith for tomato seed. This project initially was funded by the Biotechnology and Biological Sciences Research Council Grant 83/P01834, and, currently, G.B. is a Science and Technology Agency/Royal Society research fellow.
ABBREVIATIONS
- BL
brassinolide
- BR
brassinosteroid
- GC
gas chromatography
- MS
mass spectrometry
- 6-deoxoCS
6-deoxocastasterone
- D
tomato Dwarf gene
- dx
extreme dwarf allele
- CS
castasterone
- 6-OHCS
6α-hydroxycastasterone
- CN
campestanol
References
- 1.Phinney B O, Spray C. In: Plant Growth Substances. Wareing P F, editor. London: Academic; 1982. pp. 101–110. [Google Scholar]
- 2.Li J, Nagpal P, Vitart V, McMorris T C, Chory J. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 3.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei G P, Nagy F, Schell J, Koncz C. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 4.Choe S, Dilkes B P, Fujioka S, Takatsuto S, Sakurai A, Feldmann K A. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomura T, Nakayama M, Reid J B, Takeuchi Y, Yokota T. Plant Physiol. 1997;113:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell J W, Mandava N, Worley J F, Plimmer J R, Smith M V. Nature (London) 1970;225:1065–1066. doi: 10.1038/2251065a0. [DOI] [PubMed] [Google Scholar]
- 7.Grove M D, Spencer G F, Rohwedder W K, Mandava N, Worley J F, Warthen J D, Steffens G L, Flippen-Anderson J L, Cook J C. Nature (London) 1979;281:216–217. [Google Scholar]
- 8.Fujioka S, Sakurai A. Physiol Plant. 1997;100:710–715. [Google Scholar]
- 9.Yokota T. Trends Plant Sci. 1997;2:137–143. [Google Scholar]
- 10.Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S. Phytochemistry. 1998;48:595–599. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- 11.Yokota T, Nomura T, Nakayama M. Plant Cell Physiol. 1997;38:1291–1294. [Google Scholar]
- 12.Fujioka S, Li J, Choi Y-H, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Biswas M G, Chao A, Russell D W, Chory J. Proc Natl Acad Sci USA. 1997;94:3554–3559. doi: 10.1073/pnas.94.8.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop G J, Harrison K, Jones J D G. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadhzimov U K, Jupe S C, Jones M G, Scott I M. Physiol Plant. 1988;73:252–256. [Google Scholar]
- 16.Chapple C. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- 17.Fillatti J J, Kiser J, Rose R, Comai L. Bio/Technology. 1987;5:726–730. [Google Scholar]
- 18.Horsch R B, Fry J E, Hoffmann N L, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 19.Jones J D G, Shlumukov L, Carland F, English J, Scofield S R, Bishop G J, Harrison K. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 20.Urban P, Werck-Reichhart D, Teutsch H G, Durst F, Regnier S, Kazmaier M, Pompon D. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- 21.Clark B J, Waterman M R. Methods Enzymol. 1991;206:100–108. doi: 10.1016/0076-6879(91)06081-d. [DOI] [PubMed] [Google Scholar]
- 22.Frances S, White M J, Edgerton M D, Jones A M, Elliot R C, Thompson W F. Plant Cell. 1992;4:1519–1530. doi: 10.1105/tpc.4.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pompon D, Louerat B, Bronine A, Urban P. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 24.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chory J, Nagpal P, Peto C A. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azpiroz R, Wu Y, LoCascio J C, Feldmann K A. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujioka S, Noguchi T, Takatsuto S, Yoshida S. Phytochemistry. 1998;49:1841–1847. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- 28.Winter J, Schneider B, Strack D, Adam G. Phytochemistry. 1997;45:233–237. [Google Scholar]
- 29.Guengerich F P, Brian W R, Sari M-A, Ross J T. Methods Enzymol. 1991;206:130–145. doi: 10.1016/0076-6879(91)06085-h. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez F J, Aoyama T, Gelboin H V. Methods Enzymol. 1991;206:85–92. doi: 10.1016/0076-6879(91)06079-i. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez F J, Kimura S, Tamura S, Gelboin H V. Methods Enzymol. 1991;206:93–99. doi: 10.1016/0076-6879(91)06080-m. [DOI] [PubMed] [Google Scholar]
- 32.Porter T D, Larson J R. Methods Enzymol. 1991;206:108–116. doi: 10.1016/0076-6879(91)06082-e. [DOI] [PubMed] [Google Scholar]
- 33.Zuber M X, Simpson E R, Waterman M R. Science. 1986;234:1258–1261. doi: 10.1126/science.3535074. [DOI] [PubMed] [Google Scholar]
- 34.Cabello-Hurtado F, Zimmerlin A, Rahier A, Taton M, DeRose R, Nedelkina S, Batard Y, Durst F, Palett K E, Werck-Reichhart D. Biochem Biophys Res Commun. 1997;230:381–385. doi: 10.1006/bbrc.1996.5873. [DOI] [PubMed] [Google Scholar]
- 35.Bak S, Kahn R A, Olsen C E, Halkier B A. Plant J. 1997;11:191–201. doi: 10.1046/j.1365-313x.1997.11020191.x. [DOI] [PubMed] [Google Scholar]
- 36.Oritz de Montellano P R. In: Cytochrome P450 Structure, Mechanism, and Biochemistry. Oritz de Montellano P R, editor. New York: Plenum; 1995. pp. 279–286. [Google Scholar]
- 37.Shafiee A, Trzaskos J M, Paik Y-K, Gaylor J L. J Lipid Res. 1986;27:1–10. [PubMed] [Google Scholar]
- 38.Lambeth J D, Kitchen S E, Farooqui A A, Tuckey R, Kamin H. J Biol Chem. 1982;257:1876–1884. [PubMed] [Google Scholar]
- 39.Fishman J. Cancer Res. 1982;42:3277s–3280s. [PubMed] [Google Scholar]