Abstract
To acquire iron, many plant species reduce soil Fe(III) to Fe(II) by Fe(III)-chelate reductases embedded in the plasma membrane of root epidermal cells. The reduced product is then taken up by Fe(II) transporter proteins. These activities are induced under Fe deficiency. We describe here the FRO1 gene from pea (Pisum sativum), which encodes an Fe(III)-chelate reductase. Consistent with this proposed role, FRO1 shows similarity to other oxidoreductase proteins, and expression of FRO1 in yeast conferred increased Fe(III)-chelate reductase activity. Furthermore, FRO1 mRNA levels in plants correlated with Fe(III)-chelate reductase activity. Sites of FRO1 expression in roots, leaves, and nodules were determined. FRO1 mRNA was detected throughout the root, but was most abundant in the outer epidermal cells. Expression was detected in mesophyll cells in leaves. In root nodules, mRNA was detected in the infection zone and nitrogen-fixing region. These results indicate that FRO1 acts in root Fe uptake and they suggest a role in Fe distribution throughout the plant. Characterization of FRO1 has also provided new insights into the regulation of Fe uptake. FRO1 expression and reductase activity was detected only in Fe-deficient roots of Sparkle, whereas both were constitutive in brz and dgl, two mutants with incorrectly regulated Fe accumulation. In contrast, FRO1 expression was responsive to Fe status in shoots of all three plant lines. These results indicate differential regulation of FRO1 in roots and shoots, and improper FRO1 regulation in response to a shoot-derived signal of iron status in the roots of the brz and dgl mutants.
Iron is required for many functions in plants, including heme and chlorophyll biosynthesis, photosynthesis, and as a component of Fe-S cluster containing enzymes (Marschner, 1995). Iron is also vital for the establishment and function of symbiotic root nodules of legumes involved in nitrogen fixation (Udvardi and Day, 1997). Although abundant in the environment, iron is often a limiting nutrient for plant growth due to the low solubility of the oxidized form of Fe, Fe(III), at near neutral soil pH. Thus, plants have evolved with efficient mechanisms of Fe acquisition that are directed at solubilizing Fe. Strategy II plants, which includes all of the grasses, release Fe(III)-binding compounds called phytosiderophores into the surrounding soil that bind iron and are then taken up into the roots (Marschner, 1995). Strategy I plants (dicots and non-Graminaceous monocots) obtain Fe from the rhizosphere by first reducing Fe(III) to Fe(II) through the action of membrane-bound Fe(III)-chelate reductases. Iron reduction is then followed by uptake of Fe(II) into root cells by metal ion transporters. Reductase and transporter activities are inducible in roots under Fe deficiency. Furthermore, the roots of strategy I plants release more protons when Fe deficient, thereby lowering the rhizosphere pH and increasing Fe solubility.
Many of the components of the Strategy I Fe acquisition system have recently been identified in Arabidopsis. The IRT1 and IRT2 genes encode Fe(II) transporters that appear to be largely responsible for root iron uptake (Eide et al., 1996; Vert et al., 2001). IRT1 and IRT2 are expressed only in roots, and their mRNA levels increased in Fe-deficient plants. Expression of IRT1 and IRT2 in yeast (Saccharomyces cerevisiae) confers increased iron uptake in a yeast mutant defective for Fe uptake. More recently, an IRT1 mutant line of Arabidopsis was identified and found to be unviable without high iron supplements (Vert et al., 2002). Thus, IRT1 and probably IRT2 play important roles in iron uptake in Arabidopsis. Three transporter proteins unrelated to IRT1 and IRT2, designated AtNramp1, 3, and 4, have also been implicated in Fe(II) uptake by the root epidermal cells. AtNramp1, 3, and 4 are capable of iron uptake when expressed in yeast, and their expression in plant roots is induced by iron deficiency (Curie et al., 2000; Thomine et al., 2000). The FRO2 gene, encoding a Fe(III)-chelate reductase required for generating the Fe(II) substrate for these transporters, has also been identified (Robinson et al., 1999). Like the transporters, FRO2 is expressed in roots and its mRNA levels are induced by iron deficiency. Loss-of-function mutations in FRO2 result in decreased Fe(III)-chelate reductase activity, chlorosis, and poor growth under low-iron conditions. FRO2 encodes an integral membrane protein similar to the FRE1 and FRE2 Fe(III)-chelate reductases of S. cerevisiae and the human phagocytic NADPH gp91phox oxidoreductase. These enzymes transfer electrons from cytosolic NADPH to FAD and then, through two bound heme groups in the reductase, to electron acceptors on the extracellular surface.
Pea (Pisum sativum) has been used in studies of plant iron nutrition and physiology for a number of years (Grusak et al., 1999). Two pea mutants that have proven to be invaluable in the study of iron homeostasis are brz and dgl. These mutants have defective regulation of iron uptake, i.e. both show constitutive root Fe(III)-chelate reductase activity (Grusak et al., 1990; Grusak and Pezeshgi, 1996), and overaccumulate Fe (Kneen et al., 1990). It is intriguing that whereas brz seeds have normal Fe levels (Grusak, 1994), dgl seeds have been shown to overaccumulate Fe (Marentes and Grusak, 1998). In addition, both mutants are defective in nodulation (Kneen et al., 1990). An analysis of iron physiology in wild-type plants and these mutants using the tools provided by molecular genetics promises to provide great advances to our understanding of Fe acquisition and its regulation in plants. Toward this end, an IRT1 ortholog called RIT1 was recently cloned from pea (Cohen et al., 1998). The objective of the study described in this report was to characterize the gene encoding the root Fe(III)-chelate reductase from pea. Our results provide new molecular insight into the regulation of Fe reduction, potential mechanisms of distribution in plants, and the defect(s) in these processes that result from the brz and dgl mutations.
RESULTS
Membrane-bound oxidoreductases such as the human phagocyte NADPH gp91phox oxidoreductase, the yeast FRE2, and the Arabidopsis FRO2 Fe(III)-chelate reductases transfer electrons from cytosolic NADPH via enzyme-bound FAD and heme groups to electron-accepting substrates on the opposite side of the membrane. This shared mechanism of action dictates the conservation of cofactor binding sites in the amino acid sequences of the proteins. We took advantage of these conserved motifs to identify Fe(III)-chelate reductase genes of pea. In an approach similar to that used to clone Arabidopsis Fe(III)-chelate reductases (Robinson et al., 1999), a fragment of an Fe(III)-chelate reductase cDNA from pea was isolated using PCR and degenerate oligonucleotide primers designed to hybridize to sequences conserved in these reductase proteins. Those sequences were LQWHPFT, including the FAD-binding site, and EGPYGP, a sequence associated with the NADPH-binding site. 5′- and 3′-RACE was then used to isolate a full-length cDNA clone. We have designated the corresponding gene as FRO1. We concluded that this cDNA clone contains the full length FRO1 open reading frame. The cDNA contained several in-frame stop codons upstream of the proposed ATG. Following the stop codon, there was an untranslated region followed by a 17-base poly(A) tail.
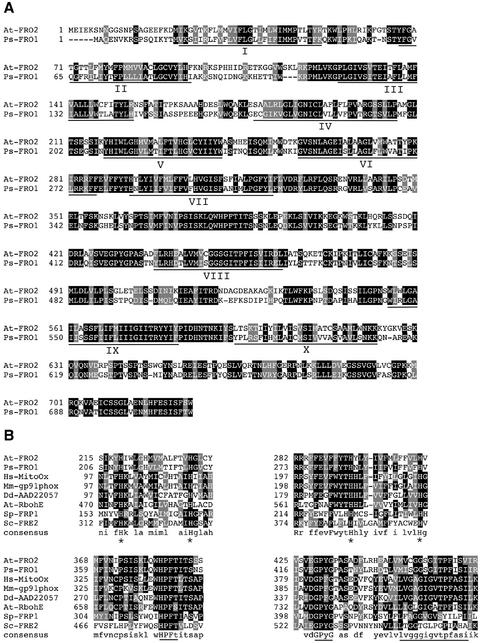
The predicted amino acid sequence of FRO1 (Fig. 1A) is 712 residues in length with a molecular mass of 80,500 D. The FRO1 amino acid sequence is shown aligned with the Arabidopsis FRO2 Fe(III)-chelate reductase. These two proteins are closely related, with an overall similarity of 74% and an identity of 55%. Figure 1B shows alignments of conserved motifs from several membrane-bound oxidoreductases, i.e. FRO1, Arabidopsis FRO2, human mitogenic oxidase, mouse gp91phox, Dictyostelium discoideum superoxide-generating NADPH oxidoreductase heavy chain, Arabidopsis RbohE, S. cerevisiae FRE2, and Schizosaccharomyces pombe FRP1. Four invariant His residues are likely to be involved in heme binding in these proteins (H210, H224, H284, and H297 of FRO1). Other conserved features include the FAD-binding motif (HPFT), the NADPH-binding motif (GPYG), and a signature sequence associated with NAD(P) H oxidoreductases (MISGGSGITPFISI in FRO1) identified by PFAM. Among this more diverse group of proteins, there is little sequence conservation outside of these motifs.
Figure 1.
Characterization of the FRO1 amino acid sequence. A, Alignment of Arabidopsis FRO2 (At-FRO2) and pea FRO1 (Ps-FRO1). Positions of amino acid identity are shaded in black, and similar residues are shaded in gray. The locations of the 10 potential transmembrane domains are underlined and numbered I-X. B, Conservation of specific motifs within FRO1 and related oxidoreductases. FRO1 and FRO2 sequences are aligned with human mitogenic oxidase (Hs-MitoOx, accession no. AF127763), mouse gp91phox (Mm-gp91phox, accession no. U43384), Dictyostelium discoideum superoxide-generating NADPH oxidase heavy chain (Dd-AAD22057, accession no. AF123275), Arabidopsis RbohE (At-RbohE, accession no. AF055356), S. pombe FRP1 (Sp-FRP1, accession no. L07749), and S. cerevisiae FRE2 (Sc-FRE2, accession no. Z28220). Conserved histidines involved in heme binding are indicated with asterisks, and the conserved FAD-binding motif (HPFT), the NAD-binding motif (GPyG), and the oxidoreductase signature sequence are underlined.
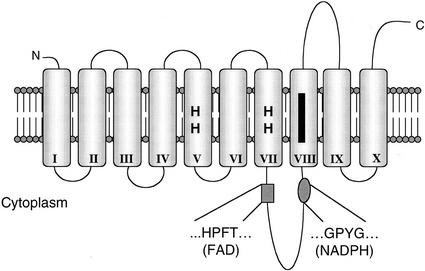
Several transmembrane domain prediction programs were used to analyze the FRO1 amino acid sequence, resulting in predictions ranging from seven to 11 membrane-spanning domains. The model based on TMAP (Milpetz et al., 1995) was largely in agreement with most predictions and is presented in Figure 2. This model describes 10 transmembrane domains. The amino and carboxy termini are predicted to be on the extracellular surface of the membrane, and the loop containing the FAD- and NADPH-binding sites are cytoplasmic. The heme-binding histidines are located in transmembrane helices V and VII, whereas the oxidoreductase signature sequence is located in transmembrane domain VIII.
Figure 2.
Predicted membrane topology of FRO1. The cylinders represent transmembrane domains I-X and the heme-binding histidines are shown. The black bar denotes the location of the oxidoreductase signature sequence.
A cDNA fragment corresponding to the carboxy-terminal 344 amino acids and 199 bp of 3′-untranslated region was used to probe a genomic Southern blot of DNA isolated from Sparkle, brz, and dgl. High-stringency wash conditions revealed a single band in BamHI and EcoRI digests, and two bands in a HindIII digest (data not shown). Consistent with these results, a HindIII restriction site is found in the FRO1-coding region and BamHI and EcoRI sites are not present. A Southern blot of a HindIII digest washed under low-stringency conditions revealed only one additional faint band. These data suggest that there are few, if any, other closely related Fe(III)-chelate reductase genes in the pea genome. Bands were detected in equivalent positions for all three genotypes, indicating that the brz and dgl mutations are not the result of easily detectable chromosomal rearrangements at the FRO1 locus.
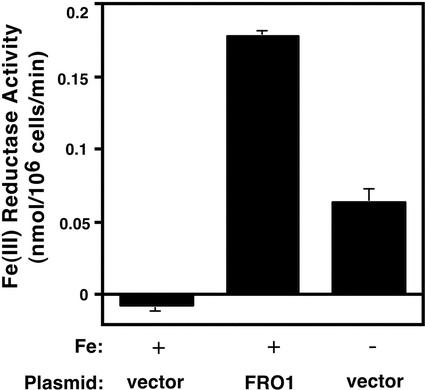
The close sequence similarity of FRO1 to the Arabidopsis FRO2 Fe(III)-chelate reductase and the conservation of functionally important conserved motifs found in other oxidoreductases strongly suggested that FRO1 is a Fe(III)-chelate reductase. To address this hypothesis directly, we tested the ability of FRO1 to confer Fe(III)-chelate reductase activity when expressed in yeast. Wild-type yeast cells were transformed with a plasmid containing the FRO1 coding sequence under the control of the GAL1 promoter (pYES2.0-FRO1) or the pYES2.0 expression vector alone. These cells were then grown in a minimal medium supplemented with 10 μm FeCl3 to repress expression of the endogenous yeast Fe(III) reductase activities. Although activity in the vector-only control was undetectable, the FRO1-expressing cells had high reductase activity (Fig. 3). For comparison, vector-only control cells were assayed in medium made iron limiting by adding 1 mm EDTA. Vector-transformed cells grown with EDTA had Fe(III) reductase activity of approximately one-third that of FRO1-expressing cells. Thus, expression of FRO1 from the GAL1 promoter in yeast conferred an even higher level of reductase activity than did induction of the yeast reductases by iron deficiency. The ability of FRO1 to confer increased reductase activity when expressed in yeast is consistent with this gene encoding a Fe(III)-chelate reductase in plants.
Figure 3.
FRO1 is a functional Fe(III)-chelate reductase when expressed in yeast. Wild-type yeast (DY1457) transformed with the vector or the FRO1-expressing plasmid pYES2.0-FRO1 (FRO1) were grown in iron-replete medium (synthetic dextrose [SD] medium supplemented with 10 μm FeCl3) (+) or in iron-limiting medium (SD medium supplemented with 1 mm EDTA) (-) and assayed for Fe(III)-chelate reductase activity. Reductase activity values were obtained by first subtracting out reduction occurring in a no-cell background control, so small negative values are possible for strains with no detectable activity.
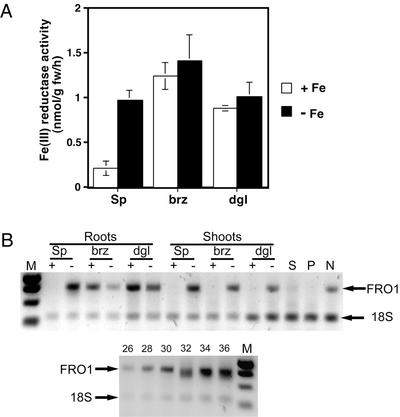
To determine if FRO1 encoded the Fe(III)-chelate reductase active in the roots of iron-limited plants, we examined the correlation between root reductase activity and FRO1 expression. Sparkle plants grown in iron-replete conditions had low root Fe(III)-chelate reductase activity (Fig. 4A). This activity was 4.5-fold higher in iron-limited roots. Reductase activity was constitutively expressed in brz and dgl roots regardless of Fe supply. These data are similar to previous reports examining reductase activity in these plant lines. To estimate FRO1 mRNA levels, semiquantitative RT-PCR was performed with RNA from Fe-deficient and replete roots of Sparkle, brz, and dgl. Consistent with the reductase activity data, FRO1 mRNA was not detectable in Fe-replete Sparkle roots, but was expressed at high levels in the Fe-deficient roots (Fig. 4B). Also consistent with reductase activity, FRO1 expression in brz and dgl was high in the roots of both Fe-replete and -deficient plants. Although there was some variability in band intensity, repeated RT-PCR analysis of mRNA isolated from different plants revealed similar levels of FRO1 mRNA in Fe-replete and -deficient brz and dgl roots. Thus, FRO1 expression in roots correlates with reductase activity in wild-type and these mutant plants.
Figure 4.
Regulation of Fe(III)-chelate reductase activity and FRO1 mRNA levels. A, Reductase activity of whole roots. Pea plants of the indicated genotype were grown hydroponically for 12 d in nutrient solution with 5 μm Fe(III)-EDDHA (+Fe) or without added Fe (−Fe). Fe(III)-chelate reductase activity was assayed with the bathophenanthrolinedisulfonic acid (BPDS) assay. B, Quantitative reverse transcriptase (RT)-PCR detection of FRO1 mRNA. Total RNA from roots and shoots of plants grown as described in A. RNA was also isolated from whole seeds (S) and pods (P) of soil-grown Sparkle plants during active seed fill, and from established Sparkle nodules (N). RNA (0.1 μg) was used as template for RT-PCR. All samples were subjected to 30 cycles PCR except for seed, pod, and nodule samples, which were subjected to 35 cycles. The lower panel shows a control indicating the quantitative nature of the assay. Numbers above indicate number of PCR cycles. A negative control without RT showed no bands, indicating the absence of genomic DNA contamination. M, Mr markers, from top to bottom, 1,000, 750, 500, and 250 bp.
To examine expression of FRO1 in other plant tissues, we assessed mRNA accumulation in shoots, seeds, pods, and nodules. In Sparkle shoots, FRO1 is expressed and its mRNA levels are iron-regulated as was observed in the roots (Fig. 4B). It is surprising that whereas brz and dgl result in constitutive root expression, iron-responsive regulation in the shoots was unaffected in these mutants. Little if any expression was detected in seeds or pods, but FRO1 mRNA was detected in nodules. It should be noted that the seeds, pods, and nodules analyzed in this experiment were obtained from iron-replete Sparkle plants. We were unable to assess expression of FRO1 in these structures in iron-deficient plants because they failed to develop properly under these conditions.
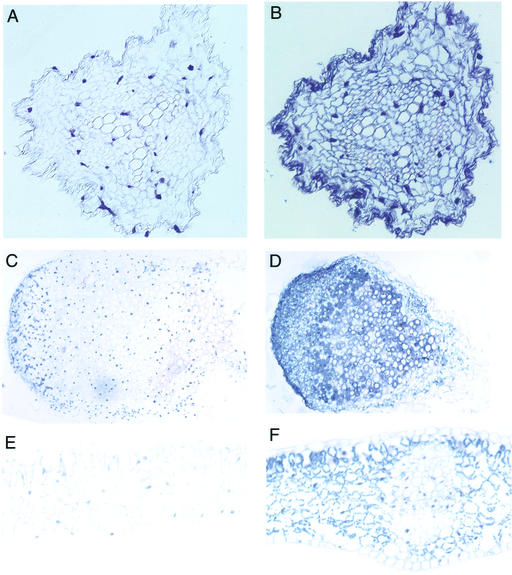
Expression of FRO1 in roots, shoots, and nodules suggested that this reductase may be important for root iron uptake and may play other roles in iron distribution elsewhere in the plant. To assess these various roles, we examined the tissue-specific expression of FRO1 by in situ hybridization. In all tissues examined, the negative control sense probe showed little staining (Fig. 5, A, C, and E). In contrast, hybridization of the antisense probe in roots (Fig. 5B), nodules (Fig. 5D), and leaves (Fig. 5F) detected FRO1 mRNA. In roots, FRO1 mRNA was observed in cells throughout the tissue, including the vascular cylinder and cortex. FRO1 expression appeared most intense in the outer epidermal cell layer. FRO1 expression was also found throughout nodules. Cells in the nodule meristem region stained intensely, suggesting high FRO1 expression. A region located approximately one-third of the distance from the apex to the base of the nodule also showed intense staining; this region is most likely the zone of infection where cells are first colonized by Rhizobium leguminosarum. Less staining was observed in the region of cell senescence at the base of the nodule. In leaves, FRO1 expression was observed in mesophyll and parenchyma cells. Little FRO1 mRNA was observed in leaf vascular cells or epidermis.
Figure 5.
Localization of tissue-specific FRO1 expression by in situ hybridization. Thin sections of plant tissues were probed with sense (A, C, and E) or antisense (B, D, and F) biotinylated FRO1 probes. Probe hybridization was detected with a streptavidin-alkaline phosphatase conjugate and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate. Fe-deficient Sparkle root (A and B), soil-grown Sparkle nodule (C and D), and Fe-deficient Sparkle leaf (E and F) cross sections are shown.
DISCUSSION
The data presented here strongly support the hypothesis that the pea FRO1 gene encodes a Fe(III)-chelate reductase. FRO1 is closely related to FRO2 of Arabidopsis, which has been established genetically to be an Fe(III)-chelate reductase required for reduction of rhizosphere Fe(III) (Robinson, et al., 1999). Transmembrane domains are especially conserved between FRO1 and AtFRO2, as are functionally important motifs that are also shared among many other membrane-bound oxidoreductases. The topology model presented for FRO1 predicts that the amino- and carboxy-terminal regions are extracellular. This topology differs from that proposed for AtFRO2 where both ends of the protein were predicted to be cytoplasmic (Robinson et al., 1999). Further evidence that FRO1 is a Fe(III)-chelate reductase comes from its functional expression in yeast. This result argues that other subunits are not required for FRO1 activity in plants. Support for the role of FRO1 in root Fe uptake comes from the correlation of whole root Fe(III) reduction with FRO1 mRNA accumulation in response to Fe status and genotype. Whenever elevated root ferric reductase activity is present (Fe-deficient Sparkle, brz, and dgl under either condition), FRO1 mRNA is detectable (Fig. 4B). Measurement of apparent ferric reductase activity despite the lack of detectable FRO1 mRNA in Fe-replete Sparkle roots may indicate that FRO1 is expressed at a low level not detected with our RT-PCR assay conditions, or that there is another reductase responsible for noninduced Fe acquisition. In an alternate manner, the reduction of Fe(III) EDTA in the assay buffer may result from the nonspecific action of other systems.
We have also addressed the function of FRO1 by determining the sites of its expression in whole plants. The expression of FRO1 mRNA within root tissue is consistent with its proposed role in reduction of rhizosphere Fe(III). FRO1 mRNA levels were high in the outer epidermal cells, i.e. the cells that would be in contact with the soil solution. This expression pattern was also observed in brz and dgl mutants (data not shown). Expression of the Arabidopsis IRT1 and IRT2 iron transporters was recently localized to the outer epidermal cells (Vert et al., 2001; Vert et al., 2002). FRO1 and one or more iron transporters, perhaps related to IRT1 and IRT2, probably comprise the major Fe uptake system in pea roots. The pea IRT1/IRT2 ortholog called RIT1 (Cohen et al., 1998) may serve as the Fe(II) transporter for this system.
FRO1 is expressed in nodules, which require large quantities of Fe for leghemoglobin synthesis and for Fe-dependent enzymes, such as nitrogenase, that function in nodules (Udvardi and Day, 1997). An Fe(III)-chelate reductase activity was previously found to be associated with the plant-derived peribacteroid membrane in soybean (Glycine max) nodules (LeVier et al., 1996). This reductase activity was suggested to be important for movement of Fe into the symbiosomes. FRO1 may play this role in pea, and its localization in the infection zone and nitrogen-fixing region of the nodule supports this hypothesis. We have not yet assessed if nodule expression of FRO1 is regulated in response to Fe status.
FRO1 mRNA levels were low in Fe-sufficient shoots, but were elevated in shoots of Fe-deficient plants. These results suggest a role for Fe(III) reduction in leaf tissues. Although some studies have indicated that reduction of Fe(III) is necessary for uptake of Fe by leaf cells, controversy over this issue still remains (for review, see Schmidt, 1999). To be specific, some studies failed to detect Fe(III)-chelate reductase activity in leaf disc samples. Our results show that an induction of FRO1 mRNA does occur in Fe-deficient shoots, which would likely result in higher levels of Fe(III)-chelate reductase protein in the leaves. The localization of FRO1 mRNA in leaves was confined to mesophyll and parenchyma cells and was not seen in the epidermal cells. Therefore, previous experiments that did not detect Fe(III) reduction by leaf discs may be explained by the localization of FRO1 protein in the interior cells of the leaf and not on the surface. Iron is thought to be transported in the xylem as a Fe(III)-citrate complex (Tiffin, 1966). Leaf cells may need to reduce Fe(III) from this complex to Fe(II) before uptake, and FRO1 may be involved in this reduction step. It is notable that FRO1 mRNA was not highly expressed in the vascular bundles of leaves, indicating that other proteins or transport mechanisms may be at work mobilizing xylem Fe(III). A Fe(III)-chelate reductase activity has also been suggested to aid uptake of intracellular Fe by chloroplasts (Bughio et al., 1997) and, given its expression in chloroplast-containing cells, FRO1 could conceivably play that role as well. It is interesting to note that although we have detected FRO1 in multiple tissues and have not found other ferric reductase genes, the Arabidopsis genome contains at least five FRO family members, each of which may carry out specific functions in different organs or cell types (Robinson et al., 1999).
FRO1 mRNA was detected in Fe-deficient shoots and was undetectable in Fe-replete shoots. Moreover, this regulation was unaffected by the brz and dgl mutations. These results are in marked contrast to the accumulation of FRO1 mRNA in roots. Like in shoots, wild-type Sparkle roots showed Fe-responsive expression of FRO1. However, FRO1 expression was constitutive in the roots of brz and dgl. These results demonstrate that expression of FRO1 in shoots and roots is affected by different signals or Fe-sensing mechanisms. Regulation of FRO1 in shoots appears to respond to the Fe status of these tissues per se. Several studies have indicated that a shoot-derived signal of Fe status influences root Fe(III)-chelate reductase activity (Maas et al., 1988; Romera et al., 1992; Grusak, 1995; Grusak and Pezeshgi, 1996), and a mutant with roots unable to detect a shoot-derived Fe signal has been described (Brown et al., 1971; Ling et al., 1996). It appears that brz and dgl inappropriately generate or transmit a shoot-derived signal of Fe status to the roots. This hypothesis is consistent with grafting experiments showing that the genotype of the shoot and not the root dictates the phenotypic effects of these mutations in roots (Grusak and Pezeshgi, 1996). In contrast, Schikora and Schmidt (2001) showed that root Fe(III)-chelate reductase activity and transfer cell formation were differentially regulated in brz and dgl: i.e. reductase activity was high regardless of Fe supply, whereas transfer cell formation was responsive to nutrient solution Fe concentration. Thus, separate signals may regulate root Fe reductase and other Fe-deficiency responses.
MATERIALS AND METHODS
Plant Growth Conditions
Pea (Pisum sativum) seeds of the parental genotype Sparkle and the mutant genotypes brz (Sparkle [brz, brz]) and dgl (Sparkle [dgl, dgl]) were obtained from Dr. Michael Grusak (U.S. Department of Agriculture-Agricultural Research Service, Children's Nutrition Research Center, Houston, TX). For hydroponic studies, seeds were imbibed overnight, and then placed between layers of germination paper and held upright between plastic trays. After 4 d of germination, seedlings were planted into plastic cups with a single hole in the bottom and were covered with black plastic beads. The cups were fitted into lids of plastic pots filled with 700 mL of nutrient solution, with one plant per pot. The nutrient solution contained 1.2 mm KNO3, 0.8 mm Ca(NO3)2, 0.1 mm NH4H2PO4, 0.2 mm MgSO4, 25 μm CaCl2, 25 μm H3BO3, 2 μm MnSO4, 2 μm ZnSO4, 0.5 μm CuSO4, 0.5 μm Na2MoO4, and 0.1 μm NiCl2 (Grusak et al., 1990). The nutrient solution was buffered to pH 5.0 with 1 mm MES and, where indicated, Fe was supplied as Fe(III)-EDDHA at a concentration of 5 μm. For production of seeds, pods, and nodules, imbibed seeds were inoculated with Rhizobium leguminosarum and were planted into commercial potting mix. Nodules were harvested with forceps. Observation of nodules by microscopy indicated that non-nodule root tissue accounted for 1% (v/v) or less of total tissue designated nodule. Plants were grown in a growth chamber programmed for a 16-h photoperiod (600 μmol photons m−2 s−1) at 20°C and 8 h of darkness at 15°C, 70% relative humidity.
DNA Manipulations
A full-length FRO1 cDNA fragment was isolated as follows. Roots of 14-d-old Fe-deficient Sparkle were harvested, and RNA was extracted (McClure et al., 1990). Poly(A)+ mRNA was selected from total RNA with an oligo dT cellulose column (Molecular Research Center, Cincinnati) and was subsequently used for RT-PCR. The degenerate primers used in the first PCR reaction were DF3 and DR3 (Table I). Three resulting PCR products were cloned into the vector pBluescript SK (Stratagene, La Jolla, CA) and sequenced. A BLAST (Altschul et al., 1997) search using the sequence of one PCR product of 165 bp revealed a high degree of similarity to the Arabidopsis Fe(III)-chelate reductases FRO1 and FRO2. The DNA sequence of this clone was used to design nested gene-specific primers. The forward gene-specific primer set (GSP1 and GSP3) was used in conjunction with the Universal Adapter Primer in a 3′-RACE PCR (Invitrogen, Carlsbad, CA). A single PCR product was cloned, sequenced, and confirmed to overlap the original PCR product. The reverse gene-specific primers (GSP2 and GSP4) were used in conjunction with another forward degenerate primer, DF5, based on the amino acid sequence IKYHIWLG, which is conserved in the Arabidopsis FRO proteins. This resulted in a single PCR product that overlapped the original clone. From the DNA sequence of this clone, a nested reverse gene-specific primer set was designed (GSP5 and GSP6). Nested PCR using this primer set was performed with a pea root nodule cDNA library (gift of Dr. Maria Fedorova, University of Minnesota, St. Paul) as template. The vector T3 primer was used as a forward primer. By this method, a single PCR product was obtained that was cloned and sequenced (GenBank accession no. AF405422). Forward (FROF2) and reverse (FROR) gene-specific primers were used to amplify the entire FRO1 coding sequence.
Table I.
Primers used in this study
| Primer Name | Sequence (5′–3′)a |
|---|---|
| DF3 | CGCGAATTCYTICARTGGCAYCCITTYAC |
| DR3 | CGCAAGCTTGGICCRTAIGGICCYTC |
| DF5 | CGCGAATTCATHAADTAYCAYATHTGGYTIGG |
| GSP1 | GGCATCCGTTCACCATTACTTCT |
| GSP2 | GCCCTCGACGGATATTTGAAG |
| GSP3 | AGTAACTTGGAGCAAGATAAGCTAAGTG |
| GSP4 | GGCGAAGGATTTGAAAGCAAC |
| GSP5 | TCTAAGTTTTGGAATGGTTGC |
| GSP6 | TCCAATAGATAATATAACAAACTCC |
| FROF2 | TCGAAACAGTTCAATTTACAGACAAAACCC |
| FROR | CGAACGAGTAAAATCACCAGGTAAAACTGATTG |
| SEQ1 | GCAAAACACCAAACATTGTTC |
| T3 | AATTAACCCTCACTAAAGGG |
Y = C or T; R = A or G; D = A, G, or T; H = A, C, or T; I = deoxyinosine.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Sequence Analysis
The deduced amino acid sequence of FRO1 was used in BLAST (Altschul et al., 1997) and Pfam database (Bateman et al., 1999) searches to find related proteins and motifs. Multiple sequence alignments were performed using CLUSTALW software (Thompson et al., 1994) and BOXSHADE, and blocks of similar sequence were found using BLOCKMAKER (Henikoff et al., 1995). Secondary structure predictions were performed using TMAP (Milpetz et al., 1995) with a multiple sequence alignment of pea FRO1, Arabidopsis FRO1, and Arabidopsis FRO2 as input. Protein characteristics were predicted by PROTPARAM (Appel et al., 1994).
Southern-Blot Analysis
Genomic DNA was harvested from leaves as described by Dellaporta et al. (1983), digested (40 μg) with BamHI, EcoRI, or HindIII, and electrophoresed on a 1% (w/v) agarose gel. The DNA was blotted to a nylon membrane (BioTrans; ICN, Costa Mesa, CA) by capillary transfer and was fixed by UV light (120,000 μJ cm2) and baking for 20 min at 80°C. Blots were prehybridized and hybridized using PerfectHyb Plus buffer (Sigma, St. Louis) following the manufacturer's instructions. The probe template was a 1,228-bp PCR product containing the 3′ portion of FRO1 cDNA. Probes were labeled using the Megaprime DNA labeling system (Amersham Biosciences, Piscataway, NJ) and were allowed to hybridize overnight. For low stringency, blots were washed with 0.1% (w/v) SDS and 2× SSC at 65°C for 5 min, followed by four additional washes under the same conditions for 25 min each. For high stringency, one 5-min wash at 65°C in 0.1% (w/v) SDS and 2× SSC was followed by two 20-min washes in 0.1% (w/v) SDS and 0.5× SSC and one 20-min wash in 0.1% (w/v) SDS and 0.1× SSC. Blots were then exposed to x-ray film at −80°C for appropriate times.
Expression of FRO1 in Yeast (Saccharomyces cerevisiae)
The full-length FRO1 coding sequence was subcloned from pGEM-T Easy (Promega, Madison, WI) into the EcoRI site of vector pYES2.0 (Invitrogen) to generate pYES2.0-FRO1. Plasmids were transformed into a S. cerevisiae wild-type strain, DY1457 (MATα ade6 can1 his3 leu2 trp1 ura3), using the lithium acetate method (Schiestl and Gietz, 1989). DY1457 transformed with the vector alone or pYES2.0-FRO1 were grown in SD medium (Rose et al., 1990) supplemented with 2% (w/v) Gal to induce expression, all necessary auxotrophic supplements (less uridine), and 10 μm FeCl3 to repress endogenous yeast Fe(III)-chelate reductase activities. Cells were also grown in the presence of 1 mm EDTA (+10 μm ZnCl2 to aid growth) to induce endogenous reductase activities. Cells were harvested in mid-log phase, and Fe(III)-chelate reductase activity was determined as described (Eide et al., 1992).
Root Fe(III)-Chelate Reductase Activity Assay
Fe(III)-chelate reductase activity was determined for whole intact root systems as described previously (Romera et al., 1996). In brief, roots were rinsed and submerged in 70 mL of assay solution [0.2 mm CaSO4, 5 mm MES at pH 5.5, 0.1 mm Fe(III)-EDTA, and 0.2 mm BPDS]. After 20 min, an aliquot of the assay solution was removed and A535 was determined. Fe(II)-BPDS concentration was calculated using the extinction coefficient of 22.14 mm−1 cm−1.
RT-PCR
Plant tissues were harvested, and RNA was extracted with Trizol Reagent (Invitrogen) and quantified by UV spectrophotometry. Total RNA (0.1 μg) was used for each RT reaction, which was carried out in a volume of 20 μL according to manufacturer's instructions (GeneAmp RNA PCR; Perkin Elmer, Foster City, CA). To each RT reaction, 30 μL of PCR solution was added, which consisted of 2 mm MgCl2, 1× PCR buffer, 2.5 units of Taq DNA polymerase (Promega), 2 μm SEQ1 primer, 2 μm FROR primer, and 4 μL of QuantumRNA 18S primers and competimers at a 1:9 ratio (Ambion, Austin, TX). QuantumRNA primers/competimers attenuate the amplification of 18S ribosomal RNA, allowing it to be used as an internal control. The thermalcycler program was 94°C for 5 min, the specified number of cycles at 94°C (45 s), 55°C (45 s), and 72°C (45 s), followed by a final 6-min 72°C incubation. PCR products were electrophoresed on a 1.5% (w/v) agarose gel, stained with ethidium bromide, and photographed using a CCD camera. The identity of the FRO1 PCR product was confirmed by DNA sequencing.
In Situ Hybridization
Root and leaf tissue harvested for in situ hybridization was taken from Sparkle plants grown in conditions described above to generate Fe-deficient plants. Tissues from brz and dgl were harvested from plants grown in Fe-replete conditions. Ferric reductase assays were carried out on individual plants to confirm ferric reductase activity status. Lateral root tissue (excluding the apical 3 cm) was cut in approximately 1-cm-long segments. Leaf tissue was harvested from young leaves with a paper-hole punch. Nodule tissue was harvested from soil-grown Sparkle plants as described above. Plant tissues were harvested and immediately fixed in 10% (w/v) buffered formalin for 6 h to overnight. Tissues were processed in an ethanol dehydration series followed by xylene treatment, and were embedded in paraffin. Five-micrometer sections were mounted on positively charged microscope slides. Biotin-labeled sense and antisense FRO1 probes were generated using the MAXIscript (Ambion) and Biotin RNA Labeling Mix (Roche Molecular Biochemicals, Summerville, NJ) kits according to the manufacturer's instructions. The probe template, pGEM-T Easy carrying the FRO1 cDNA, was linearized with XmnI, resulting in a fragment containing the vector with 352 bp of 5′ FRO1-coding sequence and 210 bp of 3′ FRO1-coding sequence. In vitro transcription with SP6 RNA polymerase produced the antisense probe, and in vitro transcription with T7 RNA polymerase generated the sense probe. Tissues were incubated with the probes using the mRNAlocator-hyb kit (Ambion), and were washed and detected using the mRNAlocator-Biotin kit (Ambion) according to manufacturer's instructions.
ACKNOWLEDGMENTS
We thank Mary Lou Guerinot (Dartmouth College, Hanover, NH) for critical reading of the manuscript and helpful discussions, Jill Gruenkemeyer and staff at the University of Missouri Veterinary Histopathology Laboratory for processing and sectioning of plant tissues, Michael Grusak (U.S. Department of Agriculture-Agricultural Research Service, Houston, TX) for supplying brz and dgl seeds, and Maria Fedorova (University of Minnesota, St. Paul) for supplying the nodule cDNA library.
Footnotes
This work was supported by the Plant Science Unit and by the National Institutes of Health (grant no. 1RO1–GM58265).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010829.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel RD, Bairoch A, Hochstrasser DF. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy S, Finn RD, Sonnhammer ELL. Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Chaney RL, Amber JE. A new tomato mutant inefficient in the transport of iron. Physiol Plant. 1971;25:45–53. [Google Scholar]
- Bughio N, Takahashi M, Yoshimura E, Nishizawa NK, Mori S. Characteristics of light-regulated iron transport system in barley chloroplasts. Soil Sci Plant Nutr. 1997;43:959–963. [Google Scholar]
- Cohen CK, Fox TC, Garvin DF, Kochian LV. The role of iron-deficiency stress responses in stimulating heavy-metal transport in plants. Plant Physiol. 1998;116:1063–1072. doi: 10.1104/pp.116.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Echker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Davis-Kaplan S, Jordan I, Sipe D, Kaplan J. Regulation of iron uptake in Saccharomyces cerevisiae: The ferrireductase and Fe(II) transporter are regulated independently. J Biol Chem. 1992;267:20774–20781. [PubMed] [Google Scholar]
- Grusak MA. Iron transport to developing ovules of Pisum sativum: seed import characteristics and phloem iron-loading capacity of source regions. Plant Physiol. 1994;104:649–655. doi: 10.1104/pp.104.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA. Whole-root iron(III)-reductase activity throughout the life cycle of iron-grown Pisum sativum L. (Fabaceae): relevance to the iron nutrition of developing seeds. Planta. 1995;197:111–117. [Google Scholar]
- Grusak MA, Pearson JN, Marentes E. The physiology of micronutrient homeostasis in field crops. Field Crops Res. 1999;60:41–56. [Google Scholar]
- Grusak MA, Pezeshgi S. Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 1996;110:329–334. doi: 10.1104/pp.110.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, Welch RM, Kochian LV. Physiological characterization of a single-gene mutant of Pisum sativum exhibiting excess iron accumulation. Plant Physiol. 1990;93:976–981. doi: 10.1104/pp.93.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences, Gene-COMBIS. Gene. 1995;163:17–26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Kneen BE, LaRue TA, Welch RM, Weeden NF. Pleiotropic effects of brz. Plant Physiol. 1990;93:717–722. doi: 10.1104/pp.93.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVier K, Day DA, Guerinot ML. Iron uptake by symbiosomes from soybean root nodules. Plant Physiol. 1996;111:893–900. doi: 10.1104/pp.111.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Pich A, Scholz G, Ganal MW. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol Gen Genet. 1996;252:87–92. doi: 10.1007/BF02173208. [DOI] [PubMed] [Google Scholar]
- Maas FM, van de Wetering DAM, van Beusichem ML, Bienfait HF. Characterization of phloem iron and its possible role in the regulation of Fe-efficiency reactions. Plant Physiol. 1988;87:167–171. doi: 10.1104/pp.87.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marentes E, Grusak MA. Iron transport and storage within the seed coat and embryo of developing seeds of pea (Pisum sativum L.) Seed Sci Res. 1998;8:367–375. [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. Ed 2. San Diego: Academic Press; 1995. [Google Scholar]
- McClure BA, Gray JE, Anderson M, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- Milpetz F, Argos P, Persson B. TMAP: a new email and WWW service for membrane-protein structural predictions. Trends Biochem Sci. 1995;20:204–205. doi: 10.1016/s0968-0004(00)89009-x. [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Romera FJ, Alcantera E, de la Guardia MD. Role of roots and shoots in the regulation of the Fe efficiency responses in sunflower and cucumber. Physiol Plant. 1992;85:141–146. [Google Scholar]
- Romera FJ, Welch RM, Norvell WA, Schaefer SC. Iron requirement for and effects of promoters and inhibitors of ethylene action on stimulation of Fe(III)-chelate reductase in roots of strategy I species. BioMetals. 1996;9:45–50. [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded DNA as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schikora A, Schmidt W. Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol. 2001;125:1679–1687. doi: 10.1104/pp.125.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W. Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol. 1999;141:1–26. [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin LO. Iron translocation: plant culture, exudate sampling, iron citrate analysis. Plant Physiol. 1966;45:280–283. doi: 10.1104/pp.41.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Day DA. Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:493–523. doi: 10.1146/annurev.arplant.48.1.493. [DOI] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C. Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J. 2001;26:181–189. doi: 10.1046/j.1365-313x.2001.01018.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldecham F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron update from the soil and plant growth. Plant Cell (in press) [DOI] [PMC free article] [PubMed]