Abstract
Enteric bacteria possess multiple fimbriae, many of which play critical roles in attachment to epithelial cell surfaces. SEF14 fimbriae are only found in Salmonella enterica serovar Enteritidis (S. enteritidis) and closely related serovars, suggesting that SEF14 fimbriae may affect serovar-specific virulence traits. Despite evidence that SEF14 fimbriae are expressed by S. enteritidis in vivo, previous studies showed that SEF14 fimbriae do not mediate adhesion to the intestinal epithelium. Therefore, we tested whether SEF14 fimbriae are required for virulence at a stage in infection after the bacteria have passed the intestinal barrier. Polar mutations that disrupt the entire sef operon decreased virulence in mice more than 1,000-fold. Nonpolar mutations that disrupted sefA (encoding the major structural subunit) did not affect virulence, but mutations that disrupted sefD (encoding the putative adhesion subunit) resulted in a severe virulence defect. The results indicate that the putative SEF14 adhesion subunit is specifically required for a stage of the infection subsequent to transit across the intestinal barrier. Therefore, we tested whether SefD is required for uptake or survival in macrophages. The majority of wild-type bacteria were detected inside macrophages soon after i.p. infection, but the sefD mutants were not readily internalized by peritoneal macrophages. These results indicate that the potential SEF14 adhesion subunit is essential for efficient uptake or survival of S. enteritidis in macrophages. This report describes a role of fimbriae in intracellular infection, and indicates that fimbriae may be required for systemic infections at stages beyond the initial colonization of host epithelial surfaces.
Fimbriae play a critical role in virulence by allowing bacteria to interact with host cells and other solid substrates (1, 2). The distribution of fimbrial operons among enteric bacteria suggests a role for fimbriae in pathogenesis; broadly distributed fimbrial operons may provide general adhesive functions, but fimbriae whose distribution is limited may provide specific functions required in virulence. For example, the common type I fimbriae found in many Gram-negative bacteria mediates adherence to the pharynx, intestinal epithelium, and bladder, whereas plasmid encoded fimbriae found only in Salmonella bind specifically to M cells in the intestine (1, 3).
Most fimbriae (with the notable exception of type IV pili) have a conserved mechanism of translocation to the bacterial surface. Fimbrial proteins are secreted into the periplasm by means of the general secretory system. Two accessory proteins assist in construction of the fimbrial shaft. First, the fimbrial subunits are bound by a chaperone in the periplasm to prevent premature aggregation and then the subunits are translocated across the outer membrane by an usher protein. Recently, the crystal structures of FimC–FimH and PapD–PapK complexes have been solved, demonstrating an elegantly simple mechanism for the interactions between the subunit and chaperone and between pairs of subunits (4, 5). In all fimbrial systems that have been studied, the genes encoding the chaperone and usher are located in the same operon as the major subunit. Cursory examination of the Escherichia coli, Yersinia pestis, Salmonella enterica serovar Typhimurium (S. typhimurium), and Salmonella Typhi (S. typhi) genomes identified about one dozen chaperone–usher-dependent fimbrial operons per genome, although only a handful of these have currently been characterized (R.A.E., B. M. Matlock, and S.R.M., unpublished observations). It is expected that Salmonella enteritidis contains a similar number of fimbrial operons to these other enteric bacteria. The S. enteritidis fimbriae (SEF14) is restricted to S. enteritidis and other closely related group D Salmonella (7). Therefore, an analysis of SEF14 fimbrial function may provide insight into the unique aspects of virulence that distinguish this group of Salmonella.
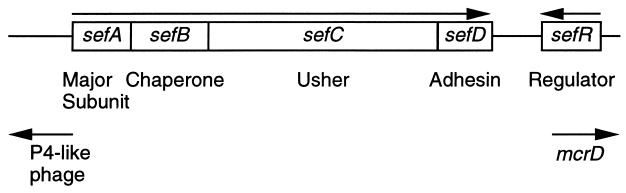
The sef operon is located on a small pathogenicity island. The operon contains four structural genes (sefABCD) required for the translocation and biogenesis of SEF14 fimbriae: sefA encodes the major subunit, sefB and sefC encode the chaperone and usher, respectively, and sefD encodes the putative adhesin. Adjacent to sefD, there is an AraC-like regulatory protein (encoded by sefR) that activates transcription of the sef genes (6) (Fig. 1). Some evidence suggests that SEF14 fimbriae may play a role in pathogenesis. For example, immunization of mice with purified SEF14 subunits induces a strong T lymphocyte response, and the mice present a delayed-type hypersensitive response to whole S. enteritidis, demonstrating that these fimbriae are expressed in vivo and stimulate cell-mediated immunity (8). Furthermore, pretreatment of mice with anti-SEF14 antibodies protects mice from S. enteritidis infection (9). However, other studies on the effect of sef mutants on virulence showed conflicting results (10, 11).
Figure 1.
The sef pathogenicity island. Organization of the sef genes, direction of transcription (arrows), and predicted function of the ORFs.
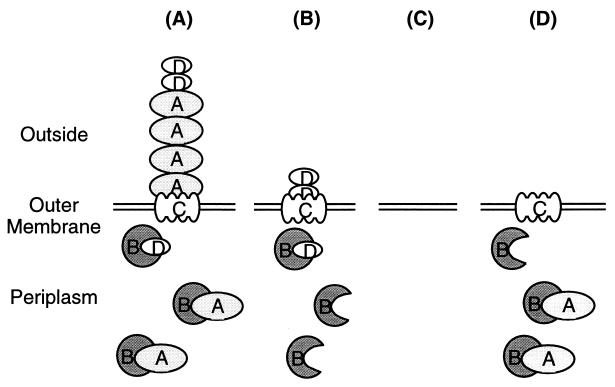
Because discrepancies in earlier studies may be the result of genetic differences in the mutants used, we constructed several defined sef mutants (R.A.E., B. M. Matlock, and S.R.M., unpublished observations; L. H. Keller, R. C. Boston, and D.M.S., unpublished observations; ref. 13). Like many other fimbrial operons, the sefABCD genes are cotranscribed so that insertion of a kanamycin cassette into sefA eliminates transcription of sefD, and translocation of SefD across the outer membrane is a prerequisite for translocation of SefA (6). Based on these studies, we predict that the SEF14 fimbrial proteins displayed on the surface of the sef mutants is as shown in Fig. 2. In this work, we assay the role of the various sef mutants in virulence and show that SEF14 fimbriae mediate interactions with phagocytes in the peritoneal cavity, which allows S. enteritidis to survive the macrophage assault. This role for fimbriae in vivo is in contrast to the routine modus operandi of previously characterized fimbriae involved in binding to host epithelial surfaces.
Figure 2.
Predicted export of SEF14 fimbrial subunits from the periplasm in the mutants used in this study. Note that all subunits contain signal sequences, suggesting that they are exported across the cytoplasmic membrane in a secretory-dependent manner. Wild-type SEF14 fimbriae (A) consist of the major subunit, SefA. Based on sequence homology, SefB is the chaperone that binds subunits in the periplasm and prevents premature aggregation. SefC is the membrane-located usher through which the subunits are translocated. SefD is probably a tip-located adhesin. Export of SefD occurs in the absence of SefA (ΔsefA; B) and may form a fibrillar structure on the cell surface. The polar sefA∷Kan strain (C) does not produce any subunits. Export of SefA does not occur in the absence of SefD (ΔsefD; D).
Materials and Methods
Bacterial Strains and Growth Conditions.
All strains used in this study were derivatives of S. enteritidis LK5 (14) (Table 1). This prototrophic, phage type 8, S. enteritidis strain was isolated as a chicken pathogen. For routine growth, all strains were grown overnight at 37°C with vigorous aeration in LB broth supplemented with kanamycin sulfate (Kan) at a final concentration of 50 μg ml−1 when required (15). Prior to infection of animals, bacteria were grown in conditions that optimize SEF14 fimbrial expression, colonization factor antigen (CFA) media buffered with 0.1 M Mes to pH 6.0 (16). The ΔsefA in-frame deletion removes the central 37 amino acids of the 176-aa polypeptide. The ΔsefD in-frame deletion removes the central 78 amino acids of the 149-aa polypeptide. Construction of the sef mutants and analysis of the exported proteins are described elsewhere (R.A.E., B. M. Matlock, and S.R.M., unpublished observations; L. H. Keller, R. C. Boston, and D.M.S., unpublished observations, ref. 13). ΔsefA and ΔsefD strains were marked with zjg∷MudJ transduced from JS107 (17). This kanamycin-resistant insertion has been extensively tested and does not affect virulence of Salmonella in mice.
Table 1.
LD50 values of wild-type and sef mutants
| Strain | sef genotype* | IP LD50† | Oral LD50† |
|---|---|---|---|
| LK5 | sef+ | <10 | >104 |
| TYT3230 | ΔsefA | <10 | >104 |
| LK19 | sefA∷Kan | >104 | >107 |
| TYT3150 | ΔsefD | >104 | >107 |
*Only the sef genotype is shown.
† To reduce the number of mice required, we only tested two mice for each dilution of S. enteritidis; therefore, all LD50 values are approximate.
Mouse Studies.
Female BALB/c mice between 6 and 8 wk old were purchased from Harlan Breeders (Indianapolis) and housed at the University of Illinois according to local Laboratory Animal Care Advisory Committee guidelines. Overnight bacterial cultures grown in CFA media (pH 6.0) were diluted 5 × 105 in sterile 0.85% NaCl to yield approximately 104 colony-forming units (CFU) per ml, and 0.1 ml of the diluted culture was injected into the peritoneal cavity (i.p.). For competition assays, strains were grown separately overnight, diluted by 5 × 105, and then mixed in a 50:50 ratio. To estimate the i.p. LD50, 10-fold serial dilutions in 0.85% NaCl were prepared to result in a range from 107 to 100 bacteria per ml, and 0.1 ml of these samples were injected into the peritoneal cavity. To estimate the oral LD50, a similar range of dilutions was prepared in PBS. The mice were anaesthetized by brief exposure to CO2, and oral inoculations were performed with a feeding needle. All inocula were quantitated by plating serial 10-fold dilutions of the cultures onto LB plates and counting viable colonies. For mixed infections, the samples were replica plated onto LB and LB-Kan to quantitate total and mutant CFU. LD50 values were estimated by using the method of Reed and Muench (18).
At appropriate times following injection, mice were sacrificed by CO2 asphyxiation, and organs were aseptically removed and placed in 2 ml ice-cold, sterile, 0.85% NaCl. Tissues were homogenized, and then aliquots were plated onto LB plates. For mixed infections, the samples were replica plated onto LB and LB-Kan after overnight growth to allow quantitation of wild-type and mutant bacteria. The liver and spleen were routinely assayed for bacterial load; however, to study tissue tropism, other tissues were also examined (including the lungs, heart, pancreas, intestine, and kidneys).
Internalization Assays.
To quantitate the internalization of S. enteritidis in vivo, overnight cultures were diluted in 0.85% NaCl, and then approximately 2,000 bacteria were injected into the peritoneal cavity of live mice. At the indicated times after infection, mice were sacrificed by CO2 asphyxiation, surface sterilized in 70% ethanol, and the skin over the abdomen was removed leaving the peritoneum intact. An amount of 5 ml ice-cold RPMI 1640 tissue culture media (BioWhittaker) was injected into the peritoneal cavity. After 1 min, the tissue culture media was aspirated and immediately placed on ice. Two aliquots of this peritoneal lavage were compared. Gentamicin was added to a final concentration of 10 μg/ml to one aliquot, and after a 30-min incubation on ice both samples were washed by centrifugation and resuspended in sterile 0.85% NaCl and then plated on LB agar. Viable counts from the sample that was not treated with gentamicin gave the total number of bacteria, and viable counts from the gentamicin-treated sample gave the number of internalized bacteria. There was no difference in sensitivity between any of the strains used in this study to gentamicin in vitro (data not shown).
Results
Virulence of a sefA∷kan Mutant Is Attenuated in Vivo.
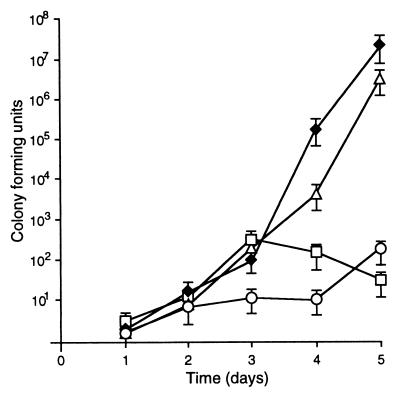
To decipher the role of SEF14 fimbriae in virulence, we examined the growth of wild-type S. enteritidis and defined sef mutants in vivo. The i.p. dose of wild-type S. enteritidis required to cause a lethal infection in 50% (LD50) of BALB/c mice was <10 bacteria (Table 1). S. enteritidis with a polar mutation in sefA (sefA∷Kan) was severely attenuated in vivo, resulting in an approximate 1,000-fold increase in LD50. In contrast, a strain with a nonpolar, in-frame deletion in sefA (ΔsefA) had a similar LD50 to wild-type bacteria. To investigate the cause of this difference in LD50, we followed the kinetics of S. enteritidis infections in vivo. Following i.p. injection, S. enteritidis is rapidly taken to the target organs (liver and spleen) where the bacteria multiply such that at 5 days postinoculation these organs contain between 107 and 108 bacteria. At this time, the mice show symptoms of advanced salmonellosis, and ultimately the mice succumb to infection approximately 6 days postinoculation. Competition assays between wild-type and mutant bacteria provide a sensitive method for analyzing the growth of Salmonella strains in vivo without requiring large numbers of animals. Equal numbers of wild-type and sef mutants were injected i.p. The liver and spleen were rapidly colonized; however, sefA∷Kan mutants were unable to persist as efficiently as wild-type bacteria (Fig. 3A). In contrast, ΔsefA mutants colonized the liver and spleen as efficiently as the wild-type bacteria, indicating that the sefA gene product is not required for virulence (Fig. 3B). In contrast, competition assays between wild-type bacteria and S. enteritidis with mutations in other fimbrial systems (i.e., PEF, LPF, FIM, AGF) showed no effect on virulence when bacteria were administered by means of the i.p. route, and there was no difference during in vitro growth between any of the strains tested (data not shown).
Figure 3.
Competition between wild-type and sefA mutants of S. enteritidis. Square symbols represent data from the spleens, and diamonds represent data from the livers. (A) Wild-type (closed symbols and solid lines) vs. sefA∷Kan (open symbols and dashed lines). (B) Wild-type (closed symbols and solid lines) vs. ΔsefA (open symbols and dashed lines). All data are the average of at least three independent samples, and error bars represent the SEM.
The SefD Subunit Is Required for Virulence.
Because the nonpolar ΔsefA mutation did not affect virulence but the polar sefA∷Kan reduced virulence, we examined the effects of nonpolar mutations in other sef genes on virulence. The time course of infection of the liver and spleen was determined for each strain (Fig. 4). The results confirm that the polar sefA∷Kan and nonpolar ΔsefD mutations attenuate growth in vivo but the nonpolar ΔsefA mutation has no effect on virulence. To confirm these results, approximate oral and i.p. LD50 values were measured. The LD50 values for the wild-type and ΔsefA strains are comparable, but both the oral and i.p. virulence of mutants lacking SefD (sefA∷Kan and ΔsefD) are greatly reduced (Table 1). Transposon mutations in sefB and sefC also showed reduced virulence as expected because these mutations are also polar on expression of sefD (data not shown). Thus, although SefA and SefD are components of the same fimbriae (6), these results show that only the SefD subunit is required for virulence.
Figure 4.
Comparison of virulence of individual S. enteritidis strains in the liver. ⧫, Wild-type S. enteritidis. ▵, ΔsefA mutants. ○, sefA∷Kan mutants. □, ΔsefD mutants. Similar data were obtained from the spleen but are not shown. All data are the average of at least three independent samples, and error bars represent the SEM.
Tissue Tropism Is Not Affected by sef Mutations.
Previous studies on the role of fimbriae in virulence have shown that different fimbriae bind to specific types of host cells (1, 2, 19, 20). Thus, it seemed possible that the SEF14 fimbriae were required for binding to different tissues during the course of infection. Examination of the tissue distribution of mutant and wild-type bacteria indicated that there was no difference in the ability of wild-type and ΔsefA strains to colonize any tissues. Although much lower numbers of sefA∷Kan and ΔsefD mutants were recovered than the wild-type bacteria, equally low numbers were recovered from all tissues and there did not appear to be any difference in the colonization of any tissue examined (data not shown). Hence, SEF14 fimbriae are probably not responsible for tissue tropism.
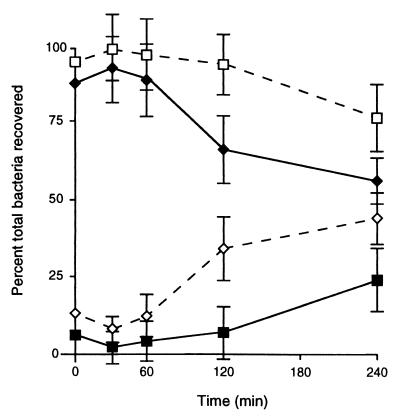
SefD Is Required for Efficient Internalization into Macrophages.
Soon after injection of bacteria into the peritoneal cavity, they are engulfed by phagocytic cells as part of the host immune response. Phagocytes kill most bacteria, however Salmonella have evolved elaborate mechanisms to evade cytotoxicity and can persist inside the phagocyte. To investigate the uptake of S. enteritidis by macrophages in vivo, we compared the localization of wild-type bacteria and ΔsefD mutants in the peritoneal cavity at different times following injection (Fig. 5). The majority of wild-type bacteria are internalized rapidly after injection (within 10 min; Fig. 5), and subsequently the total number of bacteria in the peritoneum declines as bacteria are taken from the peritoneal cavity to the liver and spleen. In contrast, the ΔsefD mutants are not internalized as efficiently and the majority of viable mutant bacteria remain extracellular. The ΔsefA mutant behaved like wild-type, remaining intracellular. In contrast, the sefA∷Kan mutant behaved like the ΔsefD mutant, remaining extracellular (data not shown).
Figure 5.
Location of S. enteritidis in the peritoneal cavity. Solid lines and solid symbols represent intracellular bacteria, and dashed lines and open symbols represent extracellular bacteria. Diamonds represent wild-type bacteria, and squares represent ΔsefD mutants. Each time point is an average from at least three independent samples, and error bars represent the SEM.
Discussion
These results indicate that the SEF14 fimbriae are essential for full virulence of S. enteritidis in vivo, and that the fimbriae mediate interactions between the bacteria and host phagocytes. Using a genetic approach with well defined, isogenic mutants, we have shown that the minor subunit, SefD, is essential for SEF14 fimbrial function in vivo. Based on these results and sequence homology between SefD and other fimbrial systems, we propose that SefD encodes the adhesin subunit of SEF14 fimbriae. The translocation of SefD is a prerequisite for the export of SefA across the outer membrane (R.A.E., B. M. Matlock, and S.R.M., unpublished observations); thus, SefD is probably located at the tip of the fimbrial shaft. The major SEF14 subunit (SefA) was not required for the virulence of S. enteritidis, indicating that the tip of the fimbrial structure composed of SefD subunits is probably sufficient for successful interactions with phagocytes. It is possible that either the major subunit is redundant and SefD is presented on another fimbrial shaft, or that SefD tip structures can successfully fulfill all the functions required of the SEF14 fimbriae in the absence of SefA. When there is insufficient SefA, SefD may form a fibrillar structure on the bacterial cell surface (Fig. 2B) similar to that described for other fimbriae (21).
The role of SEF14 fimbriae in virulence has been controversial. For example, it has been shown that SEF14 fimbriae are not involved in primary attachment to intestinal epithelia (22), but SEF14 fimbriae can illicit a strong, protective, immune response (8, 11). The results presented here explain this paradox: SEF14 fimbriae are essential for proper interactions with macrophages subsequent to the colonization and penetration of the intestinal barrier.
In addition to demonstrating a role for SEF14 fimbriae in virulence, the results provide insight into the growth of Salmonella in vivo. It has previously been shown that rapidly after infection S. enteritidis transit from the peritoneum to the liver and spleen (23, 24), where they infect Kupffer cells, macrophages associated with liver sinusoids (25). For approximately 2–3 days, the number of bacteria increase slowly. During this time, Salmonella multiply inside the Kupffer cells, and any free bacteria released into the sinusoids by macrophage lysis are killed by neutrophils (25). The number of bacteria seen over the first 3 days presumably represents the equilibrium established between growth inside Kupffer cells and bactericidal activity of neutrophils (26). This cycle of infection of macrophages, release into the environment, and uptake into either neutrophils (lethal) or macrophages (safe havens) is thought to allow Salmonella to persist for up to 48 hr at the site of infection (27, 28). During this time, the infective foci include a large number of neutrophils and a small number of macrophages. After 48–72 hr, the neutrophils in the infective foci are replaced with activated macrophages, which in a normal host would be able to ultimately clear the infection (29, 30). However, BALB/c mice contain a mutation in NRAMP-1 (natural resistance associated macrophage protein) that makes them susceptible to Salmonella infections by reducing the efficacy of the activated macrophage onslaught (31). The data shown in Figs. 3 and 4 demonstrate that S. enteritidis persists in the liver and spleen for the first 3 days but is not able to grow rapidly. However, as resident macrophages and neutrophils are replaced by activated macrophages, it is clear that S. enteritidis begins growing at an almost exponential rate until the mice succumb. In contrast, S. enteritidis that contain either the sefA∷Kan or the ΔsefD mutations are no longer able to compete once the neutrophils are replaced by activated macrophages. Unless the immune system is overwhelmed with very large numbers of bacteria, mutant bacteria are eventually cleared and the mice survive the infections. The difference between wild-type and mutant bacteria becomes accentuated starting 3 days postinfection (Fig. 4), indicating that SefD is required for S. enteritidis to survive the assault by activated macrophages.
An initial step in virulence of Salmonella in mice involves attachment to the phagocyte surface before the bacteria are ingested and transported by the lymph system to the liver and spleen (23). SEF14 fimbriae could be involved in either attachment to the macrophage surface or survival after uptake. Attachment mediated between adhesins on the bacterial surface and receptors on the phagocyte surface has been characterized in many bacteria (32–34). SefD may bind to a receptor on the macrophage surface and alter the uptake of S. enteritidis into the phagocyte so that S. enteritidis can survive in the intracellular environment. In contrast, mutants lacking sefD may be taken up by pathways that result in a hostile response from the phagocyte and lead to bacterial cell death. Alternatively, SefD may be required subsequent to uptake into the phagosome in a similar manner to that which has been proposed for the Y. pestis (pH 6) antigen (6, 12).
This is the first Salmonella fimbriae that has been shown to play an essential role in interactions with macrophages. All other Salmonella fimbriae that have been shown to be required for virulence are involved in binding to epithelial surfaces. Since S. typhimurium does not contain the sef operon, it seems likely that one or more of the currently uncharacterized fimbrial operons, or another adhesive structure (such as an afimbrial adhesin) may perform the functions in S. typhimurium that SEF14 fimbriae perform in S. enteritidis. The specific interactions between fimbrial adhesins and macrophages may be important determinants in the different etiology and epidemiology of the variety of S. enterica serovars.
Acknowledgments
We thank D. Guiney, K. Hughes, M. Laird, S. Libby, J. L. Puente, and R. K. Taylor for comments on the manuscript; and S. Libby and F. Fang for experimental advice. This work was supported in part by grants from the National Institutes of Health and the Illinois Council for Food and Agriculture Research.
Abbreviations
- SEF
Salmonella enteritidis fimbriae
- Kan
kanamycin sulfate
- CFA
colonization antigen factor
- CFU
colony-forming unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Low D, Braaten B, van der Woude M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 146–157. [Google Scholar]
- 2.Edwards R A, Puente J L. Trends Microbiol. 1998;6:282–287. doi: 10.1016/s0966-842x(98)01288-8. [DOI] [PubMed] [Google Scholar]
- 3.Bäumler A J, Tsolis R M, Heffron F. Adv Exp Med Biol. 1997;412:149–158. [PubMed] [Google Scholar]
- 4.Sauer F G, Futterer K, Pinkner J S, Dodson K W, Hultgren S J, Waksman G. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 5.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S D. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 6.Lindler L E, Tall B D. Mol Microbiol. 1993;8:311–324. doi: 10.1111/j.1365-2958.1993.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 7.Clouthier S C, Muller K H, Doran J L, Collinson S K, Kay W W. J Bacteriol. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogunniyi A, Manning P A, Kotlarski I. Infect Immun. 1994;62:5376–5383. doi: 10.1128/iai.62.12.5376-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peralta R C, Yokoyoma H, Ikemori Y, Kuroki M, Kodama Y. J Med Microbiol. 1994;41:29–35. doi: 10.1099/00222615-41-1-29. [DOI] [PubMed] [Google Scholar]
- 10.Thorns C J, Turcotte C, Gemmell C G, Woodward M J. Microb Pathog. 1996;20:235–246. doi: 10.1006/mpat.1996.0022. [DOI] [PubMed] [Google Scholar]
- 11.Ogunniyi A D, Kotlarski I, Morona R, Manning P A. Infect Immun. 1997;65:708–717. doi: 10.1128/iai.65.2.708-717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Merriam J J, Mueller J P, Isberg R R. Infect Immun. 1996;64:2483–2489. doi: 10.1128/iai.64.7.2483-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards R A, Keller L H, Schifferli D M. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 14.Keller L H, Benson C E, Garcia V, Nocks E, Battenfelder P, Eckroade R J. Avian Dis. 1993;37:501–507. [PubMed] [Google Scholar]
- 15.Maloy S R, Stewart V J, Taylor R K. Genetic Analysis of Pathogenic Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 16.Müller K H, Collinson S K, Trust T J, Kay W W. J Bacteriol. 1991;173:4765–4772. doi: 10.1128/jb.173.15.4765-4772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann B A, Slauch J M. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed L J, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 19.Bäumler A J, Tsolis R M, Heffron F. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bäumler A J, Tsolis R M, Bowe F A, Kusters J G, Hoffmann S, Heffron F. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones C, Pinkner J, Roth R, Heuser J, Nicholes A, Abraham S, Hultgren S. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dibb-Fuller M P, Allen-Vercoe E, Thorns C J, Woodward M J. Microbiology. 1999;145:1023–1031. doi: 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- 23.Buchmeier N A, Heffron F. Infect Immun. 1991;59:2232–2238. doi: 10.1128/iai.59.7.2232-2238.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small P L. Infect Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nnalue N A, Shnyra A, Hultenby K, Lindberg A A. Infect Immun. 1992;60:2758–2768. doi: 10.1128/iai.60.7.2758-2768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conlan J W. Infect Immun. 1997;65:630–635. doi: 10.1128/iai.65.2.630-635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conlan J W. Infect Immun. 1996;64:1043–1047. doi: 10.1128/iai.64.3.1043-1047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunlap N E, Benjamin W H, Jr, McCall R D, Jr, Tilden A B, Briles D E. Microb Pathog. 1991;10:297–310. doi: 10.1016/0882-4010(91)90013-z. [DOI] [PubMed] [Google Scholar]
- 29.Richter-Dahlfors A, Buchan A M J, Finlay B B. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassiloyanakopoulos A P, Okamoto S, Fierer J. Proc Natl Acad Sci USA. 1998;95:7676–7681. doi: 10.1073/pnas.95.13.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Govoni G, Vidal S, Cellier M, Lepage P, Malo D, Gros P. Genomics. 1995;27:9–19. doi: 10.1006/geno.1995.1002. [DOI] [PubMed] [Google Scholar]
- 32.Krukonis E S, Dersch P, Eble J A, Isberg R R. J Biol Chem. 1998;273:31837–31843. doi: 10.1074/jbc.273.48.31837. [DOI] [PubMed] [Google Scholar]
- 33.Ishibashi Y, Arai T. Microb Pathog. 1996;21:435–446. doi: 10.1006/mpat.1996.0074. [DOI] [PubMed] [Google Scholar]
- 34.Baorto D M, Gao Z, Malaviya R, Dustin M L, van der Merwe A, Lublin D M, Abraham S N. Nature (London) 1997;389:636–639. doi: 10.1038/39376. [DOI] [PubMed] [Google Scholar]