Abstract
Expression of the fis1 gene from flax (Linum usitatissimum) is induced by a compatible rust (Melampsora lini) infection. Infection of transgenic plants containing a β-glucuronidase (GUS) reporter gene under the control of the fis1 promoter showed that induction is highly localized to those leaf mesophyll cells within and immediately surrounding rust infection sites. The level of induction reflects the extent of fungal growth. In a strong resistance reaction, such as the hypersensitive fleck mediated by the L6 resistance gene, there is very little fungal growth and a microscopic level of GUS expression. Partially resistant flax leaves show levels of GUS expression that were intermediate to the level observed in the fully susceptible infection. Sequence and deletion analysis using both transient Agrobacterium tumefaciens expression and stable transformation assays have shown that the rust-inducible fis1 promoter is contained within a 580-bp fragment. Homologs of fis1 were identified in expressed sequence tag databases of a range of plant species including dicots, monocots, and a gymnosperm. Homologous genes isolated from maize (Zea mays; mis1), barley (Hordeum vulgare; bis1), wheat (Triticum aestivum; wis1), and Arabidopsis encode proteins that are highly similar (76%–82%) to the FIS1 protein. The Arabidopsis homologue has been reported to encode a Δ1-pyrroline-5-carboxylate dehydrogenase that is involved in the catabolism of proline to glutamate. RNA-blot analysis showed that mis1 in maize and the bis1 homolog in barley are both up-regulated by a compatible infection with the corresponding species-specific rust. The rust-induced genes homologous to fis1 are present in many plants. The promoters of these genes have potential roles for the engineering of synthetic rust resistance genes by targeting transgene expression to the sites of rust infection.
For successful parasitism of plants by fungal pathogens, particularly in the case of obligate, biotrophic phytopathogens such as rust fungi, it is apparent that the pathogen has evolved strategies for manipulating host cellular metabolism, morphology, and development (Clancy and Coffey, 1980; Sutton and Shaw, 1982, 1986; Scholes, 1992; Roy, 1993; Chou et al., 2000; Hall and Williams, 2000). Successful infection of the host plant by the pathogen requires the induction of a subset of pathogen genes that are essential for pathogenicity (Pieterse et al., 1993; Talbot et al., 1993; Rogers et al., 1994; Hwang et al., 1995; Hahn and Mengden, 1997; Hahn et al., 1997). Less clear is the role and expression of specific host genes that may be required for successful pathogen infection of the plant. Identification of host genes induced during a successful pathogen infection may provide insight into the compatible host/pathogen interaction at a molecular level.
The fis1 gene of flax (Linum usitatissimum) was identified as a host gene up-regulated during a successful or compatible flax rust (Melampsora lini) infection (Roberts and Pryor, 1995). RNA-blot analysis of fis1 demonstrated that this gene was induced during a compatible flax rust infection, but its induction was not detectable during an incompatible infection when rust growth was halted by the action of the L6 resistance gene. Homology searches suggested that the fis1 gene encoded an aldehyde dehydrogenase enzyme of unknown substrate specificity (Roberts and Pryor, 1995). Deuschle et al. (2001) recently have isolated a gene from Arabidopsis, AtP5CDH, which encodes a protein with 82% identity to the predicted FIS1 protein. The AtP5CDH gene was isolated by screening an Arabidopsis cDNA library in a yeast (Saccharomyces cerevisiae) mutant deficient for the enzyme Δ1-pyrroline-5-carboxylate dehydrogenase. The AtP5CDH enzyme catalyzes the second step in the degradation of Pro to Glu, specifically the conversion of pyrroline-5-carboxylate to Glu (Deuschle et al., 2001). Pyrroline-5-carboxylate is potentially a toxic intermediate and thus the Δ1-pyrroline-5-carboxylate dehydrogenase enzyme may provide protection from Pro toxicity (Deuschle et al., 2001).
In this report, we describe the characterization of the promoter and expression pattern of the fis1 gene in response to rust infection and the identification of related genes in a diverse range of plant species. Homologs of the fis1 gene in both maize (Zea mays) and barley (Hordeum vulgare) are also shown to be up-regulated in response to rust infection, demonstrating the generality of this response. The highly localized up-regulation of this promoter in response to a compatible rust infection makes it a candidate for controlling the expression of synthetic rust disease resistance genes. In addition, the similarity of the protein products of these genes to Δ1-pyrroline-5-carboxylate dehydrogenase suggests that Pro catabolism plays an as yet undefined role in the interaction between rust fungi and the host plant.
RESULTS
Cellular Localization of the Rust-Dependent Induction of the fis1 Promoter
Previous RNA-blot analysis had indicated that expression of the fis1 gene was induced in flax leaves in response to infection with a compatible race of flax rust (Roberts and Pryor, 1995). To determine the cellular localization of the expression of this gene in response to rust infection, the promoter of the fis1 gene was used to express a β-glucuronidase (GUS) reporter gene in transgenic flax plants infected with rust.
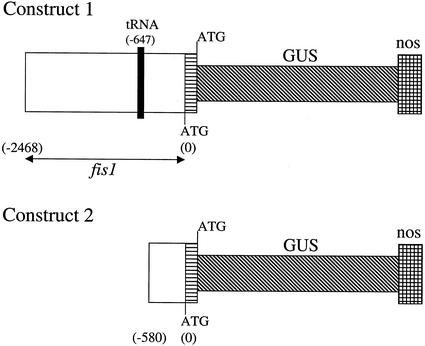
The promoter of the fis1 gene was obtained from a lambda genomic clone of the fis1 gene. Partial sequence analysis of this clone identified a 2.5-kb BamHI fragment that contained the 5′-flanking sequence and 5′-untranslated region of the gene, in addition to encoding the first 33 amino acids of the FIS1 protein. This BamHI fragment, called fis1-2.5P, was used to produce a promoter-GUS fusion construct (Fig. 1, construct 1). This construct encodes a fusion protein of the first 33 amino acids of the FIS1 protein fused in frame to the GUS protein, expressed by the putative fis1-2.5P promoter.
Figure 1.
Design of gene constructs used for flax transformation. Construct 1, fis1-2.5P-GUS consists of a 2.5-kb DNA fragment that contains the fis1 promoter and 5′-untranslated region (white box underlined with an arrow), in addition to the first 33 codons of the fis1 open reading frame (ORF; horizontally hatched box) fused in frame to a GUS reporter gene (obtuse hatched box). Also encoded on this 2.5-kb fragment is a tRNA-Ala gene depicted as a black box and described in the text. An Agrobacterium tumefaciens nopaline synthase (nos) 3′ transcription terminator is present at the 3′ end of this construct (checkered box). Construct 2, fis1-0.6P-GUS differs from fis1-2.5P-GUS in that 1,889 bp of extreme 5′ sequence has been deleted from the 2.5-kb fragment encoding the fis1 promoter. This construct uses the first 580 bp of sequence immediately upstream of the translation initiation site of the fis1 ORF for expression. On each map, numbering refers to the distance in bp from the translation start site of the fis1 ORF.
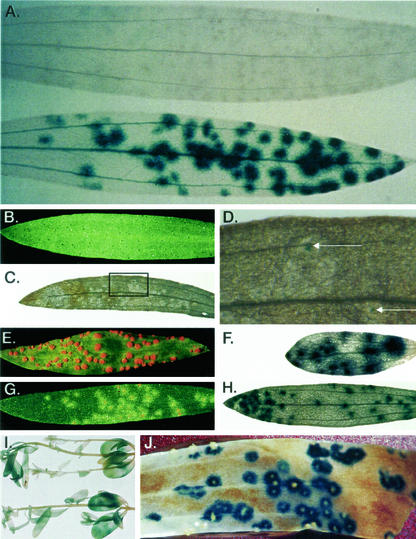
The fis1-2.5P-GUS construct (Fig. 1, construct 1) was introduced into the flax varieties Hoshangabad and Ward and plants containing the promoter-GUS transgene inoculated with flax rust race CH5, which established a compatible rust infection upon these plants. Transgenic plants were stained for GUS activity 6 d postinfection, by which time developing rust pustules were clearly observed within the infected leaf. A highly localized pattern of GUS expression in leaf mesophyll cells was observed around the rust infection sites (Fig. 2A, lower leaf). Weaker GUS expression was also present in the plant vascular tissue (Fig. 2A, lower leaf) that was not observed in untransformed plants (Fig. 2A, upper leaf). No GUS expression was observed in untransformed flax plants that had also been infected with flax rust strain CH5, demonstrating that GUS activity was not derived from the fungus (Fig. 2A, upper leaf). Plants containing the fis1-2.5P-GUS transgene that were not infected with rust showed the weaker GUS expression within the vascular tissue; however, no GUS expression was observed elsewhere within the leaf (not shown). It was concluded that the abundant and highly localized host expression of the GUS reporter gene around sites of rust infection was in response to the rust pathogen.
Figure 2.
Phenotypes of rust-infected fis1-GUS transgenic flax plants. A, Expression of the fis1-2.5P-GUS transgene in flax plants infected with a compatible race of rust. Both leaves have been infected with flax rust for 6 d, stained for GUS activity, and cleared. The upper leaf is derived from a nontransgenic control plant, whereas the lower leaf is from a flax plant containing the fis1-2.5P-GUS transgene. Each GUS-staining spot on the lower leaf corresponds precisely to a rust infection site. Small brown dots visible on the upper leaf were developing rust pustules. B through F, Expression of the fis1-2.5P-GUS transgene in flax plants during a highly incompatible rust infection. B, Leaf from a flax plant containing an fis1-2.5P-GUS transgene and an endogenous L6 resistance gene, 8 d after infection with rust strain CH5 that contains the corresponding A-L6 avirulence gene. A second rust infected leaf from the same plant was GUS stained (C) and only a small amount of GUS expression was detected around infection sites (D). The boxed region in C is enlarged in D. As a control, the same plant line was infected with a fully compatible rust race (E) and abundant GUS expression was detected around infection sites (F). G and H, Expression of the fis1-2.5P-GUS transgene during an intermediate resistance response. G, Leaf from a flax plant containing an fis1-2.5P-GUS transgene and an endogenous P3 resistance gene, 8 d after infection with rust strain CH5 which contains the corresponding A-P3 avirulence gene. A second rust infected leaf from this plant was GUS stained and is shown in H. I, Two flax seedlings GUS stained and cleared, 3 d after infiltration with an A. tumefaciens strain containing a GUS reporter gene under the control of a 35S promoter. An intron was incorporated into the GUS gene to prevent A. tumefaciens expression of a functional GUS protein. J, Flax leaves that had been infected with flax rust for 5 d were vacuum infiltrated with an A. tumefaciens strain containing the fis1-2.5P-GUS transgene. Three days postinfiltration leaves were stained for GUS activity and cleared. GUS activity is observed exclusively around rust infection sites and orange rust pustules can be seen inside these GUS-staining regions.
Expression of the fis1 Gene in Response to Incompatible Flax Rust Infections
To determine if the fis1 promoter up-regulates gene expression during an incompatible flax rust infection, a transgenic plant homozygous for the fis1-2.5P-GUS construct was crossed to flax lines Birio and Leona, which are homozygous for resistance genes L6 and P3, respectively. Progeny derived from these crosses were hemizygous for the fis1-2.5P-GUS transgene and heterozygous for the indicated rust resistance gene. The same fis1-2.5P-GUS line was used as a parent for each cross, thereby eliminating potential transgene position effects. Hybrid plants were challenged with rust race CH5, which is recognized by each resistance gene and gives an incompatible rust infection in each case. Infected plants were then analyzed for GUS gene expression 8 d postinoculation.
The resistance response mediated by the L6 rust resistance gene is a hypersensitive fleck that halts fungal growth completely and prevents fungal sporulation (Fig. 2B). Eight days after infection with rust strain CH5, no clear GUS activity was observed on plants containing the L6 resistance gene and construct 1 (Fig. 2C). However, microscopic analysis identified distinct small regions of GUS activity (Fig. 2D). Previous RNA-blot analysis of the fis1 gene could not identify induction of the fis1 promoter in the L6/A-L6-incompatible interaction (Roberts and Pryor, 1995). This discrepancy is likely to reflect the relative insensitivity of a total leaf RNA assay when analyzing such a highly localized gene induction event. When F1 plants derived from the same cross were challenged with a rust race that is not recognized by the L6 resistance gene, a compatible interaction was observed (Fig. 2E), which gave rise to a highly localized induction of GUS gene expression 8 d postinoculation (Fig. 2F).
Unlike L6, the resistance response mediated by the P3 resistance gene leads to partial resistance. Small pustules surrounded by chlorotic cells develop on plants carrying this resistance gene when challenged with an avirulent race of flax rust containing the corresponding A-P3 avirulence gene (Fig. 2G). Plants carrying the P3 gene in addition to the fis1-2.5P-GUS construct showed significant GUS activity surrounding the sites of rust infection 8 d postinoculation when challenged with strain CH5, which carries the A-P3 avirulence gene (Fig. 2H). This level of GUS expression, however, was less than that observed when plants were challenged with a fully virulent race of rust (Fig. 2F). The amount of fis1 promoter induction during incompatible interactions, therefore, appears to be correlated with the extent of fungal growth permitted by the plant host.
Deletion Analysis of the fis1 Promoter Using a Transient Expression Assay
To expedite analysis of the fis1 promoter, a transient expression assay was developed based upon infiltration of A. tumefaciens cultures into rust-infected flax plants. A number of plant species have been shown to transiently express a 35S-intron-GUS reporter gene when vacuum infiltrated with A. tumefaciens strains carrying this reporter gene in a binary transformation vector (Kapila et al., 1997). Substantial transient GUS expression is also observed in flax plants when infiltrated with an A. tumefaciens strain carrying a 35S-intron-GUS reporter gene within a binary vector (Fig. 2I).
Flax plants that been infected with a virulent rust race were vacuum infiltrated with an A. tumefaciens strain containing the fis1-2.5P-GUS construct (Fig. 1). The pattern of transient GUS expression observed in these plants 3 d postinfiltration was similar to that observed for stably transformed flax plants containing the same gene construct (Fig. 2J). Transient expression of the fis1-2.5P-GUS reporter gene was specifically localized around the foci of flax rust infection (Fig. 2J). Unlike stably transformed plants, however, not all rust pustules were surrounded by GUS activity, consistent with the patchy expression typically observed in transient vacuum infiltration assays (Kapila et al., 1997). In addition, vascular expression of the fis1 promoter was rarely observed, presumably reflecting an inability of the A. tumefaciens to successfully infect this tissue via the infiltration process. No GUS gene expression was observed in A. tumefaciens-infiltrated plants that had not been infected with flax rust (not shown). Therefore, this transient expression assay parallels the expression of the fis-2.5P-GUS construct in stably transformed flax and provides a useful tool for the molecular analysis of the fis1 promoter.
From the above analysis, it is apparent that the 2.5-kb BamHI fragment used in the fis1-2.5P-GUS construct (Fig. 1) contains the rust-inducible fis1 promoter. This BamHI fragment was subsequently sequenced (accession no. AF467540) and compared with the GenBank and EMBL databases by a BLAST search. A 76-bp sequence was identified 647 bp upstream of the translation start site of the FIS1 protein, which shows 92% DNA sequence identity to tRNA-Ala genes from both potato (Solanum tuberosum) and Arabidopsis (accession nos. Z28961 and X17757; Fig. 1, construct 1). From these sequence data, it was predicted that the fis1 promoter was contained within the first 647 bp of DNA upstream from the translation initiation site of the fis1 ORF.
To test this hypothesis, an fis1-2.5P-GUS deletion construct (Fig. 1, construct 2) was made using a PstI site located 580 bp upstream of the fis1 translation start site. This generated a construct containing 580 bp of sequence 5′ of the fis1 translation initiation site, with the first 33 amino acids of the fis1 ORF fused in frame to a GUS reporter gene. This construct was named fis1-0.6P-GUS. An A. tumefaciens strain containing fis1-0.6P-GUS was vacuum infiltrated into mature flax plants that had established an 8-d-old rust infection. Analysis of these plants 3 d postinfiltration identified a pattern of GUS expression indistinguishable to that observed for transient expression of the fis1-2.5P-GUS construct (not shown). This transient assay indicates that a functional fis1 promoter lies within the first 580 bp of DNA sequence that is upstream from the translation start site of the fis1 ORF.
Isolation and Analysis of a Related fis1 Gene from Maize, mis1
A previous database search identified some sequence similarity between the fis1 ORF and a number of genes encoding aldehyde dehydrogenase enzymes (Roberts and Pryor, 1995). Protein sequence alignment identified four amino acid motifs that are characteristic of aldehyde dehydrogenase enzymes and that occur within the FIS1 protein. These motifs have been described previously as the GPL, GSG, GQC, and EEP motifs, respectively (Roberts and Pryor, 1995). Degenerate PCR primers were designed based upon the amino acid sequence of the GPL and GQC motifs present in the FIS1 protein. These primers, when used for reverse transcriptase (RT)-PCR of maize leaf RNA amplified a 434-bp fragment that was cloned and partially sequenced. This cloned fragment showed 72% DNA sequence identity to the flax fis1 cDNA sequence and encoded an ORF of 108 amino acids that is 87% identical and 94% similar to the FIS1 protein. This cloned PCR fragment therefore was derived from a maize gene related to the flax fis1 gene, which we have named mis1.
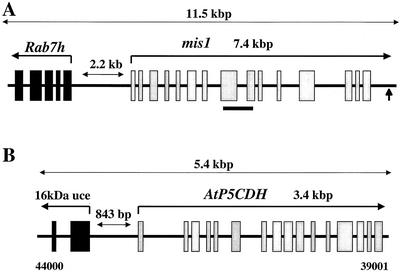
A maize cDNA library derived from leaf tissue infected with maize rust (Puccinia sorghi) was screened with the cloned mis1 PCR product and a number of cDNA clones identified, six of which were subsequently sequenced. A maize genomic library was also screened with the cloned mis1 PCR product and a genomic clone of the mis1 gene isolated. From a comparison between an 11.5-kb sequenced region of this genomic clone and the isolated cDNA clones, it was concluded that the mis1 gene contains 15 exons and 14 introns (Fig. 3A) and is predicted to encode a 549-amino acid protein that is 77% identical and 82% similar to the FIS1 protein (accession no. AF467541).
Figure 3.
Structure of the mis1 and AtP5CDH genes. A, Schematic diagram (not to scale) of the mis1 gene and flanking sequence. The 15 exons of the mis1 gene (transcribed from left to right) are represented as gray boxes with the polyadenylation site positions indicated with a vertical arrow. Upstream of the mis1 gene is part of a second predicted gene encoding a maize Rab7 homolog, labeled Rab7h. The first five exons of the predicted Rab7h gene are shown as black boxes with the direction of transcription shown as a bent arrow. The small line beneath the figure delineates the 434-bp RT-PCR fragment from the mis1 gene (excluding the intervening intron) used as a probe in Figures 4 and 6. Horizontal arrows indicate distances in kb. B, Schematic diagram of the AtP5CDH gene and flanking regions. The 16 exons of the gene, which are transcribed from left to right, are shown as gray boxes. A second gene encoding a 16-kD ubiquitin-conjugating enzyme (16 kD uce) is located 843 bp 5′ of the first exon of the gene. The first exon and part of the second exon of this gene are shown as black boxes. The direction of transcription of the ubiquitin conjugating enzyme gene is shown as a bent arrow. Horizontal arrows indicate distances in kb and bp. Numbers beneath the figure refer to the nucleotide position of this sequence on Arabidopsis bacterial artificial chromosome (BAC) clone K19B1 (GenBank accession no. AB015469).
Sequence analysis of the genomic mis1 lambda clone identified a second gene located 2.2 kb 5′ of the mis1 translation initiation site (Fig. 3A). Theoretical translation of this region of sequence identifies five potential exons that encode for a 138-amino acid protein that shows 85% and 83% identity and 92% and 91% similarity to the RAB7 proteins of Lotus japonicus (accession no. CAA98168) and soybean (Glycine max; accession no. Q43463), respectively. It is not known if this region of the maize genome encodes a full-length RAB7 protein because the lambda sequence analysis encompassed only the 5′ end of this gene. The Rab7 gene encodes a member of a superfamily of small GTP-binding proteins that have been implicated in targeting of membrane vesicles (Nuoffer and Balch, 1994). Because the mis1 and Rab7h genes are transcribed in opposite directions, the promoter elements and 5′-untranslated regions of both these genes presumably occur within the 2.2 kb of intervening sequence (Fig. 3A).
fis1-/mis1-Related Genes Occur in a Range of Plant Species, Including Wheat (Triticum aestivum) and Barley
DNA-blot analysis using the cloned 434-bp RT-PCR product from the mis1 gene as a probe (Fig. 3A) identifies related sequences in nine grass species examined, suggesting the presence of functional mis1 equivalents in these species (Fig. 4). BLAST searches of expressed sequence tag (EST) databases with the fis1 cDNA sequence identified highly homologous, expressed genes in a range of plant species encompassing monocot, dicot, and gymnosperm taxa (Table I). These EST sequences were derived from diverse tissue sources including root nodules, fruits, flowers, and xylem demonstrating a broad expression pattern of these fis1/mis1 homologs (Table I). Maize ESTs of the mis1 gene are also present in cDNA libraries derived from ear tissue, leaf primordial, and early embryo tissue (accession nos. AI691509, AI629459, and AW225209). The identification of related fis1 transcripts in all of these tissues may simply parallel the vascular expression of fis1 in flax or alternatively may be a consequence of fis1-like gene expression in other cell types in these species.
Figure 4.
Grass species contain sequences homologous to mis1. DNA blot showing hybridization of the 434-bp mis1 probe to NcoI-digested total DNA isolated from pearl millet (Pennisetum glaucum), panicum (Panicum miliaceum), maize, sugarcane (Saccharum officinarum), barley, wheat, oats (Avena sativa), Aegilops tauschii, sorghum (Sorghum bicolor), and rice (Oryza sativa; lanes 1–10, respectively). Mr sizes are indicated on the figure.
Table I.
EST homologs of the fis1 gene
| Species | Accession No. | Tissue | DNA Identity | Region of Protein Homology | Protein ID | Protein Similarity |
|---|---|---|---|---|---|---|
| % | % | |||||
| Soybean | AI973927 | Root nodules | 75 | 317–369 | 85 | 94 |
| Cotton (Gossypium hirsutum) | AI727249 | Fiber | 75 | 360–550 | 83 | 88 |
| Tomato (Lycopersicon esculentum) | AW44168 | Fruit | 76 | 196–210 | 81 | 87 |
| AW217685 | Flower bud | |||||
| Rice | C72049 | Panicle | 84 | 1–62 | 69 | 74 |
| Wheat | *ITEC 12262 | Rust infected | 73 | 1–170 | 60 | 64 |
| Barley | *ITEC 6057 | Leaf | 73 | 323–517 | 77 | 84 |
| Loblolly pine (Pinus taeda) | AA556272 | Xylem | 84 | 206–304 | 72 | 78 |
| Arabidopsis | T04386 | Mixed tissue | 74 | 294–375 | 82 | 88 |
Each EST was translated and the similarity and identity to the predicted FIS1 protein determined. The region of the FIS1 protein that was homologous to these translated ESTs is also indicated. This table is not exhaustive and homologues were identified in a no. of other plant species. *Wheat and barley ESTs were identified in the International Tritici EST Consortium database.
Wheat and barley EST clones homologous to mis1 were obtained as described in “Materials and Methods” and sequenced in full. From this sequence analysis, it was determined that a gene in wheat, wis1, expresses a 2-kb transcript encoding a 551-amino acid protein that is 76% and 91% identical and 81% and 93% similar to the predicted FIS1 and MIS1 proteins, respectively (accession no. AF467542). A homologous gene in barley, bis1, expresses a 2-kb transcript encoding a 551-amino acid protein that is 76%, 90%, and 98% identical and 79%, 93%, and 99% similar to FIS1, MIS1, and WIS1, respectively (accession no. AF467539).
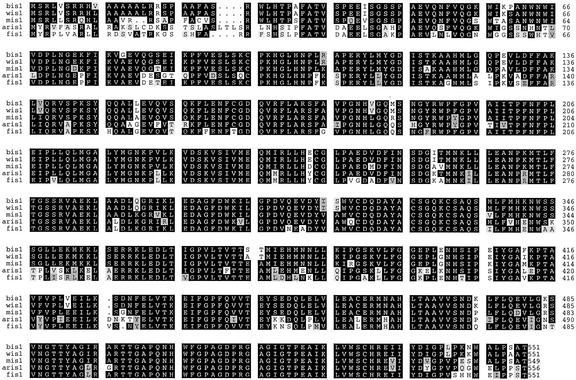
Deuschle et al. (2001) have recently isolated an Arabidopsis homologue of the flax fis1 gene and shown that the protein encoded by this gene has Δ1-pyrroline-5-carboxylate dehydrogenase activity. Independently, we have also characterized the Arabidopsis equivalent of the fis1 gene by sequencing Arabidopsis EST clone 36F11T7 (accession no. T04386), characterizing the 5′ region of this mRNA by RT-PCR and analyzing a genomic Arabidopsis BAC clone homologous to this sequence (BAC clone K19B1; accession no. AB015469). The cDNA sequence determined from this analysis was identical to a published Arabidopsis cDNA sequence (accession no. AY065072). Our molecular analysis of this gene essentially confirms that of Deuschle et al. (2001), although 18 base differences were observed in the ORFs of these genes. The predicted protein from our cDNA analysis differs from Deuschle et al. (2001) in that an S to A substitution occurs at amino acid 48 and a D to E substitution occurs at position 462. The Columbia ecotype was used for our analysis, whereas Landsberg was used by Deuschle et al. (2001), which may explain these differences. The Arabidopsis homolog of fis1, AtP5CDH, consists of 16 exons and 15 introns (Fig. 3B) and encodes a predicted 555-amino acid protein that is 82%, 77%, 76%, and 77% identical and 86%, 81%, 81%, and 82% similar to the FIS1, MIS1, BIS1, and WIS1 proteins, respectively. A PileUp alignment between these predicted proteins shows that most of the sequence differences between these conserved proteins resides in the NH2 terminus (Fig. 5).
Figure 5.
A PileUp sequence alignment of the predicted BIS1, WIS1, MIS1, AtP5CDH, and FIS1 proteins. Accession numbers for these sequences are as follows: AAL70106, AAL70109, AAL70108, AAK73756, and CAA60412, respectively. The AtP5CDH protein is labeled ′aris1′ in this alignment. Identical amino acids are shaded in black, whereas similar amino acids are highlighted in gray. Numbers on the right hand side indicate amino acid positions.
The AtP5CDH protein is targeted to the mitochondrion and probably localized in the mitochondrial matrix (Deuschle et al., 2001). Analysis of the amino-terminal 35 amino acids of the FIS1, MIS1, WIS1, BIS1, and AtP5CDH proteins using programs Target P, version 1.0, and PSORT predicted that all five proteins were targeted to the mitochondrion and localized in either the mitochondrial matrix or intermembrane space. Thus, the FIS1, MIS1, WIS1, and BIS1 proteins may also function as Δ1-pyrroline-5-carboxylate dehydrogenases given their predicted mitochondrial location and high similarity to the AtP5CDH protein.
Comparison of the predicted intron/exon structure of the AtP5CDH gene with the characterized mis1 gene also shows a high degree of similarity. Twelve of the 16 exons of the AtP5CDH gene are identical in size to the equivalent exons of the mis1 gene (Table II). These two genes differ in that exon 8 of the mis1 gene, which encodes 73 amino acids, contains an additional intron in the AtP5CDH gene, giving rise to two exons that encode for 35 and 38 amino acids, respectively. Exons 1 and 13 of mis1 and AtP5CDH also differ in size (Table II). Whereas exon size and intron position are conserved between mis1 and the AtP5CDH gene, intron size is highly variable with introns in the maize gene generally being larger than those of the Arabidopsis gene (not shown).
Table II.
Comparison of exon sizes between the AtP5CDH gene and mis1 gene
| Exon | AtP5CDH | mis1 |
|---|---|---|
| 1 | 30 | 24 |
| 2 | 27 | 27 |
| 3 | 34 | 34 |
| 4 | 23 | 23 |
| 5 | 19 | 19 |
| 6 | 38 | 38 |
| 7 | 28 | 28 |
| 8 | 35 | 73 |
| 9 | 38 | – |
| 10 | 44 | 44 |
| 11 | 30 | 30 |
| 12 | 30 | 30 |
| 13 | 71 | 70 |
| 14 | 37 | 37 |
| 15 | 27 | 27 |
| 16 | 45 | 45 |
The no. of codons in each exon is given. Exon 8 of the mis1 gene appears to be a composite of exons 8 and 9 of the AtP5CDH gene.
Analysis of sequence 5′ of the AtP5CDH gene identifies a second gene encoding a 16-kD ubiquitin-conjugating enzyme (Sullivan et al., 1994; Fig. 3B). Nucleotides 43,315 through 44,000 of BAC clone K19B1 contain two regions of 182 and 28 bp that are 100% homologous to exon 1 and part of exon 2 of the published Arabidopsis ubiquitin-conjugating enzyme sequence (accession no. L19352), whereas the intervening intron sequence is identical to intron 1 of the published sequence (Sullivan et al., 1994) except that it contains an additional 70 bp. Because the 16-kD ubiquitin-conjugating enzyme in Arabidopsis is encoded by a small multigene family (Sullivan et al., 1994), this difference in intron size is likely due to these two genes being different members of the same multigene family. Nucleotides 43,315 through 44,000 of clone K19B1, therefore, encode exon 1, intron 1, and part of exon 2 of a 16-kD ubiquitin-conjugating enzyme (Fig. 3B). The translation initiation site of the AtP5CDH gene and the first exon of the 16-kD ubiquitin-conjugating enzyme are separated by 843 bp. Because these two genes are transcribed in opposite directions, the promoters of both genes presumably occur within this 843-bp region of sequence.
Sequence comparison between the 2.2-kb region separating the mis1 gene from a maize Rab7 homolog, the 843-bp region separating the AtP5CDH gene from a 16-kD ubiquitin-conjugating enzyme, and the 580-bp fragment containing a functional fis1 promoter, show little similarity (data not shown). Some small blocks of homology are observed between these three predicted promoter regions although the functional significance of these regions of sequence is yet to be determined. A deletion analysis of the fis1 promoter currently is being conducted to identify functional regulatory elements involved in rust-induced expression.
The mis1 Gene and Related Barley bis1 Gene Are Up-Regulated during a Compatible Rust Infection
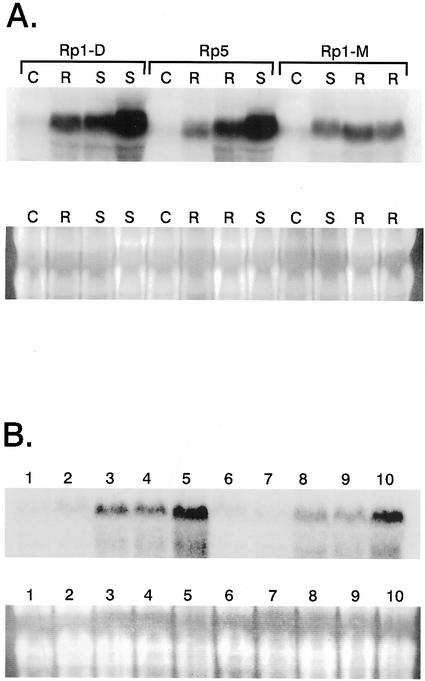
The effect of rust infection on the induction of the mis1 gene in maize was investigated in both compatible and incompatible interactions by using the mis1 434-bp PCR fragment as a hybridization probe in RNA-blot analysis (Fig. 6A). Seedlings from three near isogenic lines of maize each carrying a single rust resistance gene (Rp1-D, Rp5, and Rp1-M) were infected separately with three different races of maize rust that were capable of initiating either a compatible or incompatible infection. Total RNA was extracted 6 d after infection. In comparison to the uninfected controls, all rust-infected maize lines showed an increase in mis1 RNA levels due to rust infection and this was possibly greatest in the compatible infections (Fig. 6A). In maize and most other grasses, so-called incompatible or resistant rust infections allow some fungal growth and frequently lead to eventual, although delayed, sporulation. This is in contrast to the incompatible flax rust infection controlled by the L6 resistance gene, where rust growth was severely inhibited and fis1 induction was undetectable by RNA-blot analysis (Roberts and Pryor, 1995). However, like cereal rust resistance, the resistance response mediated by the flax P3 gene allowed pathogen growth to occur (Fig. 2G) and significant fis-2.5P-GUS transgene expression was detected (Fig. 2H).
Figure 6.
Induction of the maize mis1 and barley bis1 genes during rust infection. A, RNA-blot analysis of maize mis1 gene expression during maize rust infection. Lanes 1 through 4, 5 through 8, and 9 through 12 contain total leaf RNA isolated from three maize lines containing the Rp1-D, Rp5, and Rp1-M rust resistance genes, respectively. Lanes 1, 5, and 9, labeled C, contain RNA isolated from maize plants that had not been infected with maize rust. Lanes 3, 4, 8, and 10, labeled S, contain RNA from each maize line after infection with a compatible race of maize rust, whereas lanes 2, 6, 7, 11, and 12, labeled R, contain RNA from each maize line after infection with an incompatible rust race. A 434-bp PCR product amplified from the mis1 gene was used as a hybridization probe. Beneath the autoradiograph is shown the RNA formaldehyde gel that was subsequently transferred for the RNA-blot analysis. B, RNA-blot analysis of barley bis1 gene expression during infection with a virulent race of Puccinia graminis f.sp tritici. Total RNA was isolated from barley cv Golden Promise (lanes 1–5) and cv Morex (lanes 6–10) at 0 (lanes 1 and 6), 2 (lanes 2 and 7), 4 (lanes 3 and 8), 6 (lanes 4 and 9), and 8 (lanes 5 and 10) d post rust inoculation. The same probe as in A was used for hybridization. Shown beneath the autoradiograph is the RNA gel that was transferred for RNA-blot analysis.
Two barley cultivars, Morex and Golden Promise, were infected with a virulent race of P. graminis f. sp. tritici and RNA extracted at various time points. RNA-blot analysis using the mis1 434-bp cloned PCR product as a probe identified a transcript in both barley cultivars that is up-regulated in response to rust infection (Fig. 6B). The up-regulation of fis1-like genes in response to a compatible rust infection therefore is common to both dicot and monocot species, including at least one member of the Triticae.
DISCUSSION
Analysis of the flax fis1 promoter using a GUS reporter gene has demonstrated that the up-regulation of this promoter in response to a compatible flax rust infection is highly localized, occurring exclusively in the leaf mesophyll cells surrounding rust infection sites. As yet, we have been unable to determine whether GUS induction occurs exclusively in cells infected by rust haustoria. RNA-blot analysis has shown that related genes in maize and barley are also induced by a compatible rust infection. The fis1 homologs identified in a range of other plant species may also be up-regulated in response to rust infection. It will be of interest to determine if pathogens other than rust fungi can also induce the expression of fis1 and the homologous genes identified in other species. Microarray analysis shows that the transcript level of the Arabidopsis AtP5CDH gene that encodes a Δ1-pyrroline-5-carboxylate dehydrogenase varies little in over 230 different experimental conditions including infection with tobacco mosaic virus, Erysiphe cichoracearum, and Pseudomonas syringae (http://genome-www4.stanford.edu/MicroArray/SMD/).
The extent of fis1 expression in an incompatible interaction reflects the amount of pathogen growth. Flax plants containing resistance genes like L6, which initiates a rapid and highly effective defense response that prevents pathogen growth, show little fis1 up-regulation. Plants with genes that elicit a defense response that is less effective at preventing pathogen growth (e.g. P3) show more elevated fis1-GUS expression, whereas fully susceptible plants show the highest level of fis1 expression during rust infection. Similarly, mis1 up-regulation is observed in resistant maize lines when challenged with avirulent races of maize rust, presumably due to the significant pathogen growth that still occurs despite the resistance response.
These data are consistent with the notion that the first hours of a rust infection are similar in both the incompatible and compatible interactions. For example, in the flax-flax rust interaction, it is known that that a defense response is initiated some time after haustorial formation (Kobayashi et al., 1994). During the L6/A-L6 interaction, the first haustoria are observed within host cells 12 h postinfection, but hypersensitive cell death is not observed until some time after 12 h (Kobayashi et al., 1994). By 24 h postinfection, 60% of cells containing haustoria show development of hypersensitive cell death and this increases to 80% by 36 h postinfection (Kobayashi et al., 1994). The small amount of fis1-GUS expression observed during the L6/A-L6 response therefore may occur prior to the development of hypersensitive cell death.
The similarity between the AtP5CDH protein and FIS1, MIS1, WIS1, and BIS1 suggests that these proteins may also function as Δ1-pyrroline-5-carboxylate dehydrogenases and that Pro degradation in the host cell occurs during rust infection. The small up-regulation of fis1 during the L6/AL6-incompatible interaction suggests that this degradation appears early in the infection process before hypersensitive cell death occurs. The role of Pro catabolism in rust infection is unclear.
Pro is known to accumulate in response to a number of environmental stresses including osmotic stress, drought, salinity, temperature, UV irradiation, and heavy metals and is thought to fulfill a number of roles such as acting as an osmolyte, protecting enzymes from denaturation, scavenging free radicals, interacting with membranes, and balancing NAD to NADP ratios, in addition to serving as a precursor in protein biosynthesis (Hare and Cress, 1997; Nanjo et al., 1999). Conversely, Pro supplied externally to plants is highly toxic and the Pro degradation intermediates pyrroline-5-carboxylate and Glu semialdehyde are proposed to be responsible for this toxicity (Hellmann et al., 2000). Δ1-Pyrroline-5-carboxylate dehydrogenase may potentially protect the plant against Pro toxicity by converting pyrroline-5-carboxylate to Glu (Deuschle et al., 2001). Pyrroline-5-carboxylate has been shown to induce apoptosis in human tumor cell lines (Maxwell and Davis, 2000) and yeast deficient in Δ1-pyrroline-5-carboxylate dehydrogenase produce reactive oxygen species when exposed to Pro (Deuschle et al., 2001), whereas the Arabidopsis cpr5 mutant that constitutively produces pathogenesis-related proteins is hypersensitive to Pro application (Deuschle et al., 2001). Therefore, Pro is paradoxically associated with protection against environmental stress but at the same time is highly toxic. However, as noted by Deuschle et al. (2001), the induction of apoptosis in mammalian cells, the production of reactive oxygen species, and the Pro hypersensitivity of cpr5 potentially suggest a connection between Pro catabolism and the plant hypersensitive defense response, although it is unclear if such a relationship exists. Further work is required to determine if the products of the fis1, mis1, bis1, and wis1 genes have Δ1-pyrroline-5-carboxylate dehydrogenase activity and if so, whether Pro metabolism at infection sites is involved in rust nutrition or in modifying host defense reactions.
The highly localized pathogen-induced expression of fis1 makes the promoter of this gene ideal for expressing synthetic disease resistance genes because these gene products will be specifically targeted to the sites of rust infection. The up-regulation of the mis1 and bis1 genes in response to a compatible rust infection and the presence of an equivalent gene in wheat, wis1, suggests that such a strategy is also applicable to cereal species in which rust diseases are an agronomic problem. We currently are using these promoters to express potentially antifungal genes in cereal species as a means of generating synthetic rust disease resistance.
An alternative strategy for developing synthetic resistance using a highly localized pathogen inducible promoter is to express a phytotoxic compound as a means of mimicking the cell death observed during the hypersensitive response. Alternatively, synthetic resistance may be obtained by pathogen-localized expression of a gene encoding either an elicitor or activator of the plant defense response. For these approaches to be successful, the regulation of such a promoter needs to be precise.
The fis1 promoter shows endogenous vascular expression, suggesting that it is not yet feasible to use this promoter to regulate the expression of a plant cytotoxin, unless the phytotoxic effect is either expression level dependent or restricted to mesophyll cells. The Arabidopsis AtP5CDH gene showed a basal level of expression in all organs examined but was induced in the presence of Pro. The fis1 gene may also be up-regulated by Pro application. A more detailed deletion analysis of the fis1 promoter may uncouple pathogen-inducible expression from endogenous plant expression or other stress-induced expression as was observed for the tobacco nematode-inducible promoter TobRB7 (Opperman et al., 1994) and tomato anionic peroxidase gene (Mohan et al., 1993). However, it is also possible that a pathogen-inducible element per se is not present in the fis1 promoter and that up-regulation of this gene in response to rust infection is an indirect consequence of changes in host cell metabolism arising from the infection process. The transient assay developed for this promoter will facilitate a deletion analysis to answer this question. However, it should be noted that a truncated version of the TobRB7 promoter, 0.3 TobRB7, which has lost much of its endogenous expression, still has sufficient basal expression to prevent the generation of transgenic tobacco plants containing a P. syringae hrm elicitor gene under the control of this promoter (Shen et al., 2000).
Further analysis of host gene expression during a successful pathogen infection will provide greater insight into the molecular nature of the compatible plant-pathogen interaction and may provide mechanisms for manipulating the compatible interaction to generate synthetic disease resistance.
MATERIALS AND METHODS
Plant and Fungal Material
Flax (Linum usitatissimum) lines Birio, Leona, Ward, and Hoshangabad have been described previously by Lawrence et al. (1993) and Islam and Shepherd (1991). The line Hoshangabad contains no known functional rust (Melampsora lini) resistance genes, whereas Birio, Leona, and Ward contain resistance genes L6, P3, and L9 + M2, respectively.
Rust strain CH5 is derived from a previously described sexual cross of flax rust (Lawrence et al., 1981) and contains avirulence genes AL-6 and AP-3 but does not contain avirulence genes A-L9 and A-M2. Rust strain CH5F2-84 is a derived from self-fertilization of stain CH5 and contains the AL-6 gene but neither AL-9 nor AM-2 genes. Plants were infected with rust urediospores overnight in a humid environment and then transferred to a greenhouse, where resistant or susceptible reactions were allowed to develop.
Isolation of Nucleic Acids and Hybridization-Blot Analysis
Total RNA was isolated from flax tissue using the protocol of Hughes and Galua (1988), whereas maize (Zea mays) RNA and barley (Hordeum vulgare) RNA were isolated as described by Logemann et al. (1987). RNA was separated by electrophoresis on agarose formaldehyde denaturing gels and transferred to Hybond N+ membranes (Amersham, Buckinghamshire, UK) using 20× SSC as a transfer solvent. Membranes were hybridized under standard stringency conditions. Flax leaf DNA was isolated as described by Lawrence et al. (1993), whereas maize and barley leaf DNA was isolated as described by Sahai-Maroof et al. (1984). DNA was digested with restriction enzymes (New England Biolabs, Beverly, MA) under conditions recommended by the manufacturer. Digested DNA was separated by agarose gel electrophoresis, transferred to Hybond N+ membranes (Amersham), and hybridized under standard stringency conditions. Hybridization probes were labeled with P32-dCTP using a MegaPrime DNA labeling system (Amersham).
Isolation of Lambda Clones
Lambda cDNA clones were isolated from a total leaf and stem cDNA library derived from the flax line Forge, as previously described by Lawrence et al. (1995). A flax genomic library was constructed in a lambda EMBL3 vector, whereas a maize genomic library was constructed in a lambda GEM-11 vector (Promega, Madison, WI) and both libraries screened using Hybond N+ membranes (Amersham) under hybridization conditions described above. A rust-infected maize cDNA library was constructed using a HybriZAP II cDNA synthesis kit (Stratagene, La Jolla, CA) as recommended by the manufacturer and hybridized as above.
RT-PCR Reactions
RT-PCR reactions were carried out essentially as described by Ayliffe et al. (1999). Sequences of degenerate primers used for amplification of a 434-bp region of the mis1 transcript are as follows, with bases in parentheses representing alternative nucleotides: GPL primer, A(T/I) (A/T/C) ACICCITT(T/C) AA(T/C) TT(T/C) CCI; and GQC primer, TG(T/C)(T/A)(G/C) IGGICA(A/G) AA(G/A) TG(T/C)(A/T)(G/C) IGCI.
Cloning and Sequencing
Cloning was carried out essentially as described by Sambrook et al. (1989). DNA sequence was obtained using an ABI dye-primer sequencing kit (PE-Applied Biosystems, Foster City, CA) and ABI Prism 377 model DNA sequencer.
EST Sequences
Barley EST clone HVSMEf0011A14 (accession no. BF256795) was obtained from the Clemson Genomics Institute (Clemson, SC), whereas wheat (Triticum aestivum) EST clone SCL071.E0R990707 (accession no. BE418547) was kindly provided by Dr. Sylvie Cloutier (Cereal Research Centre, Winnipeg, MB).
Plant Transformation
Agrobacterium tumefaciens-mediated flax transformation was as previously described (Lawrence et al., 1989), except that 6-d-old hypocotyls were used as explants for A. tumefaciens infection in place of cotyledons. Transformed plants were selected on the basis of resistance to the antibiotic spectinomycin (Anderson et al., 1997).
Transient fis1-GUS Gene Expression Analysis
A. tumefaciens strain AGL1 containing fis1-GUS gene constructs within binary vectors were grown overnight at 28°C in Luria-Bertani solution containing the appropriate antibiotics. Bacterial cells were then concentrated 2-fold by resuspending in a solution of 10 mm MgCl2 containing 0.001% (w/v) SilwetL-77 (Lehle Seeds, Round Rock, TX). Cuttings of flax plants that had been previously infected with flax rust were then immersed completely into this bacterial solution and subjected to a vacuum of −85 kPa for 30 min. After infiltration, flax cuttings were rinsed in water and the cut stems then immersed in a solution of 10 mg L−1 GA3 with the leaves remaining out of solution. Three days postinfiltration, these plants were stained for GUS activity at 37°C for 16 h. Following staining, plants were cleared in 100% (v/v) ethanol at room temperature.
Data Analysis
DNA and protein comparisons were made using the programs from the University of Wisconsin Genetics Computer Group package (Devereux et al., 1984) and the National Center for Biotechnology Information suite of analysis programs (http://www.ncbi.nlm.nih.gov/). Mitochondrial targeting sequence predictions were made using the Target PV 1.0 program (Emanuelsson et al., 2000) and the program PSORT.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
ACKNOWLEDGMENTS
We wish to thank Mr. Luch Hac and Ms. Kim Newell for technical assistance, Dr. Sylvie Cloutier for provision of a wheat EST clone, and Ms. Sandie McIntosh for assistance in manuscript preparation.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010940.
LITERATURE CITED
- Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG. Inactivation of the flax rust-resistance gene Massociated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayliffe MA, Frost DV, Finnegan EJ, Lawrence GJ, Anderson PA, Ellis JG. Analysis of alternative transcripts of the flax L6rust resistance gene. Plant J. 1999;17:287–292. doi: 10.1046/j.1365-313x.1999.00377.x. [DOI] [PubMed] [Google Scholar]
- Chou H-M, Bundock N, Rolfe SA, Scholes JD. Infection of Arabidopsis thaliana with Albugo candida(white blister rust) causes a reprogramming of host metabolism. Mol Plant Pathol. 2000;1:99–113. doi: 10.1046/j.1364-3703.2000.00013.x. [DOI] [PubMed] [Google Scholar]
- Clancy FG, Coffey MD. Patterns of translocation, changes in invertase activity, and polyol formation in susceptible and resistant flax infected with the rust fungus Melampsora lini. Physiol Plant Pathol. 1980;17:41–52. [Google Scholar]
- Deuschle K, Funck D, Hellman H, Daschner K, Binder S, Frommer WB. A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001;27:345–355. doi: 10.1046/j.1365-313x.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Hahn M, Mengden K. Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol Plant-Microbe Interact. 1997;10:427–437. doi: 10.1094/MPMI.1997.10.4.427. [DOI] [PubMed] [Google Scholar]
- Hahn M, Neef U, Struck C, Gottfert M, Mengden K. A putative amino acid transporter is specifically expressed in haustoria of the rust fungus Uromyces fabae. Mol Plant-Microbe Interact. 1997;10:438–445. doi: 10.1094/MPMI.1997.10.4.438. [DOI] [PubMed] [Google Scholar]
- Hall JL, Williams LE. Assimilate transport and partitioning in fungal biotrophic interactions. Aust J Plant Physiol. 2000;27:549–560. [Google Scholar]
- Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997;21:79–102. [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant towards exogenous proline application. Plant Physiol. 2000;123:779–790. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DW, Galua G. Preparation of RNA from cotton leaves and pollen. Plant Mol Biol Rep. 1988;6:253–257. [Google Scholar]
- Hwang C-S, Flaishman MA, Kolattukudy PE. Cloning of a gene expressed during appressorium formation by Colletotrichum gloeosporioidesand a marked decrease in virulence by disruption of this gene. Plant Cell. 1995;7:183–193. doi: 10.1105/tpc.7.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MR, Shepherd KW. Present status of genetics of rust resistance in flax. Euphytica. 1991;55:255–267. [Google Scholar]
- Kapila J, De Rycke R, Van Montague M, Angenon G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997;122:101–108. [Google Scholar]
- Kobayashi I, Kobayashi Y, Hardham AR. Dynamic reorganisation of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta. 1994;195:237–247. [Google Scholar]
- Lawrence GJ, Ellis JG, Finnegan EJ, Dennis ES, Peacock WJ. Tagging rust resistance genes in flax. In: Iyama S, Takeda G, editors. Breeding Research: The Key to the Survival of the Earth. Tsukuba, Japan: Organising Committee of the Sixth International Congress Society for the Advancement of Breeding Researches in Asia and Oceania; 1989. pp. 535–538. [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ellis JG. Instability of the L6gene for rust resistance in flax is correlated with the presence of a linked Ac element. Plant J. 1993;4:659–669. doi: 10.1046/j.1365-313x.1993.04040659.x. [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Mayo GME, Shepherd KW. Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology. 1981;71:12–19. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA. 2000;97:13009–13014. doi: 10.1073/pnas.230445997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R, Vijayan P, Kolattukudy PE. Developmental and tissue-specific expression of a tomato anionic peroxidase (tap1) gene by a minimal promoter, with wound and pathogen induction by an additional 5′-flanking region. Plant Mol Biol. 1993;22:475–490. doi: 10.1007/BF00015977. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J. 1999;18:185–193. doi: 10.1046/j.1365-313x.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Balch WE. GTPases: multifunctional molecular switches regulating vesicular traffic. Annu Rev Biochem. 1994;63:949–990. doi: 10.1146/annurev.bi.63.070194.004505. [DOI] [PubMed] [Google Scholar]
- Opperman CH, Taylor CG, Conkling MA. Root-knot nematode-directed expression of a plant root-specific gene. Science. 1994;263:221–223. doi: 10.1126/science.263.5144.221. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Riach MBR, Bleker T, van den Berg-Velthuis GCM, Glovers F. Isolation of putative pathogenicity genes of the potato late blight fungus Phytophora infestansby differential hybridization of a genomic library. Physiol Mol Plant Pathol. 1993;43:69–79. [Google Scholar]
- Roberts JK, Pryor A. Isolation of a flax (Linum usitatissimum) gene induced during susceptible infection by flax rust (Melampsora lini) Plant J. 1995;8:1–8. doi: 10.1046/j.1365-313x.1995.08010001.x. [DOI] [PubMed] [Google Scholar]
- Rogers LM, Flaishman MA, Kolattukudy PE. Cutinase gene disruption in Fusarium solani fsp pisidecreases its virulence on pea. Plant Cell. 1994;6:935–945. doi: 10.1105/tpc.6.7.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy BA. Floral mimicry by a plant pathogen. Nature. 1993;362:56–58. [Google Scholar]
- Sahai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scholes JD. Photosynthesis: cellular and tissue aspects in diseased leaves. In: Ayres PG, editor. Pests and Pathogens. Oxford: BIOS Scientific Publishers Limited; 1992. pp. 85–105. [Google Scholar]
- Shen S, Li Q, He S-Y, Barker KR, Li D, Hunt AG. Conversion of compatible plant-pathogen interactions into incompatible interactions by expression of the Pseudomonas syringae pv. syringae 61 hrmgene in transgenic tobacco plants. Plant J. 2000;23:205–213. doi: 10.1046/j.1365-313x.2000.00772.x. [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Carpenter TB, Vierstra RD. Homologues of wheat ubiquitin-conjugating enzymes TaUBC1 and TaUBC4 are encoded by small multigene families in Arabidopsis thaliana. Plant Mol Biol. 1994;24:651–661. doi: 10.1007/BF00023561. [DOI] [PubMed] [Google Scholar]
- Sutton BCS, Shaw M. Changes in two ribonuclease isozymes during rust infection of flax cotyledons. Plant Physiol. 1982;69:205–209. doi: 10.1104/pp.69.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton BCS, Shaw M. Protein synthesis in flax following inoculation with flax rust. Can J Bot. 1986;64:13–18. [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]