Abstract
Transcripts of the ntp303 gene accumulate abundantly throughout pollen development, whereas the protein only accumulates to detectable levels after pollen germination. In an attempt to explain the divergence in the accumulation profiles of the mRNA and the protein, we investigated the role of the untranslated regions (UTRs) in enhancing ntp303 translation during the transition from developing to germinating pollen. Luciferase reporter gene fusion constructs containing the ntp303 5′-UTR gave rise to luciferase activity that was up to 60-fold higher during pollen tube growth than that of constructs containing different 5′-UTRs. No apparent differences in the luciferase activity of these constructs were observed during pollen development. The ntp303 5′-UTR-mediated increase in luciferase activity was not significantly influenced by coding region or 3′-UTR sequences. Furthermore, enhanced luciferase activity directed by the ntp303 5′-UTR occurred predominantly at the post-transcriptional level. A series of 5′-UTR deletion constructs was created to identify putative regulatory sequences required for the high level of translation during pollen tube growth. Two predicted stem loop structures (H-I and H-II) caused a complete inhibition of the enhanced translation after their total or partial deletion. A (GAA)8 repeat within the H-I stem loop structure was demonstrated to be important for the modulation of translation efficiency. The H-II stem loop structure was found to be essential for the determination of mRNA stability.
In plants as well as in animals, gamete development involves defined transitions of cells from one physiological state to another by mitotic and meiotic divisions. These series of events require multiple changes in gene expression. Many stages of gamete development in plant and animal species proceed almost without transcriptional activity and depend mainly upon translation of presynthesized mRNAs. Thus, in these species, post-transcriptional control of gene expression is very important for gamete development. Examples include the post-transcriptional control of genes expressed during spermatogenesis in Drosophila melanogaster (Kuhn et al., 1991; Schäfer et al., 1993) and mouse (Mus musculus; Nayernia et al., 1994; Schäfer et al., 1995) and during oocyte development in marine invertebrates (Swenson et al., 1987) and mammals (Stutz et al., 1998; Lasko, 1999).
An example of post-transcriptional regulation of gene expression during gamete development in plants is the development and germination of the male gametophyte (pollen; for review, see Mascarenhas, 1990, 1993; McCormick, 1991, 1993; Taylor and Hepler, 1997). Pollen grains consist of a small generative cell and a large vegetative cell that are formed from microspores by a mitotic division (for review, see Mascarenhas, 1989; Bedinger, 1992). During subsequent pollen development, a range of processes leads to progressive dehydration of the grain and its transition to dormancy (Lin and Dickinson, 1984; Van Aelst et al., 1993). This maturation of pollen is generally accompanied by a progressive storage of large quantities of rRNAs, tRNAs, mRNAs, and ribosomes (for review, see Mascarenhas, 1990, 1993). As soon as the pollen grain lands on a compatible stigma, an extensive rehydration of the grain occurs, leading to a rapid reactivation of the translation machinery that uses the stored RNAs. Proteins that are synthesized from these stored products are required for the progamic phase, i.e. germination and subsequent growth of the pollen tube (Mascarenhas, 1990, 1993; Muschietti et al., 1994).
Despite the importance of translation of presynthesized mRNAs in the contribution of sexual reproduction, little attention has been paid to elucidate the mechanisms underlying post-transcriptional regulation of pollen gene expression (Op den Camp and Kuhlemeier, 1998; Ylstra and McCormick, 1999; Honys et al., 2000). Many mRNA species from different eukaryotic systems can be modulated in their translation efficiency by signals encoded in the 5′- or 3′-untranslated region (UTR) (for review, see Gallie, 1993, 1996; Fütterer and Hohn, 1996; Pain, 1996; Danon, 1997; Day and Tuite, 1998; Bailey-Serres, 1999). In these cases, translation has often been found to be regulated at the level of translation initiation. This led to the hypothesis that the UTRs might play an important role in the efficient induction of translation during the transition of developing pollen to pollen in the progamic stage (e.g. Bate et al., 1996). If so, it may be assumed that specific sequences within the 5′- or 3′-UTR are prerequisite to direct the high level of translation during pollen tube growth.
To obtain more insight in the mechanism of post-transcriptional regulation of pollen gene expression during pollen tube growth, we focused on the pollen gene ntp303. Regulation of the synthesis of the NTP303 protein takes place at the post-transcriptional level (Čapková et al., 1994; Štorchová et al., 1994; Wittink et al., 2000). Transcripts of the ntp303 gene first appear after pollen mitosis I and continue to accumulate during pollen maturation and early stages of pollen tube growth (Weterings et al., 1992). In contrast, the protein only starts to accumulate at the onset of pollen rehydration (Wittink et al., 2000).
In the present study, we investigated the contribution of the UTRs of the ntp303 gene in directing pollen gene expression. Several gene fusion constructs containing different promoter and UTR combinations linked to the luciferase+ open reading frame were introduced in developing and germinating pollen by particle bombardment and their transient expression was assayed. Furthermore, several ntp303 5′-UTR deletion constructs were generated and tested to identify putative cis-acting regulatory sequences.
RESULTS
UTR Gene Fusion Constructs
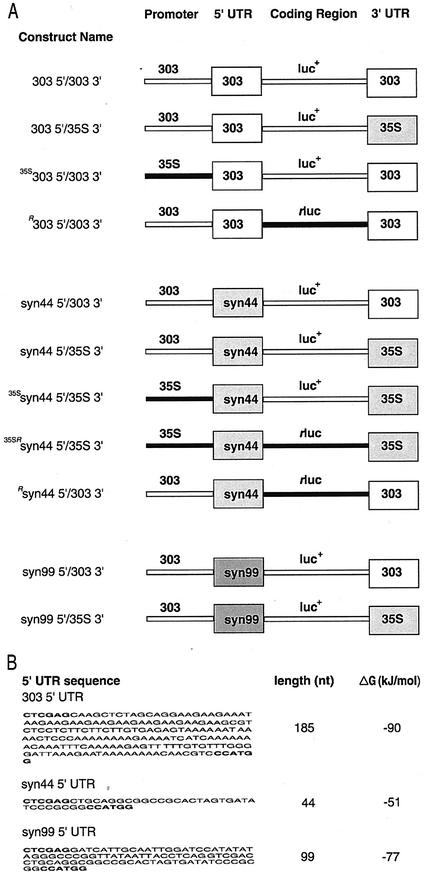
Several UTR gene fusion constructs containing the ntp303 promoter, the firefly luciferase+ reporter gene (Promega, Madison, WI), and different combinations of 5′- and 3′-UTRs were built to investigate the ability of the UTRs of ntp303 to modulate translation during pollen development and pollen tube growth (Fig. 1A). The names of the constructs refer to their 5′- and 3′-UTRs (5′-UTR/3′-UTR). The abbreviation “35S” or “R” that is given in uppercase letters before a construct name indicates that the construct contains the CaMV 35S instead of the ntp303 promoter or the R. reniformis luciferase instead of the firefly luciferase coding region, respectively.
Figure 1.
UTR gene fusion constructs used in the present study. A, Schematic representation and names of the UTR gene fusion constructs. The constructs were driven by the ntp303 (303) or the cauliflower mosaic virus (CaMV) 35S (35S) promoter. The coding regions that were used in the constructs are firefly luciferase (luc+) or Renilla reniformis luciferase (rluc). The left or right box in the construct represents the 5′- or 3′-UTR, respectively. UTR abbreviations: 303, ntp303 5′- or 3′-UTR; syn44, synthetic 44 5′-UTR; syn99, synthetic 99 5′-UTR; 35S, CaMV 35S 3′-UTR. The construct name refers to the 5′- and 3′-UTRs (5′-UTR/3′-UTR). “35S” or “R” in uppercase before a construct name means that the construct contains the CaMV 35S promoter or the R. reniformis luciferase coding region, respectively. B, Sequence, length, and the calculated stability of the 5′-UTRs used in the UTR gene fusion constructs.
The different constructs were introduced into developing or mature pollen by particle bombardment. The level of expression was estimated after a period of 20 h of in vitro development or germination by measurement of luciferase activity. To correct for differences in bombardment efficiencies, a second construct was cobombarded. This construct contained the ntp303 promoter, a synthetic 5′-UTR (syn44 5′), the luciferase reporter gene from R. reniformis and the CaMV termination sequence (35S 3′; Rsyn44 5′/35S 3′). The luciferase activity value of the firefly luciferase construct was normalized to the value of the R. reniformis luciferase construct, which gave rise to the relative luciferase activity. For each construct, at least six independent bombardments were performed.
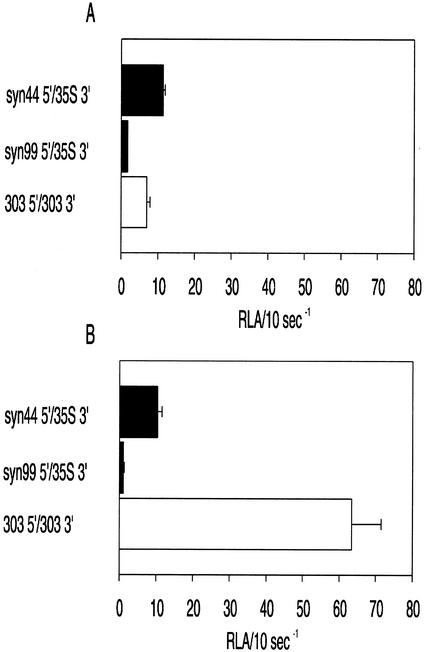
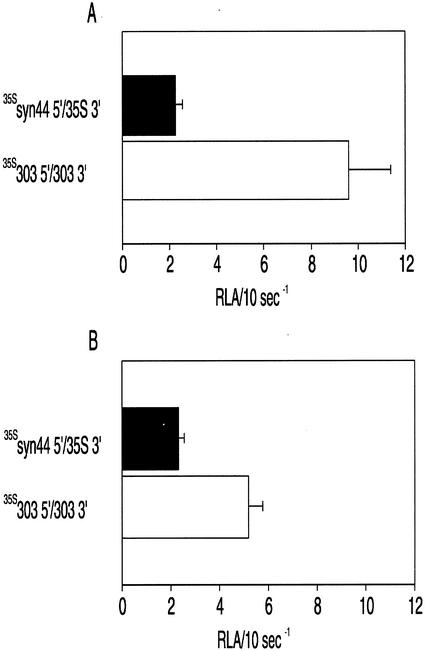
The effect of ntp303 UTRs on luciferase activity during pollen development and pollen tube growth was determined by comparing the translation level of constructs containing different combinations of ntp303 and control UTRs. For replacement of the ntp3033′ or 5′-UTR, we used the CaMV 35S termination sequence (35S 3′) or two different synthetic leader sequences designated as syn99 5′ and syn44 5′, respectively (Fig. 1B). UTR gene fusion constructs containing the synthetic 5′-UTRs have been demonstrated to be translated efficiently by Bate et al. (1996). The syn99 5′-UTR was used as a control UTR because it revealed a free energy value that was more or less comparable with that of the ntp303 5′-UTR (calculated energy values [ΔG] of −90 and −77 kJ mol−1, respectively; Fig. 1B). Lowering the potential energy (i.e. a more negative value of ΔG) of secondary structures within a 5′-UTR has a negative effect on translation (Kozak, 1989; Gallie et al., 2000). The syn44 5′-UTR was used as a positive control because its secondary structure has a relative high potential energy (ΔG of −51 kJ mol−1), and therefore a positive effect on translation compared with the ntp303 and syn99 5′-UTRs. The difference in translation efficiency of both control UTRs becomes clear in Figure 2, A and B. During pollen development and pollen tube growth, the construct containing the syn44 5′-UTR gave rise to an approximately 10-fold higher luciferase activity level compared with the syn99 5′-UTR-containing construct.
Figure 2.
Luciferase activity of gene fusion constructs in developing pollen (A) and growing pollen tubes (B) containing control (black) or ntp303 (white) UTRs. RLA/10 s−1 means the relative luciferase activity (luminescence) per 10-s measuring time after normalization with the luciferase activity of the reference construct Rsyn44 5′/35S 3′. Results are given as means ± se (n ≥ 6). For details, see “Results” and “Materials and Methods.”
The 5′-UTR, But Not the 3′-UTR, of Ntp303 Enhances Translation Specifically during Pollen Tube Growth
The effect of ntp303 UTRs on luciferase activity during pollen development and pollen tube growth was investigated by comparing the expression level of a construct containing the ntp303 UTRs (303 5′/303 3′) with that of constructs containing control UTRs (the syn99 5′- or syn44 5′-UTR and the 35S 3′-UTR). In developing pollen incubated for 20 h after bombardment, the ntp303 UTRs construct gave rise to a luciferase activity level that was approximately 4-fold higher than that of syn99 5′/35S 3′ and slightly lower than that of syn44 5′/35S 3′ (Fig. 2A). After 20 h of pollen tube growth, the luciferase activity level of 303 5′/303 3′ was approximately 60- and 6-fold higher than that of syn99 5′/35S 3′ and syn44 5′/35S 3′, respectively (Fig. 2B). The differences in the luciferase activity level could already be observed in pollen tubes 5 h after bombardment (data not shown). The luciferase activity levels of the constructs containing the control UTRs were not significantly different during pollen development or pollen tube growth (Fig. 2, A and B). This clearly illustrates that expression mediated by these control UTRs is independent of the developmental stage in which they were tested.
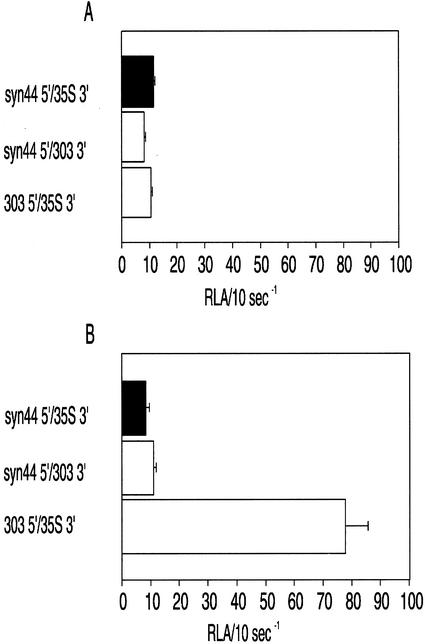
To examine whether the 5′-UTR or the 3′-UTR of the ntp303 mRNA determines the level of expression during pollen development and pollen tube growth, luciferase activity of gene fusion constructs containing the ntp303 5′- and 35S 3′-UTRs or the syn44 5′- and ntp303 3′-UTRs was compared with that of syn44 5′/35S 3′. No significant differences in the luciferase activity level of the ntp303 UTR and control UTRs containing constructs were observed during pollen development (Fig. 3A). During pollen tube growth, the ntp303 5′-UTR increased the activity of luciferase to a level that was almost 8-fold higher than that of the control 5′-UTR construct (Fig. 3B). This enhancement effect was absent in the ntp303 3′-UTR construct.
Figure 3.
Luciferase activity of gene fusion constructs in developing pollen (A) and growing pollen tubes (B) containing control (black) or combinations of control and ntp303 (white) UTRs. Activity of the firefly luciferase determined for the test constructs was normalized with the R. reniformis luciferase activity of the reference construct Rsyn44 5′/35S 3′ (RLA/10 s−1). Results are given as means ± se (n ≥ 6).
To exclude the possibility that the enhancement of luciferase activity mediated by the ntp303 5′-UTR in growing pollen tubes was the result of a specific interaction between the 5′-UTR and the firefly luciferase coding region, this coding region was replaced by the R. reniformis luciferase coding region in the constructs syn44 5′/35S 3′ and 303 5′/303 3′. The firefly and the R. reniformis luciferase mRNAs exhibit no significant sequence identity with each other. Normalization of the luciferase activity of these constructs was established by cobombardment with a construct containing the syn44 5′-UTR, the firefly luciferase coding region, and the 35S termination sequence. As shown in Figure 4, the ntp303 UTRs gave rise to a luciferase activity level that was approximately 7-fold higher than that of the control UTRs. Because this enhancement effect of the ntp303 5′-UTR was also found for firefly luciferase mRNAs, this suggests that there is no specific interaction between the ntp303 5′-UTR and the coding region.
Figure 4.
Transient expression of gene fusion constructs containing the R. reniformis luciferase coding region and control (black) or ntp303 (white) UTRs in growing pollen tubes. Activity of the R. reniformis luciferase determined for the test constructs was normalized with the firefly luciferase activity of the reference construct syn44 5′/35S 3′ (RLA/10 s−1). Results are given as means ± se (n ≥ 6).
We conclude that the 5′-UTR of the ntp303 gene is an important determinant in the high level of expression during pollen tube growth.
The 5′-UTR-Specific Enhancement during Pollen Tube Growth Is Post-Transcriptional
To investigate whether the enhancement mediated by the ntp303 5′-UTR during pollen tube growth was the result of post-transcriptional regulation or an enhanced transcript level, we determined the relative transcript and luciferase activity levels of pollen that were bombarded with the syn99 5′/35S 3′, 303 5′/35S 3′, or syn99 5′/303 3′ construct (Table I). Each pollen batch bombarded was separated in two fractions. One fraction was used for total RNA isolation and the other was used for the luciferase assay. Ten micrograms of total RNA was hybridized with a 32P-labeled luciferase probe. The relative transcript level was determined by calculation of the ratio of the hybridization signal of firefly luciferase mRNA to that of R. reniformis luciferase mRNA of the cobombarded Rsyn44 5′/35S 3′ construct. The ntp303 5′-UTR construct showed a relative transcript level that was approximately 2- to 3-fold higher after 20 h of pollen tube growth than that of the syn99 5′-UTR constructs. The construct containing the ntp303 5′-UTR exhibited a 50-fold increase in luciferase activity as compared with syn99 5′/35S 3′ (Table I). These data indicate that during pollen tube growth, chimeric luciferase transcripts containing the ntp303 5′-UTR are translated more efficiently than luciferase mRNAs containing the control 5′-UTR. Although the high luciferase activity levels are primary due to an enhanced translation efficiency, the ntp303 5′-UTR exhibits a stimulatory influence on the transcript level.
Table I.
Analysis of the relative transcript (relative luciferase mRNA abundance) and luciferase activity (relative luciferase activity/10 s−1) levels of UTR gene fusion constructs during pollen tube growth
| Construct | Relative Luc mRNA Abundance | Relative LUC Activity |
|---|---|---|
| counts | RLA/10 s−1 | |
| syn99 5′/35S 3′ | 1.36 ± 0.17 (1.00) | 1.02 ± 0.21 (1.00) |
| 303 5′/35S 3′ | 3.70 ± 0.22 (2.72) | 50.60 ± 0.22 (49.61) |
| syn99 5′/303 3′ | 1.81 ± 0.22 (1.33) | 2.50 ± 0.42 (2.45) |
The values in parentheses represent the fold increase of the transcript and luciferase activity levels compared to the levels of the syn99 5′/35 3′ construct. Results are given as means ± se (n ≥ 6). See “Results” for a description of the followed methodology. RLA, Relative LUC activity.
The 5′-UTR-Mediated Enhancement of Translation Also Occurs in Sporophytic Tissue, But Is Highest in Pollen Tubes
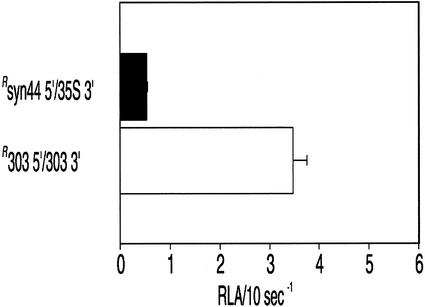
To test whether the ntp303 5′-UTR-mediated enhancement of translation in growing pollen tubes was restricted to a pollen-specific environment, the constructs syn44 5′/35S 3′ and 303 5′/303 3′ were reconstructed by replacing the ntp303 promoter with the CaMV 35S promoter. The CaMV 35S promoter is somewhat active in pollen, and highly active in sporophytic tissues (Twell et al., 1989). After bombardment of these constructs into mature pollen and young leaves followed by 20 h of in vitro incubation, luciferase activity was assayed. Normalization of the luciferase activity level of these constructs was established by cobombardment with a construct containing the CaMV 35S promoter, the syn44 5′-UTR, the R. reniformis luciferase reporter gene, and the 35S 3′-UTR. In growing pollen tubes, the ntp303 5′-UTR increased the luciferase activity approximately 5-fold (Fig. 5A). The difference in the luciferase activity level approached that of the constructs containing the same UTR combinations but linked to the ntp303 promoter (compare Fig. 5A with Fig. 2B). In young leaves, the ntp303 5′-UTR construct led to a luciferase activity level that was approximately 2-fold higher than the control UTR's construct (Fig. 5B). These data demonstrate that the ntp303 5′-UTR-mediated enhancement of translation may also occur in sporophytic cells.
Figure 5.
Luciferase activity of gene fusion constructs containing the CaMV 35S promoter in growing pollen tubes (A) and young leaves (B). Key to bars: black is the expression of 35Ssyn44/35S, and white is the expression of 35S303 5′/303 3′. RLA/10 s−1 indicates the relative luciferase activity per 10-s measuring time after normalization with the luciferase activity of the reference construct 35SRsyn44 5′/35S 3′. Results are given as means ± se (n ≥ 6).
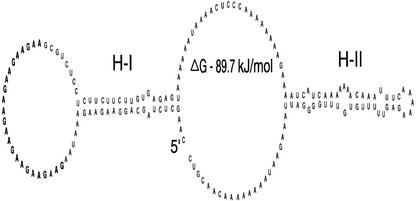
Enhancement of Translation during Pollen Tube Growth Can Be Attributed to Specific Regions within the Ntp303 5′-UTR
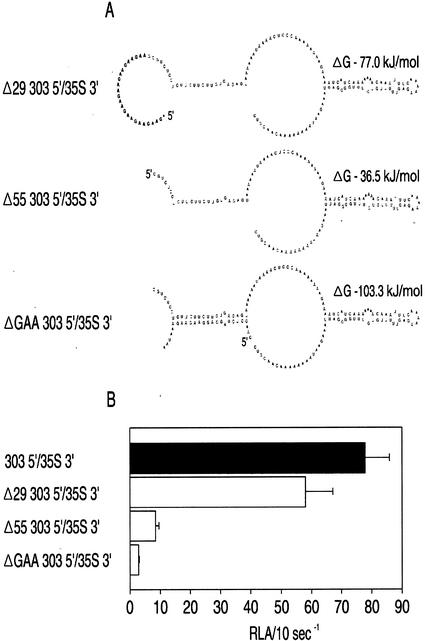
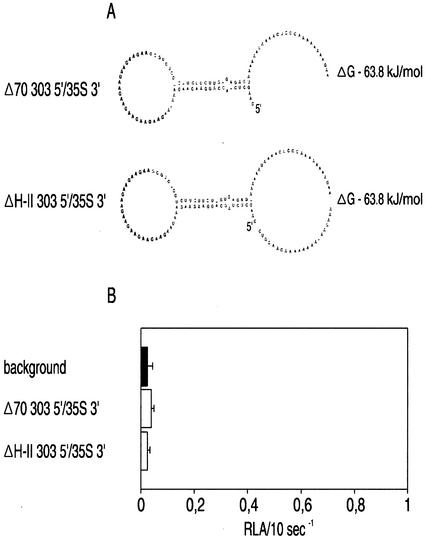
Figure 6 illustrates the predicted secondary structure of the ntp303 5′-UTR as analyzed with the RNAdraw software package (Matzura and Wennborg, 1996). There are two predicted stem loop structures designated H-I and H-II. The H-I stem loop structure is located at the 5′ terminus and has a ΔG of −64 kJ mol−1. This structure contains eight repeats of a GAA triplet in the external loop. The H-II structure is located 22 nucleotides upstream from the translation initiation site and has a ΔG of −26 kJ mol−1. The effect of sequences within the H-I and H-II structures on enhancement of translation during pollen tube growth was investigated by a series of ntp303 5′-UTR deletion constructs (Figs. 7A and 8A). These constructs were bombarded into mature pollen and the luciferase activity was assayed after 20 h of pollen tube growth (Figs. 7B and 8B). Figure 7B shows the luciferase activity of UTR gene fusion constructs with deletions within the H-I stem loop structure. The lowest level of luciferase activity was found after internal deletion of the (GAA)8 repeat (ΔGAA 303 5′/35S 3′). This luciferase activity level was comparable with the level of the control construct containing the syn99 5′-UTR (data not shown). A decrease in luciferase activity of approximately 94% occurred after deletion of the first 55 nucleotides (Δ55 303 5′/35S 3′) at the 5′ terminus including the (GAA)8 repeat. Deletion of the first 29 nucleotides at the 5′ terminus of the ntp303 5′-UTR (Δ29 303 5′/35S 3′) caused only a slight decrease in luciferase activity compared with that of the unmodified ntp303 5′-UTR. An almost complete inactivation of reporter gene activity was achieved after deletion of the last 70 nucleotides at the 3′ terminus of the ntp303 5′-UTR, which included the complete H-II structure (Δ70 303 5′/35S 3′; Fig. 8B). The same was true after internal deletion of only the H-II structure (ΔH-II 303 5′/35S 3′). In both cases, the luciferase activity values were in the same range as the background values (i.e. the measured autoluminescence of the luciferine substrate). Although the predicted stem loop structures have not been confirmed by nuclease-sensitive site mapping, these results clearly indicate that specific sequence regions within these putative structures are essential for enhancement of translation during pollen tube growth.
Figure 6.
Predicted secondary structure of the ntp303 5′-UTR. Structure prediction was performed using the RNAdraw software package (Matzura and Wennborg, 1996). H-I and H-II represents two predicted stem loop structures. The H-I structure contains eight repeats of a GAA triplet in the external loop (bold). The start of the transcription initiation site is indicated by 5′.
Figure 7.
The effect of deletions in the H-I stem loop structure of the ntp303 5′-UTR on luciferase activity. A, Graphic representation of the ntp303 5′-UTR with different H-I deletions. The secondary structures were predicted and the enthalpy calculated using the RNAdraw software package (Matzura and Wennborg, 1996). Internal deletion of 25 nucleotides including a (GAA)8 repeat gave rise to the ΔGAA 303 5′/35S 3′ construct. Deletion of the first 29 or 55 nucleotides at the proximal end of the ntp303 5′-UTR gave rise to the Δ29 303 5′/35S 3′ or Δ55 303 5′/35S 3′ constructs, respectively. The start of the 5′-UTR is indicated by 5′. B, Luciferase activity of the construct containing the ntp303 5′ and CaMV 35S 3′-UTRs (black) and the H-I deletion constructs (white) in pollen tubes. Results are given as means ± se (n ≥ 6). Details about the experimental procedure are given in “Results” and “Materials and Methods.”
Figure 8.
Effect of H-II stem loop structure deletions within the ntp303 5′-UTR on luciferase activity. A, Graphic representation of the ntp303 5′-UTR with different H-II deletions. Secondary structure prediction and enthalpy calculation was performed with the RNAdraw software package (Matzura and Wennborg, 1996). Deletion of 70 nucleotides at the 3′ terminus of the ntp303 5′-UTR gave rise to the Δ70 303 5′/35S 3′ construct. Internal deletion of the H-II structure gave rise to the ΔH-II 303 5′/35S 3′ construct. The start of the 5′-UTR is indicated by 5′. B, Autoluminescence of the luciferine substrate (black) and luciferase activity of the H-II deletion constructs (white) in pollen tubes. Results are given as means ± se (n ≥ 6). See “Results” and “Materials and Methods” for details about the experimental procedure.
The Predicted H-I and H-II Structures in the Ntp303 5′-UTR Influence Transcript Accumulation and Translation Efficiency
Whether the decrease in translation of the 5′-UTR deletion constructs was the result of a change in the transcript level or translation efficiency was investigated by measuring the relative transcript and luciferase activity levels of some of the ntp303 5′-UTR deletion constructs (Table II). The relative transcript values of the constructs containing deletions of the complete H-II structure (Δ70 303 5′/35S 3′ and ΔH-II 303 5′/35S 3′) dropped to a level that was more than 2.5-fold lower than that of the construct containing the unmodified ntp303 5′-UTR. Internal deletion of the (GAA)8 repeat (ΔGAA 303 5′/35S 3′) resulted in a relative transcript level that was somewhat lower than the transcript level of 303 5′/303 3′. In contrast to the effects of either the deletion of the (GAA)8 repeat or the H-II structure on the relative transcript level, a more drastic effect was observed for the luciferase activity levels. A drastic decrease in luciferase activity was observed after deletion of the H-II structure, and the measured values were in the range of the luciferine autoluminescence background. Deletion of the (GAA)8 repeat revealed an almost 2-fold lower luciferase activity value compared with 303 5′/303 3′. From these data, we conclude that the drop in translation observed after deletion of either the H-I or H-II structures is the result of a decrease in the transcript level, but most drastically in the translation efficiency.
Table II.
Effect of different ntp303 5′-UTR deletions on the relative transcript (relative luciferase mRNA abundance) and luciferase activity (relative luciferase activity/10 s−1) levels of different constructs during pollen tube growth
| Construct | Relative Luc mRNA Abundance | Relative LUC Activity |
|---|---|---|
| counts | RLA/10 s−1 | |
| 303 5′/303 3′ | 2.97 ± 0.09 (1.00) | 31.08 ± 5.61 (1.00) |
| ΔGAA 303 5′/35S 3′ | 2.30 ± 0.51 (0.77) | 17.49 ± 4.76 (0.56) |
| Δ70 303 5′/35S 3′ | 1.13 ± 0.06 (0.38) | 0.01 ± 0.00 (0.00) |
| ΔH-II 303 5′/35S 3′ | 0.82 ± 0.09 (0.28) | 0.01 ± 0.00 (0.00) |
The values in parentheses represent the fold increase of the transcript and luciferase activity levels compared with the levels of the 303 5′/303 3′ construct. Results are given as means ± se (n ≥ 6). For details, see “Results.” RLA, Relative LUC activity.
DISCUSSION
In the present study, we investigated the mechanism underlying 5′-UTR-mediated enhancement of translation in pollen. Although the transient expression results have to be confirmed by stable transformants, we conclude that the 5′-UTR of the ntp303 gene exhibits the capacity to enhance translation (measured as luciferase activity) in pollen tubes, where it acts as an autonomous enhancer element independent of linked promoter, coding region, or 3′-UTR sequences. Specific sequences within two predicted stem loop structures of the 5′-UTR are essential for enhanced translation. Enhancer activity mediated by the 5′-UTR is absent during pollen development and appears specifically during pollen tube growth. The enhancement effect of the ntp303 5′-UTR is not limited to pollen tubes but also can be found in sporophytic tissues.
In growing pollen tubes, the high level of translation of ntp303 transcripts is fully dependent on the 5′-UTR. The presence of the element led to activity values of two reporter genes to a level up to 60-fold higher than that of constructs containing control 5′-UTRs (syn44 5′ and syn99 5′; Fig. 2B). The enhancement effect is not the result of a structurally inefficient translation of the control constructs because in this case the ntp303 5′-UTR would lead to a luciferase activity level that is similar in developing pollen, growing pollen tubes, and leaves. The similar ratios of luciferase activity levels for the control constructs during pollen development and pollen tube growth (Fig. 2, A and B) argue also against their inefficient translation.
Comparison of the relative transcript and luciferase activity values mediated by the ntp303 5′-UTR or the control UTRs revealed that the enhancement is mainly the result of an increase in the translation efficiency (Table I). In line with this, the level of enhancement is only slightly influenced by the strength of the promoter, which is clear from the activities of the constructs containing the CaMV 35S promoter (Fig. 5A). That the 5′-UTR may be a regulatory site in the modulation of translation fits the scanning model of protein synthesis in which the pre-initiation complex scans the 5′-UTR in search of the first translation initiation codon (Kozak, 1999). Although the ntp303 5′-UTR acts mainly at the translational level, an increase in the relative transcript level was also observed. Whether the increase of the relative transcript level was the result of an increase in the transcription rate or transcript stability remains to be investigated. We assume that the 5′-UTR affects the transcript stability because the enhancement of translation is rather independent of the transcriptional activity of the linked promoter.
The regulatory effect of the ntp303 5′-UTR is also independent of the linked coding region and 3′-UTR sequences (Figs. 3B and 4). Therefore, the ntp303 5′-UTR acts as an autonomous element in the modulation of translation. Furthermore, this autonomy is also apparent in sporophytic tissue, although the difference is larger in growing pollen tubes, which argues for the involvement of pollen tube-preferential factors.
Because the action of the ntp303 5′-UTR is independent of other gene constituents, we hypothesize that the ntp303 5′-UTR contains sequence elements or secondary structures that are essential for the regulatory effect. The presence, position, and architecture of secondary structures and the nature of primary sequences have been demonstrated to modulate the level of translation efficiency of mRNAs in higher eukaryotes (Kozak, 1989; Bailey-Serres and Dawe, 1996; Klaff et al., 1996; Curie and McCormick, 1997; Gallie et al., 2000). We created several deletions within the ntp303 5′-UTR to identify putative regulatory elements in the ntp303 5′-UTR (Figs. 7A and 8A). Deletions of the predicted H-II stem loop structure in the 5′-UTR resulted in a strong decrease in luciferase activity during pollen tube growth (Fig. 8B). Determination of the relative transcript levels also revealed a strong decrease in transcript accumulation (Table II). We assume that deletion of the H-II stem loop structure predominantly affects mRNA stability because the ntp303 5′-UTR-mediated translation enhancement is mainly independent of the linked ntp303 promoter. Although the decrease in the transcript level could not completely account for the drop of translation of the H-II UTR deletion constructs, we conclude that the H-II stem loop structure contains sequence elements that are important for the determination of ntp303 mRNA stability. It is known that 5′-UTRs can modulate mRNA stability in plants (Dickey et al., 1998; Anderson et al., 1999; Nickelsen et al., 1999; Hua et al., 2001). Like the H-II deletions, removal of the predicted H-I stem loop structure also caused a strong decrease in the luciferase activity level during pollen tube growth (Fig. 7B). The strongest effect on the luciferase activity level was established after internal deletion of the (GAA)8 repeat. Unlike the H-II deletions, deletion of the (GAA)8 repeat caused only a slight decrease in the relative transcript level (Table II). From this, we conclude that the H-I stem loop structure contains sequence elements that are important for the modulation of translation efficiency. The decrease in the luciferase activity level after deletion of (parts of) the H-I stem loop structure are in contradiction to the generally accepted view that removal of secondary structures within the 5′-UTR often results in higher translation by facilitating scanning (Kozak, 1989; Gallie et al., 2000). This indicates that primary sequences within the H-I stem loop, rather than structural characteristics of the ntp303 5′-UTR, determine the regulatory effect. This conclusion is strengthened by the observation that internal deletion of the (GAA)8 repeat in the H-I structure, which causes a minor alteration in the overall ΔG value of the ntp303 5′-UTR, led to complete inhibition of the enhanced translation. The (GAA)8 repeat clearly represents a primary sequence element within the ntp303 5′-UTR that is necessary for enhancement of translation during pollen tube growth.
In contrast to the effect of the ntp303 5′-UTR in pollen tubes and sporophytic cells, no enhancement of luciferase activity was observed in developing pollen (Fig. 2A). This implies that, although ntp303 transcripts accumulate in developing pollen, the presence of the 5′-UTR is not sufficient to induce a high level of translation of these transcripts. Because the enhancement effect was already observed during early stages of pollen tube growth, it seems obvious that conditions at the start of pollen tube growth define the onset of the activity of the ntp303 5′-UTR. The temporal activity of the ntp303 5′-UTR differs from that of another pollen expressed gene, lat52. Here, the 5′-UTR increased luciferase activity already during pollen development (Bate et al., 1996).
With regard to mechanisms that account for the role of the ntp303 5′-UTR in regulation of translation of ntp303 transcripts during the transition of developing to germinating pollen, we propose the following model. Pollen development and pollen tube growth are two physiologically distinct phases in the life span of the pollen grain. In the final stage of development, progressive dehydration transforms the developing pollen grain into a dormant structure that contains a large stock of presynthesized rRNAs, tRNAs, ribosomes, and mRNAs (for review, see Mascarenhas, 1990, 1993). To avoid premature degradation, it is obvious that these stored transcripts exhibit a high degree of stability. Ntp303 transcripts have been shown to be highly stable during pollen development (Ylstra and McCormick, 1999). The drastic decrease in the transcript level after deletion of the H-II stem loop structure strongly suggests that parts of the ntp303 5′-UTR are involved in stabilization of ntp303 transcripts that are utilized during subsequent pollen tube growth. Delayed translation of stored ntp303 transcripts has been experimentally demonstrated (Čapková et al., 1994; Štorchová et al., 1994; Wittink et al., 2000). Rehydration of the mature pollen grain leads to (re-) initiation of translation. In such a case, the translation machinery must select transcripts that code for products needed for pollen germination and tube growth from the total mRNA population. It may be assumed that such a preferential translation of transcripts occurs through interaction of specific factors with selected 5′-UTRs. Such a mechanism would explain the enhancement of translation mediated by the ntp303 5′-UTR during pollen tube growth. The decrease in translation after deletion of (parts of) the H-I stem loop structure within the 5′-UTR might be due to removal of sequences that are crucial for the interaction with these “enhancer” factors. It is plausible that other pollen-expressed genes that encode for products that are needed for pollen germination or tube growth are regulated by a similar mechanism. Examination of the architecture of 5′-UTRs of other pollen genes by computer analysis is suitable to investigate this possibility. A first attempt has been made to address this question by means of computational pattern discovery (R.J.M. Hulzink, unpublished data).
The key function of the 5′-UTR of ntp303 transcripts to direct stage-dependent protein synthesis exhibits parallels with translation regulation mechanisms in reproduction processes in animals and other plant systems. Examples are enhanced translation of transcripts during spermatogenesis (Schäfer et al., 1993; Nayernia et al., 1994; Gu and Hecht, 1996), oocyte development (Lasko, 1999), and gametophyte development (Bate et al., 1996). As in pollen, changes in the activity of 5′-UTRs of genes that are under post-transcriptional control in these systems often occur during transition of tissues or cells to another physiological state. In this respect, it would be intriguing to examine whether components of the regulatory mechanisms of translation of stored pollen transcripts are conserved in other reproduction systems.
MATERIALS AND METHODS
Plant Material
Greenhouse-grown plants of tobacco (Nicotiana tabacum L. cv Petit Havana SR1) were used as the source of pollen and leaf tissue for microprojectile bombardment. To assess the translation of the different UTR gene fusion constructs during pollen development, immature pollen of the late-bicellular stage were aseptically isolated from flower buds of 35 mm in length in M1 medium as previously described (Tupý et al., 1991). Translation of the UTR gene fusion constructs during pollen tube growth was measured using mature pollen that were isolated from dehiscent tobacco flowers (Van Herpen et al., 1992). After isolation, the pollen pellet was suspended in 100 μL of M1 medium at a density of 108 cells mL−1. To fixate the pollen for particle bombardment, the pollen suspension was pipetted onto the surface of a sterile Hybond N+ membrane (Amersham, Buckinghamshire, UK) that was placed on 1% (w/v) agar solidified M1 medium. Following bombardment, the membrane containing late bicellular or mature pollen was soaked in 10 mL of M1 medium or Read medium (Read et al., 1993a, 1993b), respectively. The late bicellular pollen was incubated at 25°C in the dark at vigorous shaking. After centrifugation, the mature pollen was suspended in a 10-mL tube containing 0.5 mL of Read medium followed by a 20-h incubation in the dark at 25°C. Treatment of leaf tissue before and after bombardment was performed as described by Hamilton et al. (1992). In all cases, bombardments were performed within 60 min of placing the plant material onto the solidified medium.
Preparation of Gene Fusion Constructs Containing Different UTRs
In all constructs, either a modified version of the firefly luciferase coding region, luc+, or a luciferase coding region from Renilla reniformis (rluc) was used as the reporter gene (Fig. 1A). The luc+ coding region was amplified by PCR on the pGL3 vector (Promega) using a forward sequence-specific primer which introduced a NcoI site at the 5′ end (5′-ATATCCATGGAAGACGCC, NcoI site underlined) and a reverse sequence-specific primer that introduced a BamHI site at the 3′ end (5′-ATATGGATCCTTACACGGCGATC, BamHI site underlined). The rluc coding region was amplified by PCR on the pRL-SV40 vector (Promega) using the following sequence-specific primers: 5′-GTGTCCATGGATGACTTCGAAAG (NcoI site underlined) and 5′-GTGTGGATCCTTATTGTTCATTTTTGAG (BamHI site underlined). For construction of 35Ssyn44 5′/35S 3′, the PCR product of luc+ was digested with NcoI and BamHI and, after removal of the luciferase coding region, ligated into the NcoI and BamHI sites of pRTS2LUC (Bate et al., 1996). pRTS2LUC contains the CaMV 35S promoter (Wilkinson et al., 1997), a 44-bp-long synthetic 5′-UTR (designated as syn44 5′), the luciferase coding region, and the CaMV 35S3′-UTR. An almost identical construct was built, 35SRsyn44 5′/35S 3′, in which the luc+ coding region was replaced by the rluc coding region. To obtain a gene fusion construct containing both ntp303 UTRs and the CaMV 35S promoter (35S303 5′/303 3′), the syn44 5′-UTR was removed from 35Ssyn44 5′/35S 3′ using XhoI and NcoI restriction enzymes. The ntp303 5′-UTR was amplified by PCR on the ntp303 genomic clone (Weterings et al., 1995) using the following primers with restriction sites incorporated into the 5′ end: 5′-GTGTCTCGAGCAAGCTCTAGCAGGAAG (XhoI site underlined) and 5′-GTGTCCATGGGACGTTGTTTTTTTTATTC (NcoI site underlined). Following the PCR, the ntp303 5′-UTR was treated with XhoI and NcoI restriction enzymes and ligated in the 35Ssyn44 5′/35S 3′ construct (lacking the syn44 5′-UTR) to create the construct 35S303 5′/35S 3′. The oligonucleotides 5′-ATATGGATCCATTCTGTAATGATCAATCTG (BamHI site underlined) and 5′-ATATGAGCTCATTTAATGTTTTGTCCTA (SacI site underlined) were used to generate the ntp303 3′-UTR using the ntp303 genomic clone as a template. The PCR product was digested with BamHI and SacI and cloned into 35S303 5′/35S 3′ (replacing the CaMV 35S3′-UTR) to create 35S303 5′/303 3′.
UTR gene fusion constructs containing the ntp303 promoter were made as follows. Using the genomic clone of ntp303 as a template, a 578-bp-long promoter fragment, including the transcription initiation site (Weterings et al., 1995), was amplified with the primers 5′-ATATAAGCTTGATACACTCGCAACGTGTGT (HindIII site underlined) and 5′-ATATCTCGAGGAGCTTGCACTATTCACCAT (XhoI site underlined). The amplified ntp303 promoter fragment was digested with HindIII and XhoI and, after removal of the CaMV 35S promoter, ligated into 35S Rsyn44 5′/35S 3′, 35S303 5′/35S 3′, and 35S303 5′/303 3′ to create Rsyn44 5′/35S 3′, 303 5′/35S 3′, and 303 5′/303 3′, respectively. To obtain a construct containing the ntp303 promoter, the ntp303 UTRs and the R. reniformis luciferase coding region (R303 5′/303 3′), the luc+ coding region was digested from 303 5′/303 3′ using NcoI and BamHI, after which the rluc coding region was ligated into the NcoI and BamHI sites. All constructs that were linked with the ntp303 promoter and the luc+ coding region contained a longer version of the synthetic 5′-UTR that was used in the other constructs. This 99-bp-long synthetic 5′-UTR was obtained by PCR using the pNBL52–42 plasmid (Bate et al., 1996) as the template. This fragment, designated as syn99 5′, was amplified using the following primers: 5′-GTGTCTCGAGGATCATTGCAATTGGATCC (XhoI site underlined) and 5′-GTGTCCATGGCCGCGGG (NcoI site underlined). After removal of the ntp303 5′-UTR from 303 5′/35S 3′ and 303 5′/303 3′, the syn99 5′-UTR was cloned into the XhoI and NcoI sites, creating the syn99 5′/35S 3′ and syn99 5′/303 3′ constructs, respectively.
Constructs containing deletions in the ntp303 5′-UTR (Δ 5′-UTR) were obtained by PCR using the ntp303 5′-UTR in the 303 5′/35S 3′ construct as starting material. In the forward primers, a XhoI restriction site was incorporated, whereas the reverse primers contained a NcoI restriction site. Schematic drawings of these modified ntp303 5′UTRs are represented in Figures 7A and 8A. All fragments, which were obtained by PCR, were sequenced completely to exclude mismatches within the sequences. All constructs used for the transient expression assays were in the pUC19 plasmid.
Microprojectile Bombardment
Microcarriers, rupture discs, and macrocarriers were obtained from Bio-Rad Laboratories (Hercules, CA). Preparation and coating of the microcarriers was performed according the manufacturer's manual (Bio-Rad Laboratories). For biolistic transformation of late bicellular pollen and mature pollen, we used per bombardment 250-μg gold particles with a size of 1 and 1.6 μm, respectively. The microcarriers were coated with a total amount of 1 μg of DNA containing 0.7 μg of test construct DNA and 0.3 μg of normalization construct DNA. Test constructs containing the ntp303 promoter and the luc+ coding region, the CaMV 35S promoter, and the luc+ coding region, or the ntp303 promoter and the rluc coding region, were coprecipitated with the constructs Rsyn44 5′/35S 3′, 35SRsyn44 5′/35S 3′, and syn44 5′/35S 3′, respectively. Microprojectile bombardment was performed using the helium-driven PDS-1000/He System (Bio-Rad Laboratories). For biolistic transformation of pollen and leaves, the following bombardment parameters were used: a target distance of 6 cm, a gap distance of one-fourth inch, a macroprojectile/stopping screen distance of 8 mm, a chamber vacuum of 28 mm Hg, and a burst pressure of the rupture discs of 1,100 psi.
Total RNA Isolation and Northern-Blot Analysis
To determine both the relative transcript and luciferase activity levels, pollen were separated in two fractions. One fraction was used for total RNA isolation and the other for the luciferase assay. Total RNA was isolated as described by Van Eldik et al. (1995). Ten micrograms of total RNA was denatured for 1 h at 50°C in a glyoxal/dimethyl sulfoxide mixture (Sambrook et al., 1989). After denaturation, the total RNA samples were loaded on Hybond N+ membranes according to the manufacturer's manual (Amersham). The dot blots were hybridized with a 32P-labeled luciferase probe for 20 h at 65°C in 6× SETS buffer (20× SETS stock = 3 m NaCl, 0.4% [w/v] polyvinylpyrrolidone, and 4% [w/v] bovine serum albumin), 5× Denhardts (50× stock = 1% [w/v] Ficoll, 1% [w/v] polyvinylpyrrolidone, and 1% [w/v] bovine serum albumin), 0.1% (w/v) SDS, and 75 μg mL−1 denatured herring sperm DNA. Washings were performed for 30 min at 65°C in 2× SSC, 0.1% (w/v) SDS; 1× SSC, 0.1% (w/v) SDS; and 0.5× SSC, 0.1% (w/v) SDS. The blots were exposed to X-omat films (Eastman-Kodak, Rochester, NY) using two intensifying screens at −80°C.
Luciferase Assay
After particle bombardment and incubation of the tissues, quantitative determination of translation, as determined by the luciferase activity of the UTR gene fusion constructs, was performed using chemicals of the commercial available Dual-Luciferase Reporter Assay System (Promega). In this assay, the activities of the LUC+ and RLUC luciferases were measured sequentially from a single sample extract using a luminometer provided with two auto-injectors (Wallac 1420 VICTOR2, PerkinElmer, Boston). Preparation of the buffers used in the assay was performed according the manufacture manual (Promega). After incubation, the developing pollen were transferred into a 10-mL Greiner tube and collected by centrifugation for 2 min at 2,500 rpm. Germinating pollen were collected by centrifugation for 5 min at 1,000 rpm. In all cases, the pollen pellet was resuspended in 100 μL of 1× passive lysis buffer (Promega) and ground in liquid nitrogen. The pollen extracts were stored at −70°C until they were used for the luciferase activity assay. Extracts (10 μL) were pipetted in a microtiter plate, after which 100 μL of Luciferase Assay Reagent II (Promega) was added automatically. After 2 s, chemiluminescence was measured for 10 s, which gave rise to a value representing LUC+ activity per 10-s measuring time. After quantification of the LUC+ luminescence, the reaction was quenched and the RLUC reaction was initiated by the addition of 100 μL of Stop&Glo Reagent (Promega) to the extract. Two seconds after addition of the Stop&Glo Reagent, RLUC luminescence was measured for 10 s. This measurement represents the RLUC activity per 10-s measuring time. Variability of the translation between independent experiments was normalized by calculation of the ratio of LUC+:RLUC, which gave rise to a value representing the relative luciferase activity per 10-s measuring time (relative luciferase activity/10 s−1).
ACKNOWLEDGMENT
We gratefully thank Prof. Dr. Tom Gerats for critical reading of the manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001701.
LITERATURE CITED
- Anderson MB, Folta K, Warpeha KM, Gibbons J, Gao J, Kaufman LS. Blue light-directed destabilization of the pea Lhcb1*4 transcript depends on sequences within the 5′ untranslated region. Plant Cell. 1999;11:1579–1589. doi: 10.1105/tpc.11.8.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J. Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 1999;4:142–148. doi: 10.1016/s1360-1385(99)01386-2. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Dawe RK. Both 5′ and 3′ sequences of maize adh1 mRNA are required for enhanced translation under low-oxygen conditions. Plant Physiol. 1996;112:685–695. doi: 10.1104/pp.112.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N, Spurr C, Foster GD, Twell D. Maturation-specific translational enhancement mediated by the 5′-UTR of a late pollen transcript. Plant J. 1996;10:613–623. doi: 10.1046/j.1365-313x.1996.10040613.x. [DOI] [PubMed] [Google Scholar]
- Bedinger P. The remarkable biology of pollen. Plant Cell. 1992;4:879–887. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čapková V, Zbrožek J, Tupý J. Protein synthesis in tobacco pollen tubes: preferential synthesis of cell-wall 69-kDa and 66-kDa glycoproteins. Sex Plant Reprod. 1994;7:57–66. [Google Scholar]
- Curie C, McCormick S. A strong inhibitor of gene expression in the 5′ untranslated region of the pollen-specific Lat59 gene of tomato. Plant Cell. 1997;9:2025–2036. doi: 10.1105/tpc.9.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Tuite MF. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF. Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell. 1998;10:475–484. doi: 10.1105/tpc.10.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T. Translation in plants-rules and exceptions. Plant Mol Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. Post-transcriptional regulation of gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:77–105. [Google Scholar]
- Gallie DR. Translational control of cellular and viral mRNAs. Plant Mol Biol. 1996;32:145–158. doi: 10.1007/BF00039381. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Ling J, Niepel M, Morley SJ, Pain VM. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hecht NB. Translation of a testis-specific Cu/Zn superoxide dismutase (SOD-1) mRNA is regulated by a 65-kilodalton protein which binds to its 5′ untranslated region. Mol Cell Biol. 1996;16:4535–4543. doi: 10.1128/mcb.16.8.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DA, Roy M, Rueda J, Sindhu RK, Sanford J, Mascarenhas JP. Dissection of a pollen-specific promoter from maize by transient transformation assays. Plant Mol Biol. 1992;18:211–218. doi: 10.1007/BF00034950. [DOI] [PubMed] [Google Scholar]
- Honys D, Combe JP, Twell D, Čapková V. The translationally repressed pollen-specific ntp303 mRNA is stored in non-polysomal mRNPs during pollen maturation. Sex Plant Reprod. 2000;13:135–144. [Google Scholar]
- Hua XJ, van de Cotte B, Van Montagu M, Verbruggen N. The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J. 2001;26:157–169. doi: 10.1046/j.1365-313x.2001.01020.x. [DOI] [PubMed] [Google Scholar]
- Klaff P, Riesner D, Steger G. RNA structure and the regulation of gene expression. Plant Mol Biol. 1996;32:89–106. doi: 10.1007/BF00039379. [DOI] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Kuhn C, Börsch D, Glätzer KH, Schäfer U, Schäfer M. A cluster of four genes selectively expressed in the male germ line of Drosophila melanogaster. Mech Dev. 1991;35:143–151. doi: 10.1016/0925-4773(91)90064-d. [DOI] [PubMed] [Google Scholar]
- Lasko P. RNA sorting in Drosophila oocytes and embryos. FASEB J. 1999;13:421–433. doi: 10.1096/fasebj.13.3.421. [DOI] [PubMed] [Google Scholar]
- Lin Y-K, Dickinson DB. Ability of pollen to germinate prior to anthesis and effect of desiccation on germination. Plant Physiol. 1984;74:746–748. doi: 10.1104/pp.74.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. The male gametophyte of flowering plants. Plant Cell. 1989;1:657–664. doi: 10.1105/tpc.1.7.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. Gene activity during pollen development. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:317–338. [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput Appl Biosci. 1996;12:247–249. doi: 10.1093/bioinformatics/12.3.247. [DOI] [PubMed] [Google Scholar]
- McCormick S. Molecular analysis of male gametogenesis in plants. Trends Genet. 1991;7:298–303. doi: 10.1016/0168-9525(91)90312-E. [DOI] [PubMed] [Google Scholar]
- McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Reim K, Oberwinkler H, Engel W. Diploid expression and translational regulation of rat acrosin gene. Biochem Biophys Res Commun. 1994;202:88–93. doi: 10.1006/bbrc.1994.1897. [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Fleischmann M, Boudreau M, Rahire E, Rochaix J-D. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp RGL, Kuhlemeier C. Phosphorylation of tobacco eukaryotic translation initiation factor 4A upon pollen tube germination. Nucleic Acids Res. 1998;26:2058–2062. doi: 10.1093/nar/26.9.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- Read SM, Clarke AE, Bacic A. Requirements for division of the generative nucleus in cultured pollen tubes of Nicotiana. Protoplasma. 1993a;174:101–115. [Google Scholar]
- Read SM, Clarke AE, Bacic A. Stimulation of growth of cultured Nicotiana tabacum W38 pollen tubes by polyethylene glycol and Cu(II) salts. Protoplasma. 1993b;177:1–14. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatus T. Electrophoresis of RNA after denaturation with glyoxal and dimethyl sulfoxide. In: Ford N, Nolan C, Ferguson M, editors. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 7.40–7.42. [Google Scholar]
- Schäfer M, Börsch D, Hülster A, Schäfer U. Expression of a gene duplication encoding conserved sperm tail proteins is translationally regulated in Drosophila melanogaster. Mol Cell Biol. 1993;13:1708–1718. doi: 10.1128/mcb.13.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Nayernia K, Engel W, Schäfer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Štorchová H, Čapková V, Tupý J. A Nicotiana tabacum mRNA encoding a 69-kDa glycoprotein occurring abundantly in pollen tubes is transcribed but not translated during pollen development in the anthers. Planta. 1994;192:441–445. [Google Scholar]
- Stutz A, Conne B, Huarte J, Gubler P, Völkel V, Flandin P, Vassalli J-D. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of dormant mRNA in mouse oocytes. Genes Dev. 1998;12:2535–2548. doi: 10.1101/gad.12.16.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson KI, Borgese N, Pietrini G, Ruderman JV. Three translationally regulated mRNAs are stored in the cytoplasm of clam oocytes. Dev Biol. 1987;123:10–16. doi: 10.1016/0012-1606(87)90421-0. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Tupý J, Řihová L, Žárský V. Production of fertile tobacco pollen from microspores in suspension culture and its storage for in situ pollination. Sex Plant Reprod. 1991;4:284–287. [Google Scholar]
- Twell D, Klein TM, Fromm ME, McCormick S. Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 1989;91:1270–1274. doi: 10.1104/pp.91.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst AC, Pierson ES, van Went JL, Cresti M. Ultrastructural changes of Arabidopsis thaliana pollen during final maturation and rehydration. Zygote. 1993;1:173–179. doi: 10.1017/s096719940000143x. [DOI] [PubMed] [Google Scholar]
- Van Eldik GJ, Vriezen WH, Wingens M, Ruiter RK, van Herpen MMA, Schrauwen JAM, Wullems GJ. A pistil-specific gene of Solanum tuberosum is predominantly expressed in the stylar cortex. Sex Plant Reprod. 1995;8:173–179. [Google Scholar]
- Van Herpen MMA, De Groot PFM, Schrauwen JAM, van den Heuvel KJPT, Weterings KAP, Wullems GJ. In-vitro culture of tobacco pollen: gene expression and protein synthesis. Sex Plant Reprod. 1992;5:304–309. [Google Scholar]
- Weterings K, Reijnen W, van Aarssen R, Kortstee A, Spijkers J, van Herpen M, Schrauwen J, Wullems G. Characterization of a pollen-specific cDNA clone from Nicotiana tabacum expressed during microgametogenesis and germination. Plant Mol Biol. 1992;18:1101–1111. doi: 10.1007/BF00047713. [DOI] [PubMed] [Google Scholar]
- Weterings K, Reijnen W, Wijn G, van den Heuvel KJPT, Appeldoorn N, de Kort G, van Herpen M, Schrauwen J, Wullems G. Molecular characterization of the pollen-specific genomic clone NTPg303 and in situ localization of expression. Sex Plant Reprod. 1995;8:11–17. [Google Scholar]
- Wilkinson JE, Twell D, Lindsey K. Activities of CaMV 35S and nos promoters in pollen: implications for field release of transgenic plants. J Exp Bot. 1997;48:265–275. [Google Scholar]
- Wittink FRA, Knuiman B, Derksen J, Čapková V, Twell D, Schrauwen JAM, Wullems GJ. The pollen-specific gene Ntp303 encodes a 69-kDa glycoprotein associated with the vegetative membranes and the cell wall. Sex Plant Reprod. 2000;12:276–284. [Google Scholar]
- Ylstra B, McCormick S. Analysis of mRNA stabilities during pollen development and in BY2 cells. Plant J. 1999;20:101–108. doi: 10.1046/j.1365-313x.1999.00580.x. [DOI] [PubMed] [Google Scholar]