Abstract
A cDNA that encodes a methyltransferase (MT) was cloned from a cold-acclimated wheat (Triticum aestivum) cDNA library. Molecular analysis indicated that the enzyme WPEAMT (wheat phosphoethanolamine [P-EA] MT) is a bipartite protein with two separate sets of S-adenosyl-l-Met-binding domains, one close to the N-terminal end and the second close to the C-terminal end. The recombinant protein was found to catalyze the three sequential methylations of P-EA to form phosphocholine, a key precursor for the synthesis of phosphatidylcholine and glycine betaine in plants. Deletion and mutation analyses of the two S-adenosyl-l-Met-binding domains indicated that the N-terminal domain could perform the three N-methylation steps transforming P-EA to phosphocholine. This is in contrast to the MT from spinach (Spinacia oleracea), suggesting a different functional evolution for the monocot enzyme. The truncated C-terminal and the N-terminal mutated enzyme were only able to methylate phosphomonomethylethanolamine and phosphodimethylethanolamine, but not P-EA. This may suggest that the C-terminal part is involved in regulating the rate and the equilibrium of the three methylation steps. Northern and western analyses demonstrated that both Wpeamt transcript and the corresponding protein are up-regulated during cold acclimation. This accumulation was associated with an increase in enzyme activity, suggesting that the higher activity is due to de novo protein synthesis. The role of this enzyme during cold acclimation and the development of freezing tolerance are discussed.
Winter survival and crop productivity are influenced by many different winter stresses, such as freezing temperature, length of freezing period, ice encasement, flooding, and oxidative stress caused by low temperature (LT)-induced photoinhibition (Fowler et al., 1999). Exposure of plants to LT produces morphological, physiological, and biochemical changes that are often highly correlated with plant freezing tolerance (FT) and winter survival. These changes are regulated by LT at the gene expression level. Cold-regulated genes and their products have been identified and characterized in many species (Thomashow, 1999). The complexity of the LT response has made it difficult to separate genes responsible for LT acclimation and cold hardiness from those associated with metabolic adjustments to LT. To characterize the genetic components associated with LT response, a better understanding of the LT-responsive genes responsible for adaptation to winter stresses and their interaction with the environment is needed (Fowler et al., 1999).
Survey of the literature reveals that a large number of genes are being altered during the process of cold acclimation (CA; Thomashow, 1999). These genes could be classified into three groups based on the presumed function of the gene. The first group represents genes encoding structural proteins that may be involved in protecting the cell during LT stress. The second group represents those genes that regulate gene expression and signal transduction pathways, such as transcription factors, protein kinases, phosphatases, and the enzymes involved in phosphoinositide metabolism. The third group represents genes encoding enzymes involved in the biosynthesis of different osmoprotectants and membrane lipids and those of the antioxidative response. These studies improve our knowledge of the different metabolic pathways associated with CA, and suggest that plants seem to employ different mechanisms to ensure LT tolerance.
Among the metabolites that accumulate during the development of stress tolerance are the osmoprotectants (also termed compatible solutes). They occur in all organisms ranging from archaebacteria to higher plants and animals (Yancey et al., 1982; McNeil et al., 1999). They are highly soluble compounds that carry no net charge at physiological pH and are nontoxic at high concentrations. Osmoprotectants serve to raise osmotic pressure in the cytoplasm and can also stabilize proteins and membranes during plant growth under stressful conditions, such as freezing, drought, or high salt (Bohnert and Jensen, 1996; McNeil et al., 1999). Osmoprotectants, therefore, play important roles in the adaptation of cells to various adverse environmental conditions (Yancey, 1994). There are many amino acid (AA)-derived osmoprotectants that have been identified in plants under stressful conditions such as Pro, polyamines, and betaines (Wyn Jones and Storey, 1981; Kumar and Minocha, 1998). However, the regulation of their metabolic pathways and their function during stress tolerance are still poorly understood.
Two approaches are being used to study the regulation and the importance of a particular metabolite in stress tolerance. The first is the genetic approach using mutational analysis, gene knockout, and T-DNA tagging. The other approach is to identify the key enzymes involved and evaluate their contribution in regulating specific pathways associated with increased stress tolerance. Toward this goal, part of our functional genomic studies were designed to identify LT-regulated enzymes with the aim to understand their contribution to the enhancement of FT. Random sequencing of a cDNA library prepared from cold-acclimated winter wheat (Triticum aestivum) has provided many stress-regulated genes. One of the LT up-regulated genes exhibited a significant homology to methyltransferases (MTs). Because of the importance of methylation reactions in plant growth and development, as well as in the interactions with the environment, we conducted a detailed molecular, biochemical, and physiological characterization of this gene and its encoded protein. These analyses revealed that this gene encodes a MT that catalyzes and regulates the three-step sequential methylation of phosphoethanolamine (P-EA) to form phosphocholine (P-choline). Therefore, this enzyme was named wheat P-EA methyltransferase (WPEAMT). Substrate specificity and kinetic studies suggest that this enzyme may regulate the metabolic pathway leading to the synthesis of phosphatidylcholine (Ptd-choline), choline, and Gly betaine (GB), three important metabolites involved in LT tolerance. The molecular characterization and the putative function of this methyltransferase are discussed in relation to CA and the development of FT in cereals.
RESULTS
Molecular Analysis of the Wpeamt cDNA Sequence
Random sequencing of clones from a 1-d cold-acclimated wheat cDNA library and northern-blot analysis were used to identify LT-responsive genes. Among several LT-regulated clones, we identified a cDNA that encodes a protein with strong homology to multiple MTs. The nucleotide and the deduced AA sequences of WPEAMT are shown in Figure 1A. The longest open reading frame is 1,497 bp long and encodes a putative protein of 498 AAs with an ATG codon at nucleotide 51 and a stop codon at nucleotide 1,545. The calculated molecular mass of WPEAMT is 57 kD and its theoretical pI is 5.3.
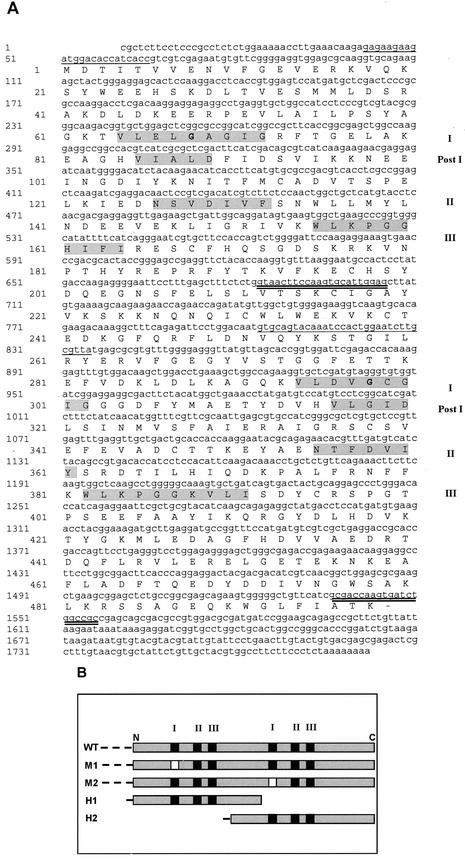
Figure 1.
Nucleotide and deduced AA sequences of Wpeamt and of mutants generated. A, Nucleotide and deduced AA sequence of Wpeamt. The open reading frame is 1,497 nucleotides. The predicted polypeptide is 498 AAs in length, with a calculated molecular mass of 57 kD and a pI of 5.3. The two putative Ado-Met-binding domains are highlighted in gray. The first domain is in the N-terminal portion of the protein from AA 64 to 164. The Ado-Met-binding domain consists of four motifs: motif I, post-I, motif II, and motif III. The second putative Ado-Met-binding domain also composed of four motifs is in the C-terminal portion of the protein from AA 294 to 391. Bold letters within each motif I indicate the site of the mutations (G–E) that gives rise to M1 or M2. The underlined nucleotides are the sequences recognized by primers to generate H1 (single) and H2 (double). GenBank accession number: AY065971. B, Schematic representation of the WT WPEAMT and its mutations and deletions. WT, WT recombinant WPEAMT; M1, recombinant WPEAMT in which motif I of the first Ado-Met-binding domain is mutated; M2, WPEAMT in which motif I of the second Ado-Met-binding domain is mutated; H1, first one-half of WPEAMT; H2, second one-half of WPEAMT. The motifs that characterize the Ado-Met-binding domain are represented by black squares (I–III). Blank squares represent the motifs in which the mutation is located. The dashed line at the beginning of each polypeptide corresponds to the His tag.
A search in the GenBank database showed that WPEAMT shares a high similarity with the recently identified proteins PEAMT from spinach (Spinacia oleracea; 86% similarity and 73% identity; accession no. AF237633) and NMT1 from Arabidopsis (88% and 77%; accession no. AAG41121). Searching the expressed sequence tag database also revealed the presence of WPEAMT homologs in other species: cotton (Gossypium hirsutum; accession no. AI731819), barley (Hordeum vulgare L. cv Winchester; accession no. BE420987), rice (Oryza sativa; accession no. BE040460), corn (Zea mays; accession no. BE344869), Physcomitrella patens (accession no. AW561535), and tomato (Lycopersicon esculentum; accession no. AW735977).
The WPEAMT sequence contains two putative Ado-Met-binding domains. The first domain is in the N-terminal portion of the protein from AA 64 to 164. Within this Ado-Met-binding domain, four consensus motifs can be identified: motif I (positions 64–72), post-I (positions 85–89), motif II (positions 126–132), and motif III (positions 155–164; Fig. 1A; Kagan and Clarke, 1994). The second putative Ado-Met-binding domain is in the C-terminal portion of the protein from AA 294 to 391. Four motifs can also be identified in this second domain: motif I (positions 294–302), post-I (positions 316–320), motif II (positions 355–361), and motif III (positions 382–391; Fig. 1A). The presence of two distinct Ado-Met-binding domains in the same protein may explain its relatively large size as compared with other typical MTs (Ibrahim and Muzac, 2000). These two sets of four motifs identified in WPEAMT are known to be relatively conserved in protein, lipid, and small molecule MTs (Kagan and Clarke, 1994).
Interestingly, plant sequence alignment using the BLAST program revealed that the highest score following spinach and Arabidopsis orthologs of WPEAMT are sterol MTs (SMTs). Sequence analysis also indicates that WPEAMT shares significant similarities with two putative MTs from the nematode Caenorhabditis elegans. The first protein temporally named ZK622.3 (accession no. T27936) is homologous with the first one-half of WPEAMT (51% similarity and 40% identity), whereas the other F54D11.1 (accession no. AAB0042824) is homologous with the second one-half of WPEAMT (50% similarity and 38% identity).
Functional Analysis of the Recombinant WPEAMT
To determine whether the Wpeamt cDNA does encode a catalytically active MT, the clone was expressed in Escherichia coli using pTrc-His B vector and the recombinant protein was purified on a His-bind column. The affinity-purified protein with an apparent molecular mass of 65 kD, containing the 8-kD histidine tag and the 57-kD MT (Fig. 2A, lane 4), was used for enzyme characterization. For antibody production, the protein was further purified on SDS-PAGE, excised from the gel, electro-eluted, and injected into a rabbit. Figure 2A shows the immunoblot of the protein at different steps of purification.
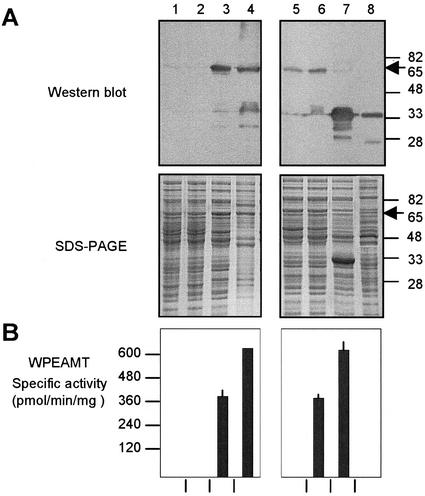
Figure 2.
Functional analysis of the recombinant WPEAMT. The WT WPEAMT, its mutants (M1 and M2), and deletions (H1 and H2) were expressed as N-terminal His tagged fusions in E. coli. All proteins (WT, M1, M2, and H1) were induced with isopropyl-β-d-thiogalactoside (IPTG) at 37°C for 3h except for H2, which was induced at 20°C for 16h. A, Upper, Immunoblot with the anti-WPEAMT antibodies. Lower, Protein pattern stained with Coomassie brilliant blue R-250. B, WPEAMT-specific activity using P-EA as substrate. The arrow indicates the location of the N-terminal fusion of the WT WPEAMT and mutants (65 kD). Lane 1, Untransformed E. coli; lane 2, transformed with WT WPEAMT before induction with IPTG; lane 3, WT WPEAMT after induction with IPTG; lane 4, purified WT WPEAMT on a His-bind resin column; lane 5, mutant M1 after induction; lane 6, mutant M2 after induction; lane 7, deletion H1 after induction; lane 8, deletion H2 after induction.
To determine the substrate specificity, the desalted bacterial lysate was assayed against the following substrates: P-EA, phosphatidylethanolamine, ethanolamine, γ-tocopherol, desmosterol, lanosterol, cholesterol, lathosterol, campesterol, 7-dihydrocholesterol, sitosterol, apigenin, and caffeic acid. Of all the substrates tested, P-EA was the only good methyl acceptor, although a low activity was obtained with lanosterol.
The recombinant WPEAMT was affinity purified and its enzymatic activity was measured following each purification step using P-EA as the substrate (Fig. 2B). Neither the untransformed nor the transformed but noninduced E. coli lysates exhibited any P-EAMT activity (Fig. 2B, lanes 1 and 2). P-EAMT activity was only detected in the transformed, IPTG-induced bacteria (Fig. 2B, lane 3). The P-EAMT activity was found to increase in the affinity-purified (His-Bind column) fraction after removing nickel ions by dialysis (Fig. 2B, lane 4). The purified enzyme has Km values of 65 and 56 μm for P-EA and Ado-Met as substrate and cosubstrate, respectively.
Contribution of the Two Ado-Met-Binding Domains to the Catalytic Activity of WPEAMT
To determine the contribution of each of the Ado-Met-binding domains to the catalytic properties of WPEAMT, we separated the N-terminal (H1) and the C-terminal domains (H2; Fig. 1B). The resulting truncated clones were expressed in E. coli as described for the intact WPEAMT. The recombinant proteins thus obtained were tested for authenticity using the India system for His tag detection and the WPEAMT polyclonal antibodies. The enzyme activities were tested as described in the experimental procedures. Removal of the first one-half of WPEAMT (H2) completely abolished P-EAMT activity, whereas deletion of the second one-half (H1) had no effect on enzyme activity (Fig. 2B, lanes 7 and 8).
Because motif I in the Ado-Met-binding domain is believed to be the most important motif involved in Ado-Met binding (Cheng et al., 1993), we mutated separately the motif I in the N-terminal (M1) and C-terminal (M2) Ado-Met-binding domain of WPEAMT. In the mutated motif, a Glu residue replaced the Gly residue at position 5 (Fig. 1). It was shown that this mutation on another MT inactivated the binding capacity for the Ado-Met molecule and, therefore, prevented substrate methylation (Wilke et al., 1988). The resulting mutated clones were also expressed in E. coli and their recombinant proteins were purified and tested for authenticity using the India system for His tag detection and the WPEAMT polyclonal antibodies. Enzyme assays demonstrated that the single point mutation in the N-terminal Ado-Met-binding domain completely abolished P-EAMT activity, whereas the mutation in the C-terminal domain did not affect the enzyme activity (Fig. 2B, lanes 5 and 6).
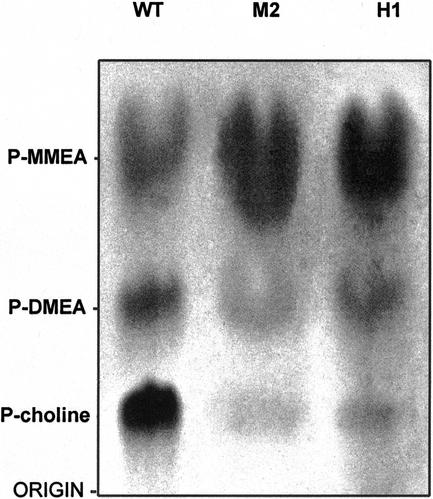
The two characterized WPEAMT orthologs from spinach and Arabidopsis were found to catalyze three consecutive methylations of P-EA leading to the tri-methylated P-choline (Bolognese and McGraw, 2000; Nuccio et al., 2000). To determine whether WPEAMT catalyzes the three steps in this reaction, enzyme activities of the wild-type (WT) WPEAMT, both deletions (H1 and H2), and both mutants (M1 and M2) were assayed for P-EAMT activity and their reaction products were subjected to thin-layer chromatography (TLC) and autoradiography. The results show that the WT WPEAMT produced three methylated products as expected (Fig. 3). These products correspond to the monomethyl- (P-MMEA), dimethyl- (P-DMEA), and trimethyl- (P-choline) ethanolamine. The mutated form (M2) and the first one-half of the enzyme (H1) also produced the three methylated products (Fig. 3), whereas the mutated form (M1) and the second one-half (H2) were inactive in the presence of P-EA as a substrate. However, both proteins (M1 and H2) were able to methylate the intermediate P-MMEA and P-DMEA (Table I). This result indicates that the N-terminal part of WPEAMT can catalyze alone the three methylation steps of P-EA to P-choline, and suggests that the C-terminal part may play a regulatory role under specific growth conditions. Results in Figure 3 indicate that the intact WT enzyme favors the accumulation of the P-choline, whereas the mutated and truncated enzyme favor the accumulation of the P-MMEA.
Figure 3.
Autoradiogram of reaction products of the P-EA methylation by WPEAMT separated by TLC. The enzyme assay as well as sample preparation, chromatography conditions, and development of TLC plates were described in “Materials and Methods,” except that [14C] Ado-Met was used as substrate and the assay time was for 2 h. Lane 1, Reaction products of WT WPEAMT. Lane 2, Reaction products of M2. Lane 3, Reaction products of H1.
Table I.
In vitro phospho-base N-methyltransferase activities
| Substrate | Activity
|
||
|---|---|---|---|
| WT | M1 | H2 | |
| P-EA | 391.5 ± 33 | 0 | 0 |
| P-MMEA | 460.3 ± 50 | 800.3 ± 45 | 195.2 ± 25 |
| P-DMEA | 56.2 ± 5 | 225.4 ± 30 | 47.5 ± 10 |
Protein crude extracts of each construct were prepared as described in “Materials and Methods.” The phosphobase N-methyltransferase assays and quantification of radiolabelled methylated phospho-base products were performed as described in “Materials and Methods.” Each measurement is a sum of all possible phospho-base products and is presented in pmol min mg−1 (mean ± se, n = 3).
Genomic Organization
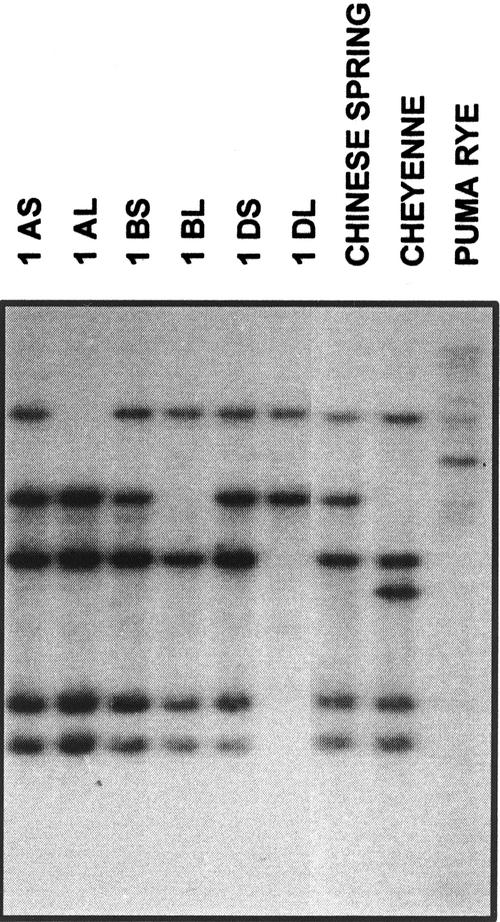
The ditelocentric (DT) series of wheat cv Chinese Spring (CS), in which one homologous pair of chromosome arms is missing in each line, was used to determine which chromosome arms carry the wpeamt genes. DNA gel-blot analysis revealed that hexaploid wheat contains five strong hybridizing fragments. With the use of the DT series, we were able to map the wpeamt gene to the short arms of the homologous group 1 chromosomes of all three genomes (A, B, and D) of hexaploid wheat (Fig. 4). These results suggest that each genome contains at least one copy of the wpeamt gene. The other Southern-blot data also show different hybridization patterns between wheat and rye species, suggesting polymorphism in the cereal family.
Figure 4.
DNA gel-blot analysis of genomic DNA using Wpeamt as a probe in DT series of wheat cv CS. The DT series of wheat cv CS, in which one homologous pair of chromosome arms is missing in each line, was used to determine which chromosome arms carry the gene. DNA gel blot of two wheat genotypes (cv CS and cv Cheyenne) and rye (Secale cereale L. cv Puma) are also shown. A, B, and D, Three genomes present in hexaploid wheat; S and L, Presence of the short or long chromosome arms, respectively. In all lanes, 1.8 μg of genomic DNA digested with XbaI was used.
Wpeamt Is Up-Regulated during CA and Salt Stress
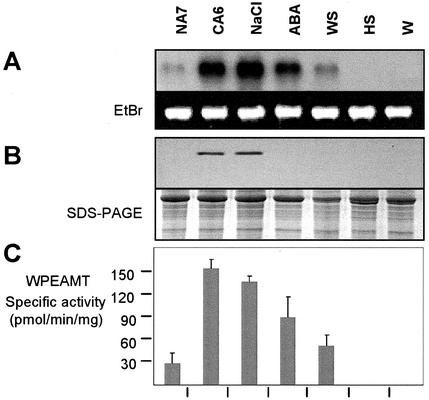
Northern analysis revealed that the Wpeamt transcripts accumulate to high levels upon exposure to LT, NaCl treatment, and to a lesser extent abscisic acid (ABA) and water stress. Heat shock and wounding have no measurable effect (Fig. 5A). The expression of Wpeamt at the protein level was also monitored by western blotting using anti-WPEAMT polyclonal antibodies. The accumulation of the WPEAMT protein follows the RNA transcript patterns for cold-stressed and NaCl-treated shoots (Fig. 5B). However, the WPEAMT protein from ABA- and water-stressed shoots does not accumulate to any detectable level. Furthermore, cellular fractionation analysis indicated that the enzyme is not associated with any cellular compartment because it was present only in the cytosolic fraction (data not shown). This suggests that the enzyme is not membrane bound.
Figure 5.
Up-regulation of WPEAMT during CA and salt stress. A, Accumulation of Wpeamt mRNAs under different stress conditions. The 28S ribosomal band stained with ethidium bromide is included to show RNA loads (7.5 μg). B, Immunoblot showing the WPEAMT accumulation. Coomassie Brilliant Blue-stained gel shows the Rubisco band as load control. C, WPEAMT activity using P-EA as substrate in soluble fractions of wheat during stress treatments. Values represent the mean ± se from four independent experiments. NA7, Nonacclimated plants grown for 7 d; CA6, 6-d cold-acclimated plants; NaCl, plants treated with 300 mm NaCl for 18 h; ABA, plants treated with 0.1 mm ABA for 18 h; WS, plants exposed to water stress for 18 h; HS, plants exposed to 40°C for 3 h (heat shock); W, wounding stress for 3 h.
The P-EAMT enzyme activity was also determined in plant tissues following the different stress treatments. The results in Figure 5C show that the P-EAMT activity follows the same pattern as the increase in mRNA level (Fig. 5A). The enzyme activity increased by 5-fold after 6 d of CA, 4-fold after NaCl addition, 3-fold after ABA treatments, and almost 1-fold after water stress. These results suggest that the increase in in vivo activity during CA and salt stress is mainly due to de novo protein synthesis during stress treatments.
Wpeamt Expression Is Associated with Increased FT
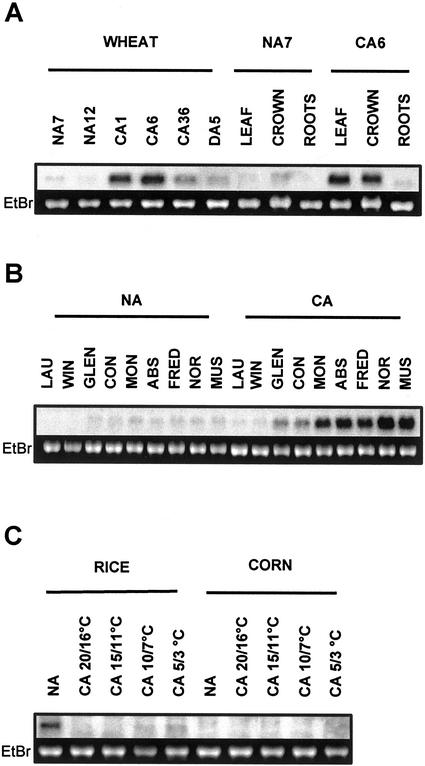
Northern-blot analysis indicates that the Wpeamt transcripts accumulate to a maximum level after 6 d of CA and then declined after 36 d. Upon de-acclimation, the level of transcripts returned to those seen in the control nonacclimated plants after 5 d (Fig. 6A). The data presented in Figure 6B also shows that the accumulation of Wpeamt mRNA correlates well with the capacity of cereal species to cold acclimate and develop FT. The accumulation was much greater in winter hardy species like wheat and rye compared with the less hardy species oat and barley. It is also interesting to note that among the different wheat cultivars used in this study, the accumulation of Wpeamt mRNA was closely related to their acclimation capacity. This may suggest that the Wpeamt transcript accumulation could be used as a marker to select for FT in cereals. The accumulation of Wpeamt transcripts was also found to be tissue specific because they were detected only in cold-acclimated leaf and crown tissues (Fig. 6A). In natural conditions, the aerial part of the plant (leaves and crown tissues) has a higher capacity to cold acclimate and develop a greater FT than the roots. Thus, the preferential expression of WPEAMT in leaves and crown tissues suggests a close correlation between the synthesis of the WPEAMT and the tissue capacity to develop a significant FT. In LT-sensitive species such as rice and corn, the Wpeamt transcripts were much less abundant compared with the LT-tolerant species (Fig. 6C). In addition, there was no accumulation upon LT exposure. This indicates that the Wpeamt homolog does not respond to LT in these sensitive species.
Figure 6.
Accumulation of Wpeamt mRNAs during CA in cereals. A, Time course and tissue specificity in winter wheat cv Norstar. NA7 and NA12, Nonacclimated plants grown for 7 and 12 d; CA1, CA6, and CA36, cold-acclimated plants for 1, 6, and 36 d; D5, cold-acclimated plants (36 d) were de-acclimated for 5 d. Exposure was for 3 h. B, Wheat genotypes and species differential accumulation. In this study, total RNA from two spring wheat genotypes (cv Glenlea [Glen], LT50 [lethal temperature that kills 50% of the seedlings] of −8°C; and cv Concorde [Con], LT50 of −8°C), four winter wheat genotypes (cv Monopole [Mon], LT50 of −15°C; cv Absolvent [Abs], LT50 of −16°C; cv Fredrick [Fred], LT50 of −16°C; and cv Norstar [Nor], LT50 of −19°C), winter rye (cv Musketeer [Mus], LT50 of −21°C), oat (Avena sativa L. cv Laurent [Lau], LT50 of −6°C), barley (cv Winchester [Win], LT50 of −7°C) were used. NA, Control plants (nonacclimated) grown for 12 d; CA, plants cold acclimated for 36 d. Exposure was for 16h. C, Accumulation in rice and corn. NA, Control (nonacclimated) plants grown under a day/night temperature of 29°C/26°C; CA, (cold-acclimated) plants grown for 24 h under the corresponding day/night temperatures. Exposure was for 3 d. In all lanes, 7.5 μg of total RNA was used. The 28S ribosomal band stained with ethidium bromide is included to show RNA loads.
DISCUSSION
In the present study, we have identified an MT from wheat. The enzyme is strongly up-regulated by LT and salt stress. The molecular and biochemical analyses indicated that the enzyme is a bipartite protein with two separate Ado-Met-binding domains, one at the N-terminal end and the second at the C-terminal end. The native enzyme catalyzed the three sequential methylations of P-EA to form P-choline, a key precursor for the synthesis of Ptd-choline and GB in plants. These two metabolites play an important role in stress response. To understand the function of each Ado-Met domain in the three-step methylation reaction catalyzed by the native enzyme, a series of deletions and mutants were generated. The data obtained indicate that the first one-half of the enzyme harboring the first Ado-Met domain is important for the three methylation reactions of P-EA to P-choline, whereas the second Ado-Met-binding domain alone did not show any methylation activity of P-EA. Mutating the first Ado-Met domain completely abolished the enzyme activity confirming the importance of the first Ado-Met domain in initiating and catalyzing the three methylation steps. On the other hand, deletion and mutation experiments showed that the second part methylates specifically the intermediates P-MMEA and P-DMEA. This observation raises the question as to the exact role of the second Ado-Met domain if the first part can catalyze the three methylation steps. Several possibilities could be proposed to explain the biochemical function of the second Ado-Met domain. The first is that the second Ado-Met domain may be required in vivo to ensure the binding and availability of the CH3 groups needed for the three methylation steps and thus increase the enzyme efficiency. This function is difficult to evaluate in an in vitro system where a saturating concentration of Ado-Met is used in the assay mixture. Moreover, the C-terminal one-half of the enzyme may only participate in the second and third methylation steps of P-EA to accelerate the reaction rate and the formation of the end product P-choline. This would be particularly important if P-MMEA and P-DMEA were eventually found to have deleterious effects on cell metabolism. It is also possible that the C-terminal part becomes more active in converting both P-MMEA and P-DMEA under stress conditions and thus accelerates the reaction rate to produce the high level of P-choline needed for both choline and GB accumulation.
The homology of the second one-half of the enzyme with SMTs from plants and fungi prompted us to test the SMT activity of the native enzyme and of its C-terminal one-half. The data showed a low SMT activity with lanosterol as the substrate. This activity was too unstable to confirm without doubt the possibility of an SMT activity. A literature search revealed that SMTs exhibit a relatively low activity compared with other MTs, which may explain the difficulty in measuring this activity (Nes et al., 1998). However, further work is required to investigate the optimum conditions for this enzyme activity.
Database searches revealed that the WPEAMT enzyme is homologous to other clones recently identified in both Arabidopsis and spinach (Bolognese and McGraw, 2000; Nuccio et al., 2000). However, biochemical analysis of the spinach P-EAMT showed that the truncated enzyme lacking the C-terminal MT domain catalyzes only the first step of P-EA methylation leading to P-MMEA (Nuccio et al., 2000). Based on this observation, the authors speculated that the C-terminal part is responsible for the second and third methylations. Two possibilities can be advanced to explain the difference in substrate specificities between the first halves of the spinach and the wheat enzymes. On one hand, these enzymes differ by the presence of both the N-terminal tag and added sequence at the C-terminal end. This may potentially lead to small differences in tertiary structure that modify substrate specificity. However, comparison of the product profiles (Fig. 3) of the wheat constructs M2 and H1 (Fig. 1B) that also contain different lengths of added sequence at the N- and C-terminal ends reveals identical substrate specificities. Therefore, this observation may not adequately explain the differences between the wheat and the spinach constructs. The second possibility is based on the slightly different primary sequences of both enzymes that may mediate a subtle distortion of the active site and thus modify substrate specificity. A close analysis of the first halves of the wheat and spinach enzymes reveals approximately 50 nonidentical AAs. Because it is known that as few as one or two AA changes can have a dramatic effect on substrate specificity (Cahoon and Shanklin, 2000), it is likely that primary sequence is the principal determinant of substrate specificity.
More in-depth molecular and biochemical analyses of P-EAMTs from different plants will certainly help to identify the determinants of substrate specificity and to understand the role of this family of enzymes in regulating the choline and Ptd-choline pathways. This is important because choline is a precursor for several important metabolites such as GB in osmoregulation, choline-o-sulfate and P-choline in the transport and detoxification of inorganic compounds (Tolbert and Weibe, 1955; Wyn Jones and Storey, 1981; Rivoal and Hanson, 1994), acetylcholine as a potential signal molecule (Tretyn and Kendrick, 1991), and Ptd-choline as the major phospholipid of plant plasma membranes (Moore Jr., 1982).
Increasing the P-choline pool during CA may have many advantageous consequences for plant FT, considering the prime importance of derivatives such as Ptd-choline and GB during environmental stresses (Harwood, 1998; McNeil et al., 1999). Ptd-choline is the major constituent of non-chloroplastic plant membranes (Moore Jr., 1982). In vitro studies on the importance of lipid structure on membrane transporter activity revealed that among all factors influencing the bilayer structure of membranes, the phospholipid head group is the most important and among the different phospholipids tested, Ptd-choline had the most effective head group (Carruthers and Melchior, 1986). It has been shown that Ptd-choline increases in quantity as well as in proportion to other lipids during CA (Horváth et al., 1980; Kinney et al., 1983; Lynch and Steponkus, 1987). Ptd-choline concentration was found to be correlated with increased FT among 13 hardened wheat cultivars (Horváth et al., 1980). It is interesting to note that in C. elegans, where homologs of the wheat WPEAMT have been identified, LT treatment increases the level of several types of phosphatidylcholines (Tanaka et al., 1999). This finding suggests that nematodes and plants may share a common pathway for Ptd-choline biosynthesis.
Other evidence that supports the importance of Ptd-choline for increased FT has been obtained from studies in which the metabolic precursor of Ptd-choline has been added to plants. In cold-sensitive cucumber (Cucumis sativus), where a chilling treatment results in loss of Ptd-choline, the addition of choline, and EA increased chilling resistance (Horváth and Van Hasselt, 1985). In wheat, treatment with 15 mm choline for 6 d increased Ptd-choline content as well as FT (Horváth et al., 1981). An increase in FT has also been observed by similar treatments of wheat suspension culture (Horváth and Vigh, 1984; Williams et al., 1987). Furthermore, nonacclimated rye protoplasts treated with exogenous Ptd-choline by liposome fusion also showed signs of increased FT along with decreased propensity of expansion-induced lysis, a form of injury associated with freeze/thaw treatments (Steponkus et al., 1988).
A breakdown of membrane phospholipids was observed in the cortical cells of poplar and black locust trees (Robinia pseudoacacia) during freezing at lethal temperature (Yoshida and Sakai, 1974). Similar results were also obtained with other plant species, and Ptd-choline was identified as the primary target of this degradation (Borochov et al., 1987; Horváth et al., 1979; Sikorska and Kacperska-Palacz, 1980). To compensate for this degradation, hardy plants have the capacity to increase the choline pool needed to replace Ptd-choline and thus ensure the integrity of the plasma membrane during freezing.
The other environmentally important product of choline metabolism is GB. GB is known to be a compatible solute that acts as a noninjurious molecule accumulating when plants and bacteria are exposed to environmental stress such as LT (Sakamoto and Murata, 2001). Recent observations of the effect of GB in plants also suggest that it may play another role at low concentration. Transgenic plants that accumulate low levels of GB show signs of increased environmental stress tolerance (Sakamoto and Murata, 2001). Because the accumulation is low, this improvement could not be explained by an osmoticum effect. Thus, GB may play another important role that elicits the development of stress tolerance. Results obtained in our laboratory suggest that GB may act as an elicitor of stress-associated genes (Allard et al., 1998). Three independent observations associate GB with FT. First, it has been shown that the capacity of accumulating GB during environmental stress among different members of the Poaceae family is highly correlated with their potential to develop FT (Hitz and Hanson, 1980; Ishitani et al., 1993; Kishitani et al., 1994; Allard et al., 1998). Second, CA, which is correlated with increased FT, is associated with GB accumulation (Naidu et al., 1991; Kishitani et al., 1994; Nomura et al., 1995; Allard et al., 1998). Third, exogenous applications of GB lead to significant increases in LT tolerance (Allard et al., 1998; Sakamoto et al., 2000).
The up-regulation of Wpeamt transcripts and the corresponding proteins during CA in cold-hardy plants may be related to their ability to accumulate GB. Overexpression of WPEAMT in GB accumulators and nonaccumulators will help to understand the exact function of this enzyme in modulating GB levels and LT tolerance in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In this study we used two spring wheat (Triticum aestivum L. cv Glenlea and cv Concorde) genotypes, four winter wheat (cv Monopole, cv Absolvent, cv Fredrick, and cv Norstar) genotypes, winter rye (Secale cereale L. cv Musketeer), oat (Avena sativa L. cv Laurent), barley (Hordeum vulgare L. cv Winchester), rice (Oryza sativa), and corn (Zea mays).
Plants were germinated in moist sterilized vermiculite for 5 d in the dark and 2 d under artificial light. The temperature was maintained at 25°C/20°C with a 15-h photoperiod under a relative humidity of 70% ± 5%. At the end of this period, plants were maintained under the same conditions of light and temperature or exposed to CA and other stresses as described previously (Danyluk et al., 1998). All plants were harvested 8 h after the beginning of the light period and immediately frozen in liquid N2 and stored at −80°C until used for molecular and biochemical analyses.
Cloning and Molecular Analyses
One hundred clones were randomly isolated from a Lambda Zap II library (Stratagene, La Jolla, CA) constructed from poly(A+) RNA that was isolated from 1-d cold-acclimated winter wheat cv Norstar (Houde et al., 1992). Terminal sequencing using the T7 sequencing kit (Amersham Biosciences, Uppsala) of both strands was first used to identify the nature of the clones. Transcript accumulation of selected clones was then analyzed on northern blots as described previously (Houde et al., 1992). Full-length cDNAs of stress-regulated clones were then isolated from the same cDNA library and their complete DNA sequences were determined from both strands. A putative MT clone containing the consensus domains for Ado-Met binding and up-regulated by both LT and NaCl was selected for detailed characterization. All other molecular biology techniques were performed using standard procedures (Sambrook et al., 1989).
Genetic Analyses
To map wpeamt to a specific chromosome, we used the DT series of wheat cv Chinese Spring (provided by the U.S. Department of Agriculture, E.R. Sears collection, Aberdeen, ID). In this series, all chromosomes are present, except that in each line one chromosome pair is represented by only the telocentric chromosome of one arm. L or S indicates the presence of long or short arm of the chromosome, respectively. Isolation of genomic DNA and Southern gel-blot analysis was as described previously (Limin et al., 1997).
Protein Expression, Purification, and Antibody Production
The Wpeamt cDNA was excised with BamHI and XhoI, subcloned in frame into the pTrc-His B vector (Invitrogen, Carlsbad, CA), and expressed as a His-tagged fusion product in Escherichia coli. Cell pellets generated from 1 L of culture (≅4 g fresh weight cells) were resuspended in 10 mL of lysis buffer (50 mm Tris-HCl, pH 7.5; 2 mm β-mercaptoethanol; 10% [v/v] glycerol; 0.1% [v/v] NP-40; and 1 mm phenylmethylsulfonyl fluoride). The resuspended cells were disrupted by sonification. Protein concentration of the supernatant was estimated using the assay kit (Bio-Rad Laboratories, Hercules, CA) and bovine serum albumin as standard. For polyclonal antibody production, the WPEAMT protein was partially purified by affinity chromatography on a His-bind resin (Novagen, Madison, WI) and then resuspended in Laemmli sample buffer (Laemmli, 1970), boiled for 10 min, and separated on 10% (w/v) SDS-PAGE. The protein band corresponding to WPEAMT was excised and electro-eluted for 3 h. The purified protein was used to raise an antibody in rabbit as previously described (Danyluk et al., 1998). Purified anti-WPEAMT antibodies did not cross react with bacterial proteins and were used for western-blot analyses as described (Danyluk et al., 1998). The anti-WPEAMT serum was diluted 1:500 (v/v). The same procedure was used with India His tag detection system except that the second antibody was omitted (Pierce Chemical, Rockford, IL).
Construction of Deleted and Base-Substituted MT
Mutations and deletions are schematically represented in Figure 1B. QuickChange site-directed mutagenesis kit (Stratagene) was used to substitute Gly by Glu in M1 and M2. Bold letters in Figure 1A represents the two mutations. The primers used for generating the mutant M1 were 5′-GCTGGAGCTCGAAGCCGGCATCG-3′ and 5′-CGATGCCGGCTTCGAGCTCCAGC-3′, and for M2 the primers used were 5′-GTGCTCGATGTAGAGTGTGGTATCGGA-3′ and 5′-TCCGATACCACACT CTACATCGAGCAC-3′. The amplified products were used to transform E. coli. The C-terminal deletion H1 and the N-terminal deletion H2 were prepared by a standard PCR approach to introduce at one end a BamHI site and on the other end an XhoI site and a stop codon. Primers used to generate H1 and H2 are single and double underlined, respectively, in Figure 1A. Primers used for H1 were 5′-GAGGATCCGATGGACACCATCACC-3′ and 5′-TAACTCGAGATTCAAGTGGATTTGTACTGCAC-3′, whereas primers for H2 were 5′-GTGGATCCCAAGTGCATTGGAG-3′ and 5′-GCCTCGAGATCACTTGGTCGC-3′. The amplified fragments were digested with BamHI and XhoI, subcloned in-frame into the pTrc-His B vector, and expressed in E. coli using standard methods. Nucleotide sequences of the constructs were confirmed by sequencing.
Enzyme Assays
Recombinant WPEAMT activities were measured under conditions in which product formation was linear to enzyme concentration and time. The standard assay mixture (100 μL) for the recombinant WPEAMT protein contained 50 mm Tris-HCl (pH 7.5), 200 μm P-EA (Sigma, St. Louis) or P-MMEA or P-DMEA (Summers and Weretilnyk, 1993), 200 μm cold Ado-Met (Sigma), 3 nm [3H] Ado-Met (0.025 μCi; 85 Ci mmol−1; American Radiolabeled Chemicals, St. Louis) and 10 to 500 μg of protein. Incubations were performed at 30°C for 30 min. Reactions were stopped by addition of 1 mL of ice-cold MilliQ water (Millipore, Bedford, MA). Each reaction was then applied to a 2-mL AG 50 (H+) cation exchanger column (Bio-Rad) and rinsed three times with 1 mL of water. Reaction products were eluted with 5 mL of 0.1 n HCl, and 1 mL was mixed with 3 mL of scintillation liquid (Eco Lite, ICN Biomedicals, Costa Mesa, CA) and counted for radioactivity. For the blank assay, P-EA was omitted. Steady-state kinetic parameters and Km and Vmax values were estimated from the Lineweaver-Burk plots by plotting reaction rates versus increasing concentrations of substrate. For the P-EA Km value, the Ado-Met concentration was held at 200 μm (saturating concentration), whereas the P-EA concentration varied. The Km value for Ado-Met was determined at saturated concentration of P-EA (200 μm). Reaction products were visualized using the method described by Smith et al. (2000) with slight modifications. Reaction products were collected from reaction with P-EA and evaporated. The dry product was solubilized in 10 μL of 0.1 n HCl and applied to a TLC plate (10 × 10 cm; 0.25-mm silica gel G, Machery-Nagel, Duren, Germany) that had been pre-equilibrated for 24 h in n-butanol:methanol:concentrated HCl:water (5:5:1:1 [v/v]). The TLC plate was developed with n-butanol:methanol:concentrated HCl:water (10:10:1:1 [v/v]) and the spots were revealed by phosphor-imaging (CS screen; Bio-Rad).
The standard SMT assay for the recombinant protein contained, in 100-μL total volume, 50 to 400 μg of protein in 50 mm Tris-HCl buffer, 100 μm lanosterol, 4 nm [14C] Ado-Met (0.025 μCi; 50 μCi mmol−1) and 0.1% (v/v) Tween 80. All samples were maintained in Tris buffer by desalting on PD-10 columns (Amersham-Pharmacia) before assay for SMT activity. Incubation was at 30°C for 30 min. The reaction was terminated with 10 μL of 20% (w/v) aqueous KOH and 80% (v/v) methanol. The methylated product was extracted with 500 μL of ether. One-half of the resulting organic layer (250 μL) was then transferred to a vial to which 3 mL of scintillation liquid was added for radioactivity measurement.
Enzyme Activity in Plant Tissues
N-MT activity in the plant extract was measured according to the method described by Summers and Weretilnyk (1993), with slight modifications. All operations were carried out at 4°C. Frozen leaf tissue (2.5 g) was coarsely chopped and ground thoroughly with sand using a mortar and pestle and extraction buffer (1 mL g−1 fresh weight) containing 0.1 m Tris-HCl (pH 7.8, 4°C), 5 mm dithiothreitol, 2 mm Na2EDTA, and 10% (v/v) glycerol. The homogenate was squeezed through four layers of Miracloth (Calbiochem, San Diego) and centrifuged at 10,000g for 10 min. The supernatant was recovered and desalted on a PD-10 column (Amersham-Pharmacia) equilibrated with 50 mm HEPES, pH 7.8; 5 mm dithiothreitol; and 1 mm Na2EDTA. The PD-10 desalted leaf extract was used for enzyme activity measurements. The assay mixture (final volume of 100 μL) contained 50 μm HEPES, pH 7.8; 1 mm Na2EDTA; 200 μm P-EA; 200 μm cold Ado-Met; and 4 nm [14C] Ado-Met (0.025 μCi; 50 μCi mmol−1; American Radiolabeled Chemicals) and 10 to 250 μg of protein. Determination of protein concentration, reaction conditions, product isolation, and counting were as described above.
ACKNOWLEDGMENTS
The authors thank Drs. Elizabeth Weretilnyk and Peter Summers (University of McMaster, Hamilton, ON, Canada) for their generous gift of P-MMEA and P-DMEA substrates.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada and Fonds pour la Formation de Chercheurs et l'Aide à la Recherche (research grants to F.S. and R.K.I.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001776.
LITERATURE CITED
- Allard F, Houde M, Kröl M, Ivanov A, Huner NPA, Sarhan F. Betaine improves freezing tolerance in wheat. Plant Cell Physiol. 1998;39:1194–1202. [Google Scholar]
- Bohnert H, Jensen RG. Strategies for engineering water stress tolerance in plants. Trends Biotechnol. 1996;14:89–97. [Google Scholar]
- Bolognese CP, McGraw P. The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine: phosphoethanolamine N-methyltransferase. Plant Physiol. 2000;124:1800–1813. doi: 10.1104/pp.124.4.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochov A, Walker MA, Kendall EJ, Pauls KP, McKersie BD. Effect of a freeze-thaw cycle on properties of microsomal membranes from wheat. Plant Physiol. 1987;84:131–134. doi: 10.1104/pp.84.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J. Substrate-dependent mutant complementation to select fatty acid desaturase variants for metabolic engineering of plant seed oils. Proc Natl Acad Sci USA. 2000;97:12350–12355. doi: 10.1073/pnas.210276297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers A, Melchior DL. How bilayer lipids affect membrane protein activity. Trends Biotechnol. 1986;11:331–335. [Google Scholar]
- Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Limin AE, Ritchie JT. Low-temperature tolerance in cereals: models and genetic interpretation. Crop Sci. 1999;39:626–633. [Google Scholar]
- Harwood JL. Engineering frost resistance in plants by genetic manipulation. In: Harwood JL, editor. Plant Lipid Biosysthesis: Fundamentals and Agricultural Application. Cambridge, UK: Cambridge University Press; 1998. pp. 155–183. [Google Scholar]
- Hitz WD, Hanson AD. Determination of glycine betaine by pyrolysis-gas chromatography in cereals and grasses. Phytochemistry. 1980;19:2371–2374. [Google Scholar]
- Horváth I, Van Hasselt PR. Inhibition of chilling-induced photooxidative damage to leaves of Cucumis sativus L. by treatement with amino alcohols. Planta. 1985;164:83–88. doi: 10.1007/BF00391029. [DOI] [PubMed] [Google Scholar]
- Horváth I, Vigh L. Self-adaptive modification of membrane lipids in cell culture of wheat (Triticum monococcum L.) In: Siegenthaler PA, Eichenberger W, editors. Structure, Function and Metabolism of Plant Lipids. New York: Elsevier Science Publishers B.V.; 1984. pp. 535–538. [Google Scholar]
- Horváth I, Vigh L, Belea VA, Farkas T. Conversion of phosphatidyl choline to phosphatidic acid in freeze injured rye and wheat cultivars. Physiol Plant. 1979;45:57–62. [Google Scholar]
- Horváth I, Vigh L, Belea VA, Farkas T. Hardiness dependent accumulation of phospholipids in leaves of wheat cultivars. Physiol Plant. 1980;49:117–120. [Google Scholar]
- Horváth I, Vigh L, Farkas T. The manipulation of polar head group composition of phospholipids in the wheat Miranovskaja 808 affect frost tolerance. Planta. 1981;151:103–108. doi: 10.1007/BF00387811. [DOI] [PubMed] [Google Scholar]
- Houde M, Dhindsa RS, Sarhan F. A molecular marker to select for freezing tolerance in Gramineae. Mol Gen Genet. 1992;234:43–48. doi: 10.1007/BF00272343. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Arakawa K, Mizuno K, Kishitani S, Takabe T. Betaine aldehyde dehydrogenase in the Gramineae: levels in leaves of both betaine-accumulating and nonaccumulating cereal plants. Plant Cell Physiol. 1993;34:493–495. [Google Scholar]
- Ibrahim RK, Muzac I. The methyltransferase gene superfamily: a tree with multiple branches. In: Romeo JT, Ibrahim R, Varin L, DeLuca V, editors. Evolution of Metabolic Pathways. London: Elsevier Science Ltd; 2000. pp. 349–383. [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kinney AJ, Clarkson DT, Loughman BC. Phospholipid metabolism in rye roots in warm and cool conditions. Biochem Soc Trans. 1983;11:390–391. [Google Scholar]
- Kishitani S, Watanabe K, Yasuda S, Arakawa K, Takabe T. Accumulation of glycinebetaine during cold acclimation and freezing tolerance in leaves of winter and spring barley plants. Plant Cell Environ. 1994;17:89–95. [Google Scholar]
- Kumar A, Minocha SC. Transgenic manipulation of polyamine metabolism. In: Lindsey K, editor. Transgenic Plant Research. Amsterdam: Harwood; 1998. pp. 187–199. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Limin AE, Danyluk J, Chauvin LP, Fowler DB, Sarhan F. Chromosome mapping of low-temperature induced Wcs120family genes and regulation of cold-tolerance expression in wheat. Mol Gen Genet. 1997;253:720–727. doi: 10.1007/s004380050376. [DOI] [PubMed] [Google Scholar]
- Lynch DV, Steponkus PL. Plasma membrane lipid alterations associated with cold acclimation of winter rye seedlings (Secale cerealeL. cv Puma) Plant Physiol. 1987;83:761–767. doi: 10.1104/pp.83.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Hanson AD. Betaines and related osmoprotectants: targets for metabolic engineering of stress resistance. Plant Physiol. 1999;120:945–949. doi: 10.1104/pp.120.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TS., Jr Phospholipids biosynthesis. Annu Rev Plant Physiol. 1982;33:235–259. [Google Scholar]
- Naidu BP, Paleg LG, Aspinall D, Jennings AC, Jones GP. Amino acid and glycine betaine accumulation in cold-stressed wheat seedlings. Phytochemistry. 1991;30:407–409. [Google Scholar]
- Nes WD, McCourt BS, Zhou WX, Ma J, Marshall JA, Peek LA, Brennan M. Overexpression, purification, and stereochemical studies of the recombinant S-adenosyl-l-methionine: delta 24(25)- to delta 24(28)-sterol methyltransferase enzyme from Saccharomyces cerevisiae. Arch Biochem Biophys. 1998;353:297–311. doi: 10.1006/abbi.1998.0665. [DOI] [PubMed] [Google Scholar]
- Nomura M, Muramoto Y, Yasuda S, Takabe T, Kishitani S. The accumulation of glycinebetaine during cold acclimation in early and late cultivars of barley. Euphytica. 1995;83:247–250. [Google Scholar]
- Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombeand characterization of the recombinant enzyme. J Biol Chem. 2000;275:14095–14101. doi: 10.1074/jbc.275.19.14095. [DOI] [PubMed] [Google Scholar]
- Rivoal J, Hanson AD. Choline-O-sulfate biosynthesis in plants. Plant Physiol. 1994;106:1187–1193. doi: 10.1104/pp.106.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Murata N. The use of bacterial choline oxidase, a glycine betaine-synthesizing enzyme, to create stress-resistant transgenic plants. Plant Physiol. 2001;125:180–188. doi: 10.1104/pp.125.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Vaverde R, Alia, Chen TH, Murata N. Transformation of Arabidopsis with cod Agene choline oxidase enhances freezing tolerance of plants. Plant J. 2000;22:449–453. doi: 10.1046/j.1365-313x.2000.00749.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. NY: Cold Spring Harbor Laboratory Press Cold Spring Harbor; 1989. [Google Scholar]
- Sikorska E, Kacperska-Palacz A. Frost-induced phospholipid changes in cold-acclimated and non-acclimated rape leaves. Physiol Plant. 1980;48:201–206. [Google Scholar]
- Smith DD, Summers PS, Weretilnyk EA. Phosphocholine synthesis in spinach: characterization of phosphoethanolamine N-methyltransferase. Physiol Plant. 2000;108:286–294. [Google Scholar]
- Steponkus PL, Uemura M, Balsamo RA, Arvinte T, Lynch DV. Transformation of the cryobehavior of rye protoplasts by modification of the plasma membrane lipid composition. Proc Natl Acad Sci USA. 1988;85:9026–9030. doi: 10.1073/pnas.85.23.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers PS, Weretilnyk EA. Choline synthesis in spinach in relation to salt stress. Plant Physiol. 1993;103:1269–1276. doi: 10.1104/pp.103.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Izuwa S, Tanaka K, Yamamoto D, Takimoto T, Matsuura F, Satouchi K. Biosynthesis of 1,2-dieicosapentaenosyl-sn-glycero-3-phosphocholine in Caenorhabditis elegans. Eur J Biochem. 1999;263:189–194. doi: 10.1046/j.1432-1327.1999.00480.x. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tolbert NE, Weibe H. Phosphorus and sulfur compounds in plant xylem sap. Plant Physiol. 1955;30:499–504. doi: 10.1104/pp.30.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyn A, Kendrick RE. Acetylcholine in plants: presence, metabolism and mechanism of action. Bot Rev. 1991;57:33–73. [Google Scholar]
- Wilke K, Rauhut E, Noyer-Weidner M, Lauster R, Pawlek B, Behrens B, Trautner TA. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988;7:2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP, Horváth I, Quinn PJ, Thomas PG, Vigh L. Freezing resistance and lipid changes in choline-treated wheat seedlings. In: Stumpf PK, Mudd JB, Nes WD, editors. The Metabolism Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 201–203. [Google Scholar]
- Wyn Jones RG, Storey R. Betaines. In: Paleg LG, Aspinall D, editors. The Physiology and Biochemistry of Drought Resistance in Plants. Sydney: Academic Press; 1981. pp. 171–203. [Google Scholar]
- Yancey P. Compatible and counteracting solutes. In: Strange K, editor. Cellular and Molecular Physiology of Cell Volume Regulation. Boca Raton, FL: CRC Press; 1994. pp. 81–109. [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sakai A. Phospholipid degradation in frozen plant cells associated with freezing injury. Plant Physiol. 1974;53:509–511. doi: 10.1104/pp.53.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]