Abstract
Switch from non-syncytium-inducing (NSI) to syncytium-inducing (SI) HIV type 1 (HIV-1) is associated with accelerated CD4+ T cell depletion, which might partially be explained by higher virulence of SI variants compared with NSI variants. Because NSI and SI variants use different coreceptors for entry of target cells, altered tropism might offer an explanation for increased pathogenesis associated with SI HIV-1 infection. To investigate whether SI and NSI HIV-1 variants infect different CD4+ T cell subsets in vivo, the distribution of SI and NSI variants over CD4+ memory (CD45RA−RO+) and naive (CD45RA+RO−) cells was studied by using limiting dilution cultures. In contrast to NSI variants that were mainly present in CD45RO+ cells, SI variants were equally distributed over CD45RO+ and CD45RA+ cells. Infection of memory cells by both NSI and SI HIV-1 and infection of naive cells primarily by SI HIV-1 corresponded closely with the differential cell surface expression of CXCR4 and CCR5. The frequency of SI-infected CD45RA+ CD4+ T cells, but not the frequency of NSI- or SI-infected CD45RO+ CD4+ T cells, correlated with the rate of CD4+ T cell depletion. Infection of naive cells by SI HIV-1 may interfere with CD4+ T cell production and thus account for rapid CD4+ T cell depletion.
In 50% of HIV-1-infected individuals, a switch occurs from the non-syncytium-inducing (NSI) phenotype to the syncytium-inducing (SI) phenotype during the course of disease (1–3). The emergence of SI variants is associated with an accelerated CD4+ T cell loss and more rapid progression to AIDS (1, 2, 4). The enhanced pathogenesis of SI HIV-1 infection may be explained by the more virulent characteristics of SI variants observed in vitro, such as enhanced cytopathicity (5) and higher replication kinetics (1, 4, 6). Because SI and NSI variants use different coreceptors for entry of target cells, altered tropism could offer an alternative (or additional) explanation for the increased pathogenicity associated with the presence of SI variants (7). The vast majority of NSI variants are restricted to the use of CCR5, while SI variants use CXCR4 and may use CCR5 and/or other chemokine receptors in addition (8–13). While CCR5 is mainly expressed on activated/memory T cells, CXCR4 is mainly expressed on cells with a resting/naive phenotype, and CCR5 is mainly expressed on activated/memory T cells (14, 15). Moreover, we recently found that within both the CD45RO− and CD45RO+ CD4+ T cell subsets of HIV-1-infected individuals, expression of CXCR4 and CCR5 is largely mutually exclusive (R. P. van Rij, H.B., J. A. Visser, M.B., R. Rientsma, S. Broersen, A. M. de Roda Husman, and H.S., unpublished results). Given the existence of two major and distinct HIV-1 target cell populations within the CD4+ T lymphocyte compartment, one expressing the NSI coreceptor and the other expressing the main SI coreceptor, the total number of potential HIV-1 target cells expands considerably after the emergence of SI variants. This could explain the higher viral load observed in individuals with SI variants compared with individuals with only NSI variants at comparable CD4+ T cell counts (16), and hence the accelerated disease progression. Next to the probable impact of an increase in the number of potential target cells, it can be envisaged that the extent to which HIV-1 infection affects the rate of CD4+ T cell decline depends on the type of cells that are infected. Recently, it was shown that both naive and memory CD4+ T cells are productively infected with HIV-1 in vivo (17). Because naive cells contribute exponentially to the memory CD4+ T cell pool, HIV-1-induced loss of naive cells is likely to have a large impact on CD4+ T cell replenishment and would consequently contribute significantly to CD4+ T cell depletion. Here, we tested the hypothesis that specifically the conversion to the SI phenotype enables HIV-1 to efficiently infect naive CD4+ T cells, thereby contributing to the increased pathogenesis associated with the presence of SI HIV-1.
Subjects and Methods
Subjects.
The study group consisted of 18 participants of the Amsterdam Cohort Studies (ACS) on AIDS in homosexual men. Patient characteristics at the moment of analysis are shown in Table 1. For 6 of the individuals, a time point was analyzed at which only NSI variants were present (N1–N6), and, for 10 individuals, a time point was analyzed at which both SI and NSI variants were present (S1–S10). For two individuals, peripheral blood mononuclear cell (PBMC) samples obtained before and after HIV-1 phenotype switch were analyzed. For the samples obtained before SI conversion, these two individuals were included in the NSI group (N7 and N8), and, for the time point after phenotype switch, these individuals were included in the SI/NSI group (N7 = S11, N8 = S12). In the SI/NSI group, the moment of analysis was at least 5 mo after SI conversion (median, 22 mo; range, 5.2–57.1 mo).
Table 1.
CD4+ T cell numbers, subsets, and virus load at moment of analysis
| Patient* | Moment of analysis, mo† | Serostatus‡ | CD4+ T cells/μl blood | CD45RO+ CD4+ T cells, % | CD45RA+ CD4+ T cells, % | Frequencies of infected cells (TCID/104 cells)
|
||
|---|---|---|---|---|---|---|---|---|
| CD4+ T cells§ | CD45RO+ CD4+ T cells | CD45RA+ CD4+ T cells¶ | ||||||
| N1 | 151 | 1 | 400 | 58 | 28 | 5 | 22 | <2 (dl) |
| N2 | 103 | 1 | 240 | 71 | 16 | 269 | 709 | 11 |
| N3 | 148 | 2 | 440 | 33 | 58 | 191 | 434 | 9 |
| N4 | 22 | 2 | 380 | 30 | 61 | 34 | 115 | 4 |
| N5 | 60 | 2 | 750 | 39 | 54 | 111 | 431 | 6 |
| N6 | 103 | 2 | 530 | 45 | 48 | 20 | 38 | 1 |
| N7 | 64 | 1 | 1020 | 47 | 45 | 53 | 178 | 4 |
| N8 | 7 | 1 | 1060 | 37 | 42 | 2 | 2 | <2 (dl) |
| S1 | 100 | 2 | 330 | 63 | 17 | 340 | 431 | 53 |
| S2 | 74 | 2 | 320 | 69 | 15 | 81 | 77 | 25 |
| S3 | 117 | 2 | 430 | 51 | 14 | 361 | 420 | 212 |
| S4 | 104 | 1 | 280 | 63 | 20 | 142 | 380 | 41 |
| S5 | 148 | 2 | 260 | 44 | 40 | NT | 68 | 190 |
| S6 | 100 | 1 | 310 | 52 | 35 | 160 | 220 | 164 |
| S7 | 148 | 2 | 70 | 85 | 9 | 42 | 109 | <19 (dl) |
| S8 | 37 | 2 | 380 | 52 | 35 | 297 | 701 | 874 |
| S9 | 120 | 1 | 540 | 28 | 67 | 5 | 3 | 4 |
| S10 | 46 | 2 | 460 | 42 | 48 | 13 | 1 | 18 |
| S11 | 94 | 1 | 290 | 39 | 37 | 103 | 282 | 39 |
| S12 | 35 | 1 | 820 | 35 | 43 | 25 | 74 | 9 |
N, patient with only NSI variants; S, patient with both SI and NSI variants; for two patients a time point before and a time point after SI switch were analyzed; N7 − S11 and N8 − S12.
† Months after seroconversion or seropositive entry in the Amsterdam Cohort Studies (ACS).
‡ HIV-1 serostatus at the moment of entry in the ACS: 1, participants who were seronegative at entry in the ACS, and seroconversion date was estimated to be the midpoint between the last HIV-1− and the first HIV-1+ visit; 2, participants who were already seropositive at entry in the ACS.
§ NT, not tested.
¶ dl, detection limit of the assay.
Isolation of CD45RA+ and CD45RO+ CD4 T Cells from PBMC.
PBMC were isolated from fresh blood and used immediately (N1, N3, N4, N6, S5, S7, S10) or after cryopreservation. PBMC (20–30 × 106) were stained with mAbs directed against CD4 [conjugated with tri-color (TC)], CD45RA [conjugated with phycoerythrin (PE)], and CD45RO (conjugated with FITC) (all mAbs from Caltag, South San Francisco, CA). CD4+CD45RA+RO− cells and CD4+CD45RA−RO+ cells were separated by sorting with a FACStar (Becton Dickinson). After cell sorting, contamination with the reciprocal CD4+ T cell subsets was on average 0.3% (CD4+CD45RO+ in CD4+CD45RA+ cell fraction) and 0.2% (CD4+CD45RA+ in CD4+CD45RO+ cell fraction).
CCR5 and CXCR4 Expression on CD45RA+ CD4 and CD45RO+ CD4 Cells.
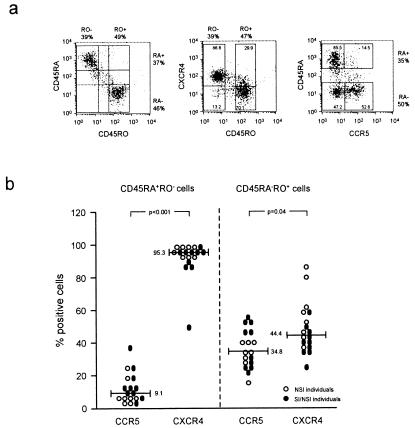
CCR5 and CXCR4 expression on the memory and naive CD4+ T cell subsets was analyzed by three-color flow cytometry. PBMC samples were obtained from time points maximally 3 mo before or after the moment at which the PBMC samples used for virus isolation were obtained. PBMC (2.5 × 105) were stained with a combination of mAbs directed against (i) CD4 (TC; Caltag), CD45RA (2H4-RD1-PE; Coulter), and CCR5 (5C7- FITC; PharMingen); (ii) CD4 (TC), CD45RO (UCHL-1-FITC; Dako), and CXCR4 (12G5-PE; PharMingen); or (iii) CD4 (TC), CD45RA (2H4-RD1-PE), and CD45RO (UCHL-1-FITC). The latter staining was performed to identify cell populations single positive for only one of the two CD45 isoforms, which was extrapolated to the CCR5/CD45RA staining and the CXCR4/CD45RO staining. Thus, CCR5 expression was analyzed on the CD45RA− (= CD45ROhigh) and CD45RAhigh (= CD45RO−) cells, and CXCR4 expression was analyzed on the CD45RO− (= CD45RAhigh) and CD45ROhigh (= CD45RA−) cells (see Fig. 3a).
Figure 3.
CCR5 and CXCR4 expression on CD45RA+ and CD45RO+ CD4 T cells. Using three-color flow cytometry, the percentages CCR5- and CXCR4-positive naive and memory cells were determined. FACScan images of one representative patient (S11) are shown (a). CD4+ lymphocytes were gated according to their forward and side scatter and CD4 staining. (Left) CD45RA vs. CD45RO staining. (Middle) CD45RO vs. CXCR4 staining. (Right) CD45RA vs. CCR5 staining. The proportion of CD45RAhigh cells corresponded with the proportion of CD45RO− cells, and the proportion of CD45RA− cells corresponded with the proportion of CD45ROhigh cells; the percentages are depicted on the outside of the graphs. The percentages of CXCR4-expressing CD45RO+ (RA−) and CD45RO− (RA+) cells and percentages of CCR5-expressing CD45RA+ (RO−) and CD45RA− (RO+) cells are depicted in the graphs. For all individuals, with NSI variants only (○) or with SI variants (●), these percentages are shown in b. The numbers and bars in the graph indicate median values.
Biological Cloning of HIV-1 and Determination of Viral Load.
CD45RA+, CD45RO+ CD4+ T cells and total PBMC were independently cocultivated with freshly phytohemagglutinin (PHA)-stimulated healthy donor peripheral blood lymphocytes (PBL) under limiting diluting conditions, as described previously (3, 16). Cultures were maintained for 4 wk, and, every seventh day, culture supernatant was analyzed for virus production in an in-house p24 capture ELISA. At 2–4 wk, the ability of biological virus clones to induce syncytia was evaluated by cocultivation with MT2 cells (18).
The frequency of productively infected cells was calculated with the formula for Poisson distribution: F = −ln(F0), in which F0 is the fraction of negative cultures, and expressed as tissue culture infectious dose (TCID) per 106 CD4+, CD45RA+CD4+, and CD45RO+CD4+ T cells. The detection limit of the assay depends on the number of patient cells available for coculture and was calculated in cases where no virus clones were obtained. The proportions of biological virus clones with NSI and SI phenotype were used to calculate the NSI virus load and the SI virus load.
CD4+ T Cell Decline.
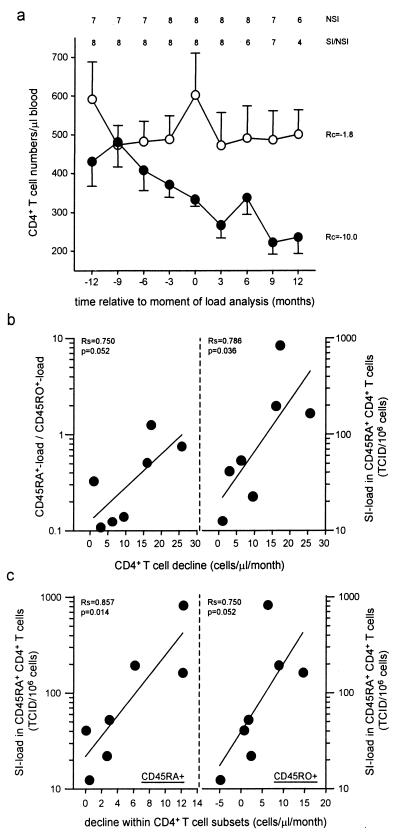
Lymphocyte immunophenotyping for CD4+ T cells was carried out by using flow cytometry at 3-mo intervals. The patients were matched for the moment of sampling at which virus load was determined (t = 0), and the group mean CD4+ T counts were calculated at each 3-mo time point a year before and after t = 0 (from t = −12 to t = 12). These mean CD4+ T cell values were used for linear regression analysis. Participants receiving anti-retroviral combination therapy at t = 0 (S5, S7, S9, and S10) were excluded from analysis. From the remaining eight NSI individuals and eight SI/NSI individuals, CD4+ T cell data obtained prior to seroconversion (time points −12 to −6; see Fig. 4a, Top) and CD4+ T cell data obtained after initiation of anti-retroviral combination therapy (time points 6 to 12; see Fig. 4a, Top) were excluded from analysis.
Figure 4.
CD4+ T cell decline and virus load for the 2-yr period spanning the moment of virus isolation and virus load determination (t = 0). The average CD4+ T cell counts per 3-mo interval were calculated for individuals with NSI variants only (○) and individuals with SI variants (●) (a). At the top of a, the number of included patients is shown. The linear regression coefficients (Rc) are depicted. Bars indicate the SEM. For the SI-carrying individuals, the correlation between CD4+ T cell decline and the relative virus load (Left) as well as the absolute SI load in the CD45RA+ cells (Right) was analyzed (b), and the correlations between the CD45RA+ SI load and the decline in CD45RA+ (Left) and CD45RO+ CD4+ T cells (Right) were analyzed (c). The Spearman correlation coefficients (Rs) and the P values are shown.
Individual decline of total CD4+ T cells and CD4+ T cell subsets during this period was determined by linear regression analysis, and the correlation with virus load was analyzed. The decline in the number of CD45RA+ and CD45RO+ CD4+ T cells was based on PBMC samples from at least five time points within the 2-yr time span.
Statistical Analyses.
The Mann–Whitney U test was used to analyze differences between individuals with only NSI variants and individuals with SI and NSI variants with respect to the CD45RA+-load/CD45RO+-load ratio. Differences between CD45RA+ cells and CD45RO+ cells within the group of individuals with SI variants were analyzed with the Wilcoxon signed rank test (i.e., proportion of biological clones with SI phenotype, and SI-load in CD45RA+ and CD45RO+ cells). CCR5 and CXCR4 expression on CD45RA+RO− and CD45RA−RO+ subsets was analyzed with the paired Student t test. Correlations of CD4+ T cell decline with virus load were determined by using the Spearman correlation coefficient.
Results
Individuals with SI Variants Have a Relatively High Frequency of Infected CD45RA+CD4+ T Cells.
The frequency of productively infected CD4+ T cells, CD45RA+CD4+ T cells, and CD45RO+CD4+ T cells (expressed per 106 cells of the respective T cell subsets) was determined for 8 individuals at a time point at which only NSI variants were present (N1–N8) and for 12 individuals at a time point at which both SI and NSI variants were present (S1–S12) (Table 1). The total virus load had a broad range and did not differ significantly between both groups (2–269 and 5–361 TCID/106 CD4+ T cells, respectively; P = 0.3, Mann–Whitney U test).
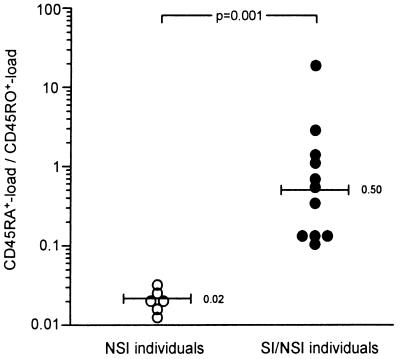
In individuals with only NSI variants, the frequency of infected CD45RA+ cells was 30- to 70-fold lower than the frequency of infected CD45RO+ cells (Table 1). This is visualized by the low ratio of CD45RA+-load to CD45RO+-load (median = 0.02; Fig. 1). The high ratio observed for individuals with both NSI and SI variants (median = 0.50) reflects a relatively high frequency of infected CD45RA+ cells (9-fold lower to 18-fold higher) compared with the frequency of infected CD45RO+ cells (Table1; Fig. 1). The CD45RA+-load/CD45RO+-load ratio differed significantly between the individuals with and without SI variants (P = 0.001; Mann–Whitney U test).
Figure 1.
Ratio of virus load in CD45RA+ and CD45RO+ CD4+ T cells. The ratio between the frequency of infected CD45RA+CD4+ T cells and the frequency of infected CD45RO+CD4+ T cells is shown for individuals with only NSI variants and individuals with both SI and NSI variants. Individuals with an undetectable virus load in the CD45RA+ cells were excluded (N1, N8, S7, S12; Table 1). The numbers and bars in the graph indicate median values.
CD45RA+ CD4+ T Cells Are Preferentially Infected with SI Variants.
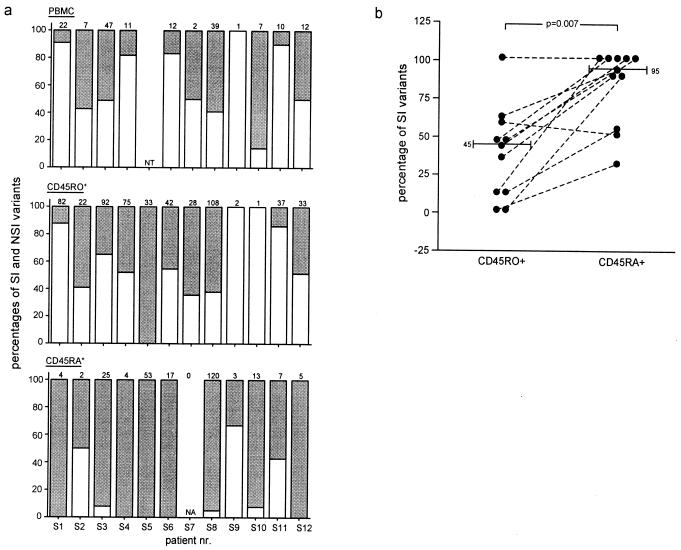
The relatively high frequencies of infected CD45RA+ cells in individuals with both NSI and SI variants compared with individuals with only NSI HIV-1 (Table 1; Fig. 1) suggested a preferential infection of CD45RA+ cells by SI variants. To verify this, we analyzed the distribution of coexisting NSI and SI HIV-1 in CD45RA+ and CD45RO+ cells from the individuals carrying both variants. Within each of the three populations of biological virus clones, obtained by cultivation of total PBMC, CD45RO+, and CD45RA+ cells, the proportion of NSI and SI viruses was calculated (Fig. 2). The distribution patterns of NSI and SI variants in total PBMC and CD45RO+CD4+ T cells were similar, yet differed in the CD45RA+CD4+ T cells (Fig. 2a). Paired analysis of CD45RA+CD4+ T cells and CD45RO+CD4+ T cells revealed a significantly higher proportion of SI variants in CD45RA+ cells (median 95% vs. 45%; P = 0.007, Wilcoxon signed rank test; Fig. 2b). Thus, while the proportions of SI and NSI variants were similar in CD45RO+ cells, CD45RA+ cells were almost exclusively infected with SI variants, explaining the higher CD45RA+ load in carriers of SI variants compared with individuals with only NSI HIV-1.
Figure 2.
Distribution of SI and NSI variants in total CD4+ T cells, CD45RA+, and CD45RO+ CD4 T cells. The total number of biological virus clones that were obtained by cocultivation with healthy donor PBL of total PBMC (a, Top), CD45RO+CD4+ T cells (a, Middle), and CD45RA+CD4+ T cells (a, Bottom) of SI carrying individuals are depicted at the top of each panel in a. The open bars represent the proportion of NSI clones, and filled bars represent the proportion of SI clones. Total PBMC from patient S5 was not analyzed (a, Top). No biological clones were obtained from CD45RA+CD4+ T cells from patient S7 (a, Bottom). The difference in the proportion of SI variants in CD45RO+ and CD45RA+ cells was analyzed for the individuals with detectable CD45RA+ load (b). The numbers and bars in the graph indicate median values. NT, Not tested. NA, not applicable.
Since both CD4+ T cell subsets were infected with SI variants, we calculated the frequencies of CD45RA+ and CD45RO+ cells infected with SI HIV-1. SI variants were equally distributed over CD45RO+ cells and CD45RA+ cells (median SI load, 53 TCID/106 cells and 41 TCID/106 cells, respectively; P = 0.48, Wilcoxon signed rank test; data not shown). In both individuals who were analyzed at a time point before and a time point after the emergence of SI variants (N7/S11 and N8/S12), an increase in the frequency of infected CD45RA+ cells and the ratio of CD45RA+-load/CD45RO+-load was observed (Table 1). In both cases, this increase could be explained by preferential infection of CD45RA+ cells by SI variants (S11, 57%; S12, 100% SI variants).
CD45RA+ CD4+ T Cells Mainly Express CXCR4, and CD45RO+CD4+ T Cells Express CXCR4 and CCR5.
To determine whether the differential distribution of HIV-1 variants correlated with the cell surface expression of CCR5 and CXCR4 in the individuals under study, we analyzed CCR5 and CXCR4 expression on CD45RA+ and CD45RO+CD4+ T cells (Fig. 3).
In agreement with Bleul et al. (14), CXCR4 was mainly present on CD45RA+RO− cells (median CXCR4 positive, 95.3%), whereas CCR5 was mainly present on CD45RA−RO+ cells (median CCR5 positive, 34.8%) (Fig. 3b). More importantly, the proportion of CXCR4-expressing cells was higher than the proportion of CCR5-expressing cells both in the naive (95.3% vs. 9.1%, respectively; P < 0.001; paired Student t test) and in the memory subset (44.4% vs. 34.8%, respectively; P = 0.04, paired Student t test). These expression patterns of CXCR4 and CCR5 on CD45RA+ and CD45RO+ cells were indeed compatible with the distribution patterns of NSI and SI variants in these T cell subsets.
Rate of CD4+ T Cell Decline Correlates with the SI Load in CD45RA+CD4+ T Cells.
To investigate the relation between the virus load in different T cells subsets and CD4+ T cell decline, we first verified whether differences in CD4+ T cell decline between individuals with and without SI variants could be observed in a short time span surrounding the moment of sampling used in the virus load analysis (t = 0). For this purpose, the individuals were matched at t = 0 and at each 3-mo time point from t = −12 to t = 12; the group mean CD4+ T cell counts were calculated and used in linear regression analysis (Fig. 4a). The mean CD4+ T cell decline in this period was about 5-fold faster in the individuals with SI variants as compared with the individuals with NSI variants only (10.0 CD4+ T cells per μl of blood per mo vs. 1.8 CD4+ T cells per μl of blood per mo, respectively).
In patients with SI variants, the rate of CD4+ T cell decline in this 2-yr period correlated with the SI load in CD45RA+CD4+ T cells (Rs = 0.79, P = 0.036) and, although not statistically significant, with the ratio of CD45RA+ load to CD45RO+ load (Rs = 0.75, P = 0.052) analyzed at t = 0 (Fig. 4b). In contrast, no correlation was observed between CD4+ T cell decline and the SI load in CD45RO+ cells (Rs = 0.36, P = 0.4; data not shown) or total CD4+ T cells (Rs = 0.18, P = 0.7; data not shown). The CD4+ T cell decline in patients with only NSI variants did not correlate with the virus load in any of the cell types (data not shown).
Analysis of CD4+ T cell subsets showed that the frequency of infected CD45RA+CD4+ T cells was associated with a decline in the numbers of CD45RA+CD4+ T cells (Rs = 0.86, P = 0.014) and, although statistically not significant, with a decline in the numbers of CD45RO+CD4+ T cells (Rs = 0.75, P = 0.052) (Fig. 4c).
Discussion
Although depletion of CD4+ T cells is one of the main features of HIV-1 infection, the mechanisms underlying this depletion are still not fully understood. Destruction of cells might occur through virus-mediated killing of infected cells or activation-induced apoptosis of uninfected cells (reviewed in ref. 19). Although recent studies have shown that in HIV-1-infected individuals CD4+ T cell turnover is maximally 2- to 3-fold increased (20, 21), limited CD4+ T cell renewal capacity in adult individuals (22, 23) might further contribute to the depletion of CD4+ T cells. In HIV-1-infected individuals, the capacity to produce T cells may be further restricted (24) by virus-induced impairment of the development of CD34+ progenitor cells (25) (D. R. Clark, S. Repping, N. G. Pakker, J. M. Prins, D. W. Notermans, F. W. N. M. Wit, P. Reiss, S. A. Danner, R. A. Coutinho, J. M. A. Lange, and F. Miedema, unpublished results). Additionally, the decrease in CD4+ T cells measured in peripheral blood might not be completely explained by actual depletion, but partly by increased trapping of cells in the lymph nodes during HIV-1 infection (26, 27).
After the emergence of SI HIV-1 variants, an accelerated loss of CD4+ T cells can be observed (1, 2, 4). This implicates that aside from the previously mentioned mechanisms, which may apply for individuals infected with NSI variants only as well as individuals with both NSI and SI variants, additional, SI HIV-1-specific mechanisms likely exist. Until recently, the accelerated loss of CD4+ T cells was attributed to the higher cytopathicity of SI variants compared with NSI variants as demonstrated in vitro (5). However, recently, it was shown that NSI variants can be equally cytopathic as SI variants and that enhanced pathogenicity of SI variants can alternatively be explained by different tropism resulting in an increased target cell population (7).
In the present study, we showed that SI HIV-1-infected individuals carry both NSI and SI variants in their CD45RO+ (memory) CD4+ T cells. However, while the CD4+ memory cells appeared to be the main T lymphocyte target cell population of NSI variants, SI variants were additionally detected in the CD45RA+ (mainly naive) CD4+ T cells. Thus, with the emergence of SI variants, the HIV-1 target population expands from almost exclusively CD45RO+CD4+ T cells to both CD45RO+ and CD45RA+ CD4+ T cells. This was indeed observed for two individuals who were analyzed at time points before and after the evolution of SI variants.
On average, the SI virus load was similar in CD45RA+ and CD45RO+ CD4+ T cells. The partly differential distribution of SI and NSI variants in the two CD4+ T cell subsets agreed with the almost exclusive presence of CXCR4-expressing cells in the CD45RA+ T cell subset and the presence of both CCR5- and CXCR4-expressing cells in the CD45RO+ T cell subset.
The frequency of CD45RA+ cells but not the frequency of CD45RO+ cells or total CD4+ T cells infected with SI HIV-1 correlated with the CD4+ T cell decline measured over a 2-yr period around the moment of viral load analysis. This suggests that not merely the expansion of the total HIV-1 target cell population, but specifically SI infection of the (largely) naive CD45RA+CD4+ T cell subset contributes to the accelerated CD4+ T cell decline in individuals harboring SI variants. Since naive cells contribute exponentially to the memory cell pool and given the intrinsically slow generation of naive cells in adults (22, 23), killing of naive cells by SI HIV-1 may have a relatively large impact on CD4+ T cell renewal. The observed association between the frequency of infected CD45RA+ cells and the decline in both the number of naive and memory cells are in support of this hypothesis. Interpretation of these results may be complicated, however, by the effect of immune activation on the naive and memory cell populations. The percentage of CD45RO+ cells was shown to increase with ongoing infection, increasing virus load and declining CD4+ T cells (28). Hence, a higher virus load may enhance the transition of naive cells into memory cells, and thus simultaneously contribute to the loss of naive cells and compensate for the loss of memory cells due to virus-induced killing.
Although especially in HIV-1-infected individuals, not all CD45RA+CD4+ T cells are naive cells (29). Ostrowski et al. (17) recently demonstrated that truly naive (CD45RA+CD62L+) CD4+ T cells are HIV-1 infected in vivo, especially in individuals harboring CXCR4 using variants. Though in the present study we cannot exclude that nonnaive cells are among the infected CD45RA+ cells, the truly naive cells are more likely to represent the major HIV-1-infected cells in the CD45RA+ population for several reasons. First, nonnaive CD45RA+CD4+ T cells are considered to belong to the memory compartment. In agreement, we recently observed that the frequency of CCR5-expressing cells among the nonnaive CD45RA+CD4+ T cells was similar to that of resting CD45RO+CD4+ T cells and intermediate to that of truly naive CD4+ T cells and activated CD45RO+CD4+ T cells (H.B. and H.S., unpublished data). The low frequency of NSI variants in the CD45RA+ cell subset would be difficult to explain if, within the CD45RA+CD4+ T cells, the nonnaive memory cells represent the major population of HIV-1-carrying cells. Furthermore, in most individuals studied here, we observed a low frequency of CCR5- and a high frequency of CXCR4-expressing cells in the CD45RA+ cell population. This expression pattern is consistent with the assumption that at the moment of analysis, the majority of the CD45RA+ cells were naive in most patients.
At this moment, it is not clear at what time point during their life cycle peripheral naive cells become infected. If infection occurred when the T cells were already mature and expressing CD45RA, HIV-1 may be present as an incomplete and labile proviral DNA species (30), in analogy to observations in quiescent cells (31–33). On the other hand, the existence of proliferating cells within the naive cell population (34) may support integration of viral DNA in part of the naive cells. In agreement, Ostrowski et al. (17) demonstrated that integrated proviral DNA is detected in naive cells, yet at lower levels compared with memory cells.
Alternatively, peripheral HIV-1-infected naive cells may derive from infected thymocytes. The existence of thymocytes that survive subsequent differentiation stages despite HIV-1 infection is suggested by the existence of productively infected CD3+CD8+CD4− thymocytes (35, 36) expressing viral mRNA (35). Thymocytes express CXCR4 during most stages of maturation and only low levels of CCR5 (37–40), compatible with preferential infection by SI variants. Although conflicting results in this respect have been obtained in vitro (36–38), SI variants were shown to be more thymocyte-tropic in vivo in a SCID-hu mice model (41).
Even though especially in SI-infected individuals, naive cells were shown to carry replication competent HIV-1, it is not known whether naive cells support HIV-1 replication in vivo. However, virus production may be supported by low levels of residual proliferation or induced upon antigen-induced stimulation, either way resulting in an increased death of naive cells. Since increased killing of CD45RA+ cells may have a large impact, even when relatively low numbers of cells are infected, the capability of SI HIV-1 to infect CD45RA+ cells next to CD45RO+ cells may represent the crucial difference between SI and NSI variants with respect to their pathogenicity.
Acknowledgments
We thank Marijke Roos and colleagues for determining CD4+ T cell counts, and Dörte Hamann, René van Lier, and Frank Miedema for critically reading the manuscript. Proleukin (rIL-2) was kindly provided by R. Rombouts, Chiron Benelux B.V., Amsterdam. We are greatly indebted to all the cohort participants for their continuous participation. This study was performed as part of the Amsterdam Cohort Studies on AIDS, a collaboration between the Municipal Health Service, The Academic Medical Centre, and the CLB.
Abbreviations
- NSI
non-syncytium-inducing
- SI
syncytium-inducing
- PBMC
peripheral blood mononuclear cell
- PBL
peripheral blood lymphocyte
- TCID
tissue culture infectious dose
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P A, III, Goudsmit J, Huisman J G, Miedema F. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M L, III, Coutinho R A, Miedema F, Schellekens P A, III, Tersmette M. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 3.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E Y, Van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connor R I, Mohri H, Cao Y, Ho D D. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier R A M, Meyaard L, Brouwer M, Hovenkamp E, Schuitemaker H. Virology. 1996;219:87–95. doi: 10.1006/viro.1996.0225. [DOI] [PubMed] [Google Scholar]
- 6.Connor R I, Ho D D. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grivel J-C, Margolis D B. Nat Med. 1999;5:344–346. doi: 10.1038/6565. [DOI] [PubMed] [Google Scholar]
- 8.Björndal Å, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, et al. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Roda Husman A M, van Rij R P, Blaak H, Schuitemaker H. J Infect Dis. 1999;180:1106–1115. doi: 10.1086/314987. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y-J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, KewalRamani V N, Moore J P. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrowski M A, Justement S J, Cantanzaro A, Hallahan C A, Ehler L A, Mizell S B, Kumar P N, Mican J, Chun T-W, Fauci A S. J Immunol. 1998;161:3195–3201. [PubMed] [Google Scholar]
- 16.Koot M, van't Wout A B, Kootstra N A, De Goede R E Y, Tersmette M, Schuitemaker H. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski M A, Chun T-W, Justement S J, Motola I, Spinelli M A, Adelsberger J, Ehler L A, Mizell S B, Hallahan C W, Fauci A S. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koot M, Vos A H V, Keet R P M, De Goede R E Y, Dercksen W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Pantaleo G, Graziosi C, Fauci A S. N Engl J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 20.Wolthers K C, Wisman G B A, Otto S A, De Roda Husman A M, Schaft N, De Wolf F, Goudsmit J, Coutinho R A, Van der Zee A G J, Meyaard L, Miedema F. Science. 1996;274:1543–1547. doi: 10.1126/science.274.5292.1543. [DOI] [PubMed] [Google Scholar]
- 21.Fleury S, De Boer R J, Rizzardi G P, Wolthers K C, Otto S A, Welbon C C, Graziosi C, Knabenhans C, Soudeyns H, Bart P-A, et al. Nat Med. 1998;4:794–801. doi: 10.1038/nm0798-794. [DOI] [PubMed] [Google Scholar]
- 22.Mackall C L, Gress R E. Immunol Rev. 1997;160:91–102. doi: 10.1111/j.1600-065x.1997.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z Q, Notermans D W, Sedgewick G, Cavert W, Wietgrefe S, Zupancic M, Gebhard K, Henry K, Boies L, Chen Z, et al. Proc Natl Acad Sci USA. 1998;95:1154–1159. doi: 10.1073/pnas.95.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellerstein M, Hanley M B, Cesar D, Siler S, Papageorgopoulos C, Wieder E, Schmidt D, Hoh R, Neese R, Macallan D, et al. Nat Med. 1999;5:83–89. doi: 10.1038/4772. [DOI] [PubMed] [Google Scholar]
- 25.Clark D R, Ampel N M, Hallet C A, Yedavalli V R K, Ahmad N, DeLuca D. J Infect Dis. 1997;176:649–654. doi: 10.1086/514086. [DOI] [PubMed] [Google Scholar]
- 26.Pakker N G, Notermans D W, De Boer R J, Roos M T L, Wolf F, Hill A, Leonard J M, Danner S A, Miedema F, Schellekens P T A. Nat Med. 1998;4:208–214. doi: 10.1038/nm0298-208. [DOI] [PubMed] [Google Scholar]
- 27.Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debré P, Leibowitch J. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 28.De Roda Husman A M, Blaak H, Brouwer M, Schuitemaker H. J Immunol. 1999;163:4597–4603. [PubMed] [Google Scholar]
- 29.Roederer M, Gregson Dubs J, Anderson M T, Raju P A, Herzenberg L A, Herzenberg L. J Clin Invest. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woods T C, Roberts B D, Butera S T, Folks T M. Blood. 1997;89:1635–1641. [PubMed] [Google Scholar]
- 31.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 32.Zack J A, Haislip A M, Krogstad P, Chen I S Y. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazenberg M D, Cohen Stuart J W T, Otto S A, Borleffs J C C, Boucher C A, De Boer R J, Miedema F, Hamann D. Blood. 2000;95:249–255. [PubMed] [Google Scholar]
- 35.Lee S, Goldstein H, Baseler M, Adelsberger J, Golding H. J Virol. 1997;71:6671–6676. doi: 10.1128/jvi.71.9.6671-6676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitchen S G, Uittenboogaart C H, Zack J A. J Virol. 1997;71:5713–5722. doi: 10.1128/jvi.71.8.5713-5722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedroza-Martin L, Gurney K B, Torbett B E, Uittenboogaart C H. J Virol. 1998;72:9441–9452. doi: 10.1128/jvi.72.12.9441-9452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaitseva M B, Lee S, Rabin R L, Tiffany H L, Farber J M, Peden K W C, Murphy P M, Golding H. J Immunol. 1998;161:3103–3113. [PubMed] [Google Scholar]
- 39.Berkowitz R D, Beckerman K P, Schall T J, McCune J M. J Immunol. 1998;161:3702–3710. [PubMed] [Google Scholar]
- 40.Kitchen S G, Zack J A. J Virol. 1997;71:6928–6934. doi: 10.1128/jvi.71.9.6928-6934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkowitz R D, Alexander S, Bare C, Linquist-Stepps V, Bogan M, Moreno M E, Gibson L, Wieder E D, Kosek J, Stoddard C L, McCune J M. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]