Abstract
Ceramide plays a crucial role as a basic building block of sphingolipids, but also as a signalling molecule mediating cell-fate decisions. Three genes, LAG1, LAC1 and LIP1, have been shown to be required for ceramide synthase activity in Saccharomyces cerevisiae [Guillas, Kirchman, Chuard, Pfefferli, Jiang, Jazwinski and Conzelman (2001) EMBO J. 20, 2655–2665; Schorling, Vallee, Barz, Reizman and Oesterhelt (2001) Mol. Biol. Cell 12, 3417–3427; Vallee and Riezman (2005) EMBO J. 24, 730–741]. In the present study, the topology of the Lag1p and Lac1p subunits was investigated. The N- and C-termini of the proteins are in the cytoplasm and eight putative membrane-spanning domains were identified in Lag1p and Lac1p by insertion of glycosylation and factor Xa cleavage sites at various positions. The conserved Lag motif, potentially containing the active site, is most likely embedded in the membrane. We also present evidence that histidine and aspartic acid residues in the Lag motif are essential for the function of Lag1p in vivo.
Keywords: ceramide synthase, LAG1, sphinganine, sphingolipid, topology, yeast
Abbreviations: endo H, endoglycosidase H; ER, endoplasmic reticulum; 5-FOA, 5-fluoro-orotic acid; fXa, factor Xa; NP40, Nonidet P40; TMD, transmembrane domain; TRAM, translocating chain-associating membrane protein
INTRODUCTION
Ceramide is a central molecule among sphingolipid precursors, but also functions in different cellular events, such as apoptosis, growth arrest, membrane traffic and stress responses [1–5]. Ceramide is synthesized mainly from the reaction of a fatty acyl-CoA with a sphingoid base by an acyl-CoA-dependent ceramide synthase [6,7]. Three genes, LAG1, LAC1 and LIP1, have been shown to be required for ceramide synthase activity in Saccharomyces cerevisiae [8–10]. Lag1p and Lac1p are homologous and functionally redundant polytopic transmembrane proteins of the ER (endoplasmic reticulum). Double deletion mutant cells, lag1Δ lac1Δ, have a strongly decreased level of sphingolipids due to loss of the acyl-CoA-dependent ceramide synthase reaction [8,9]. Purification of active ceramide synthase from yeast microsomes using tagged versions of Lac1p and Lag1p identified Lip1p (Lip1 protein) as a novel subunit of acyl-CoA-dependent ceramide synthase. Lip1p forms a heteromeric complex with Lag1p and Lac1p and is required for ceramide synthase in vivo and in vitro [10]. Interestingly, no strong homologues of Lip1p in animals or plants were found using sequence alignments. Since Lag1 homologues have been found in a wide variety of eukaryotes [11], it has been suggested that Lag1p and Lac1p will be the catalytic subunits of ceramide synthase and Lip1p could be a regulatory subunit of the complex.
Despite its crucial role in ceramide synthesis, little information on the membrane topology of ceramide synthase is available. The sphingolipid biosynthetic pathway begins with the condensation of palmitoyl-CoA and L-serine to generate 3-ketosphinganine. The reaction is catalysed by serine palmitoyltransferase, which in yeast has two major subunits named Lcb1p and Lcb2p. Protease accessibility studies suggest that the active site of serine palmitoyltransferase is cytoplasmic [12], which is consistent with the cytoplasmic localization of its substrates, serine and palmitoyl-CoA. The next enzyme of the sphingolipid biosynthesis pathway, 3-ketosphinganine reductase, which generates sphinganine, seems to have its active site localized to the cytoplasmic face of the ER [13]. On the other hand, the active site of Lcb3p, which dephosphorylates exogenous sphingoid bases, previously phosphorylated in the cytoplasm by sphingoid base kinase, has been proposed to be in the lumen of the ER [14], suggesting that one of the substrates of ceramide synthase, sphingoid base, might be made available from either side of the ER membrane. The product, ceramide, is transported to the Golgi by vesicular and non-vesicular mechanisms [15,16] where it is converted into inositolphosphorylceramide by Aur1p whose active site is localized on the luminal side of a late Golgi compartment [17], suggesting that ceramide must arrive to the luminal side at Golgi. Ceramide synthase has been proposed to have its active site on the cytoplasmic surface of the ER membrane because its activity is protease-sensitive in isolated microsomal membranes [12,18]. However, Lag1p and Lac1p span the ER membrane multiple times and their susceptibility to protease digestion cannot be taken as an indicator of the topology of the active site. Recently, the N-terminal cytoplasmic tail of Lip1p, an integral membrane protein localized in ER with one TMD (transmembrane domain), has been shown to be dispensable for ceramide synthesis activity [10], indicating that the functional region of Lip1p is localized in the lumen of the ER, or in its TMD. To determine on which side of the ER membrane the active site of ceramide synthase is located, the number and topology of transmembrane helices of Lag1p and Lac1p need to be determined.
Here, we present a detailed topology study of Lag1p and Lac1p using insertion of glycosylation sites and fXa (factor Xa)-cleavage sites in the loops between the predicted transmembrane segments. Our results are most consistent with the hypothesis that Lag1p and Lac1p possess eight TMDs each and that most of the Lag motif, which is conserved in all Lag1 homologues, is embedded in the membrane.
EXPERIMENTAL
Strains
S. cerevisiae strain RH382 (MATa his3 ura3 pep4-3 suc2Δ9 bar1-1) is a wild-type strain in our laboratory. WBY616 (lag1Δlac1Δ) was described previously [9]. WBY616 was transformed with a plasmid no. 1753 (pRS416-LAG1), creating strain RH6602.
Construction of LAG1- and LAC1-SUC2A glycosylation cassette topology reporter plasmids
LAG1 and LAC1 gene fusion alleles with the invertase glycosylation cassette (Suc2A cassette) inserted were constructed in two steps. In the first step, SacI restriction sites were inserted by using PCR at 11 positions along the LAG1 and LAC1 genes corresponding to amino acids Met1, Asn108, Thr165, Tyr208, Pro245, Thr272, Asp286, Leu298, Phe333, Gly386, and Glu411 (Lag1p) or Ile418 (Lac1p), creating plasmids 1755–1776 respectively. In the second step, the Suc2A cassette corresponding to sequences following amino acid 81 and preceding 133 and containing three potential glycosylation sites [19] was amplified from plasmid 1582 using the primers containing SacI site at the both ends (SacI-SUC2-F, 5′-CTACTTGAGCTCATTTGACTAATTGGGAAGATCAACCC, and SacI-SUC2-R, 5′-TTTCAGGAGCTCTATAAGTCCAAATCGCAACGCATC) (the SacI sites on the ends of the primers are in boldface and underlined), subcloned into pESC-URA, creating plasmid pSUC2A. The Suc2A cassette was amplified by PCR from pSUC2A, and was inserted into the SacI sites of each plasmid, creating plasmids 1777–1798. In all constructions, DNA sequences were confirmed. A list of plasmids is shown in Table 1.
Table 1. Plasmids used in the present study.
(a), Dieter Oesterhelt (Max Planck Institute for Biochemistry, Martinsreid, Germany); (b), [10]; (c), Béatrice Vallée; (Center for Molecular Biophysics, CNRS, Orleans, France) (d), the present study.
| Plasmid number | Plasmids | Inserts | Source |
|---|---|---|---|
| T1118 | pESC-URA | None | (a) |
| 1119 | pSTS30a | LAG1 (N-term., c-Myc-tagged) | (a) |
| 1121 | pSTS30b | LAC1 (N-term., FLAG-tagged) | (a) |
| 1548 | pSTS30c | LAG1 (N-term., FLAG-tagged) | (b) |
| 1582 | Full-length of SUC2 | (c) | |
| 1750 | pRS414-PT | Promoter and terminator of LAG1 | (d) |
| 1751 | pRS414-LAG1 | LAG1 (N-term., FLAG-tagged) | (d) |
| 1752 | pRS416-PT | Promoter and terminator of LAG1 | (d) |
| 1753 | pRS416-LAG1 | LAG1 (N-term., Myc-tagged) | (d) |
| 1754 | pSUC2A | SUC2 reporter cassette A | (d) |
| 1755–1765 | pESC-LAG1-N, 1–8, C | SacI site is introduced into various regions of LAG1 | (d) |
| 1766–1776 | pESC-LAC1-N, 1–8, C | SacI site is introduced into various regions of LAC1 | (d) |
| 1777–1787 | pESC-LAG1-SUC2A N, 1–8, C | SUC2A cassette is inserted into SacI sites of no. 1755–1765 | (d) |
| 1788–1798 | pESC-LAC1-SUC2A N, 1–8, C | SUC2A cassette is inserted into SacI sites of no. 1755–1765 | (d) |
| 1799–1809 | pRS414-LAG1-SUC2A N, 1–8, C | LAG1–SUC2A fusion constructs expressed under LAG1’s promoter | (d) |
| 1810 | pESC-LAG1-fXa4 | fXa cleavage site is inserted in Pro245 of LAG1 | (d) |
| 1811 | pESC-LAG1-fXa5 | fXa cleavage site is inserted in Thr272 of LAG1 | (d) |
| 1812 | pESC-LAG1-fXa6 | fXa cleavage site is inserted in Leu298 of LAG1 | (d) |
| 1813 | pESC-LAG1-fXa7 | fXa cleavage site is inserted in Phe333 of LAG1 | (d) |
| 1814 | pESC-LAG1-fXa8 | fXa cleavage site is inserted in Gly386 of LAG1 | (d) |
| 1815 | pESC-LAC1-fXa4 | fXa cleavage site is inserted in Pro245 of LAC1 | (d) |
| 1816 | pESC-LAC1-fXa5 | fXa cleavage site is inserted in Thr272 of LAC1 | (d) |
| 1817 | pESC-LAC1-fXa6 | fXa cleavage site is inserted in Leu298 of LAC1 | (d) |
| 1818 | pESC-LAC1-fXa7 | fXa cleavage site is inserted in Phe333 of LAC1 | (d) |
| 1819 | pESC-LAC1-fXa8 | fXa cleavage site is inserted in Gly386 of LAC1 | (d) |
| 1820–1824 | pRS414-LAG1-fXa4–8 | LAG1–fXa-cleavage-site fusion constructs expressed under LAG1's promoter | (d) |
| 1825 | pRS414-lag1(H255,256A) | lag1 (H255A, H256A) | (d) |
| 1826 | pRS414-lag1(L262A) | lag1 (L262A) | (d) |
| 1827 | pRS414-lag1(S266A) | lag1 (S266A) | (d) |
| 1828 | pRS414-lag1(D283A) | lag1 (D283A) | (d) |
| 1829 | pRS414-lag1(D283,286A) | lag1 (D283A, D286A) | (d) |
| 1853 | pESC-LAG1-SUC2A-4H | H255A H256A mutations were introduced into no. 1781 | (d) |
| 1854 | pESC-LAG1-SUC2A-5H | H255A H256A mutations were introduced into no. 1782 | (d) |
| 1855 | pESC-LAG1-SUC2A-6H | H255A H256A mutations were introduced into no. 1784 | (d) |
| 1856 | pESC-LAG1-SUC2A-7H | H255A H256A mutations were introduced into no. 1785 | (d) |
| 1857 | pESC-LAG1-SUC2A-4D | D283A D286A mutations were introduced into no. 1781 | (d) |
| 1858 | pESC-LAG1-SUC2A-5D | D283A D286A mutations were introduced into no. 1782 | (d) |
| 1859 | pESC-LAG1-SUC2A-6D | D283A D286A mutations were introduced into no. 1784 | (d) |
| 1860 | pESC-LAG1-SUC2A-7D | D283A D286A mutations were introduced into no. 1785 | (d) |
Determination of topology by using a glycosylation cassette
The RH382 strain transformed with the Suc2A-cassette topology reporter plasmids was grown in minimal medium containing raffinose [SRaf (−Ura)] to 5×107 cells/ml, harvested, washed with water, resuspended in minimal medium containing galactose [SGal (−Ura)] and incubated for 6 h at 30 °C. Cells (2.5×108) were lysed by vortex-mixing with glass beads in 100 μl of lysis buffer (50 mM Tris/HCl, pH 6.8, 1% SDS, 1% 2-mercaptoethanol and 2 mM PMSF) and centrifuged at 15000 g for 5 min at 4 °C to remove insoluble proteins. Aliquots (10 μl) were mixed with 15 μl of endo H (endoglycosidase H) solution [10 mM sodium citrate, pH 5.5, 0.8% Triton X-100, 2 mM PMSF, 0.001% (w/v) pepstatin A and 25 m-units of endo H or mock], incubated at 37 °C for 90 min, and proteins were resolved by SDS/12% PAGE and chimaeric proteins were visualized by immunoblotting using anti-FLAG antibody (Sigma, St. Louis, MO, U.S.A.).
Construction of LAG1- and LAC1-fXa cleavage site fusion plasmids
LAG1 and LAC1 alleles containing fXa cleavage sites were constructed by site-directed mutagenesis using PCR, creating plasmids 1810–1819. These plasmids encode proteins with eight extra amino acids that generate the tetra-residue repeat, IEGRIEGR, inserted after amino acid residue Pro245, Thr272, Leu298, Phe333 or Gly386 of Lag1p and Lac1p. In all constructs, DNA sequences were confirmed.
Yeast microsome preparation
Strains (RH382) transformed with plasmids encoding gene fusion proteins containing fXa cleavage sites were grown and protein expression was induced as described above. The 1.25×109 cells were converted into spheroplasts with Zymolyase (Seikagaku Kogyo, Tokyo, Japan). The spheroplasts were collected by centrifugation, resuspended in lysis buffer (100 mM sorbitol, 50 mM potassium acetate, 20 mM Tris/HCl, pH 7.5, 0.5 mM dithiothreitol and 2 mM PMSF), and homogenized by 20 strokes in Dounce homogenizer. The homogenate was centrifuged at 1000 g for 5 min to remove unbroken cells. The supernatant was subjected to centrifugation at 100000 g for 30 min at 4 °C, and the pelleted microsomes were resuspended in 1 ml of storage buffer (250 mM sorbitol, 50 mM potassium acetate, 20 mM Tris/HCl, pH 7.5, and 1 mM dithiothreitol). Microsome preparations were stored at −80 °C.
Cleavage with fXa protease
fXa treatment was performed essentially as described previously [19] with some modifications. Briefly, 15 μl of microsomes was diluted with 5 μl of fXa buffer (250 mM sorbitol, 100 mM NaCl, 20 mM Tris/HCl, pH 7.5, and 1 mM EDTA). Samples were mock-digested or digested with 1.1 μg of fXa protease (1.1 mg/ml; Promega, Madison, WI, U.S.A.) in the presence or absence of detergent [0.2% NP40 (Nonidet P40)] on ice for 2 h. Alternatively, we used 1% NP40, 1% digitonin, 1% Triton X-100 and 1% SDS instead of 0.2% NP40, and treated microsomes for 24 h.
Plasmids for examining complementing activity of chimaeric constructs and lag1 mutants
To check the activity of chimaeric constructs and mutants, these proteins were expressed under control of the LAG promoter and terminator. The regions of 500 bp each of 5′ upstream and 3′ downstream of LAG1 were amplified from genomic DNA by PCR using primers (SacI-prom-F, 5′-AAGCTGGAGCTCCTGCTATCTTCCTGCACTTGCTG, XbaI-prom-R, 5′-AGCTGATCTA-GAGTTGTCGTTATTTCTTCAGTTTC, XhoI-term-F, 5′-AAGTGTCTCGAGACGTATCTTAAGGAGAATACGTATC, and KpnI-term-R, 5′-GAATTGGGTACCAACATCAAAGCAGGTTAAGCCTGAC) and subcloned into SacI–XbaI and XhoI–KpnI site of pRS416 plasmid to create pRS416-PT (1752), a plasmid containing a multicloning site consisting of 69 nt between the promoter and terminator of LAG1. The SacI–KpnI fragment of pRS416-PT was subcloned into pRS414, creating plasmid pRS414-PT (1750). FLAG-tagged and Myc-tagged LAG1 was amplified from pSTS30c (1548) and pSTS30a (1119) using the primers 5′-GGGCGGCCGCACTAGTATCGATGG (for FLAG-tagged LAG1) 5′-ATATATACTAGTATGGAACAGAAGTTGATTTCCGAAG (for Myc-tagged LAG1) and 5′-CGCGCG-GGATCCTTAATTAATTATTCACACTTTTCC, and subcloned into the SpeI–BamHI site of pRS416-PT and pRS414-PT, creating plasmid pRS416-LAG1 (1753) and pRS414-LAG1 (1751) respectively. Since the plasmid expressing LAG1 under its own promoter contains excess nucleotides between its promoter and terminator, the regions of the open reading frame and 500 bp each of 5′ upstream and 3′ downstream sequences were amplified from genomic DNA using primers SacI-prom-F and KpnI-term-R, subcloned into SacI–KpnI site of pRS414, and its activity was determined by complementation of lag1Δlac1Δ cells (WBY616), which was almost same as for pRS414-LAG1 (results not shown).
Site-directed mutagenesis
The lag1 mutant alleles were generated by PCR mutagenesis. In all constructions, DNA sequences were verified.
RESULTS
Hydropathic profile analysis of Lag1p and Lac1p
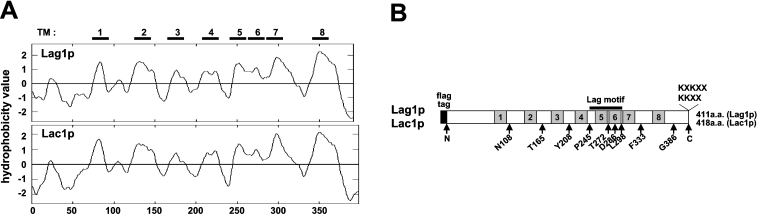
The Lag1p and Lac1p subunits of ceramide synthase are 411- and 418-amino-acid proteins respectively. Lag1p and Lac1p are 72% identical and 81% similar in amino acid sequence, and are thought to be functionally redundant proteins. Lag1p and Lac1p have the biochemical properties of integral membrane proteins [10]. Various algorithms for predicting membrane-spanning segments suggest alternative topologies for the proteins. Hydropathic profile analyses predict anywhere from four to eight potential membrane-spanning domains in the proteins (Table 2). We assumed all possible TMDs and postulated that eight TMDs might exist in Lag1p and Lac1p (Figure 1A). The ER-retrieval signals, KKXX or KXKXX, contained in the C-termini of Lag1p and Lac1p, suggest that the C-termini are exposed to the cytoplasmic side of the ER. An N-terminal FLAG-tagged fusion protein of Lac1p is protease-sensitive in the absence of detergent, which suggests that the N-termini of Lag1p and Lac1p also face the cytoplasmic side of the ER [10].
Table 2. Candidate membrane-spanning segments of Lag1p and Lac1p.
| Helix begin–end | |||||
|---|---|---|---|---|---|
| TopPredII [29] | SOSUI [30] | MEMSAT [31] | HMMTop [32] | TMPred [33] | |
| Lag1p | 83–103 | 83–100 | 84–100 | 84–102 | 85–102 |
| 131–151 | 136–158 | 133–155 | 129–146 | 135–151 | |
| 176–196 | 173–195 | 177–195 | 177–195 | 176–195 | |
| 213–233 | 222–240 | 222–239 | 218–240 | ||
| 252–272 | 249–271 | 252–269 | 252–269 | 252–272 | |
| 274–296 | 274–292 | ||||
| 297–317 | 305–327 | 301–317 | 305–323 | 298–317 | |
| 351–371 | 357–379 | 352–376 | 350–374 | 352–372 | |
| Lac1p | 82–102 | 83–99 | 84–102 | 82–98 | |
| 130–150 | 136–158 | 130–146 | 129–146 | 127–145 | |
| 176–196 | 177–195 | 177–195 | 177–195 | ||
| 223–243 | 222–240 | 222–239 | 218–240 | ||
| 252–272 | 252–274 | 252–269 | 252–269 | 252–272 | |
| 274–291 | |||||
| 298–318 | 305–327 | 301–317 | 300–317 | 298–317 | |
| 351–371 | 355–377 | 352–376 | 352–376 | 352–372 | |
Figure 1. Hydropathy profiles of Lag1p and Lac1p and positions of fusion insertions.
(A) Hydropathy plots of Lag1p and Lac1p were generated according to the algorithm of Kyte and Doolittle [29] with a window of ten amino acids. TMs 1–8 indicate predicted transmembrane segments. (B) Schematic representation of Lag1p and Lac1p; the grey boxes depict the eight predicted TMDs (TMDs 1–8). The positions of the Suc2A topological-reporter cassette and fXa-protease-cleavage site insertions are indicated by arrows. The numbers refer to the amino acid at which the insertions were introduced. The Lag motif is noted with a solid line.
Construction of Lag1p and Lac1p topology reporter proteins
To experimentally determine the topology of the yeast Lag1 and Lac1 proteins, a set of fusion proteins containing a glycosylation reporter cassette A from Suc2 (Suc2A) inserted in-frame at 11 positions along the length of Lag1p and Lac1p was constructed. The Suc2A cassette consists of a 53-amino-acid domain comprising residues 80–133 of invertase (Suc2p) that contains three NX(S/T) sites for asparagine-linked glycosylation. These sites are known to be glycosylated in the mature Suc2p protein [19]. This reporter cassette has been used in determining the topology of other membrane proteins, such as Gap1p, Lcb1p, Shr3p and Stt3p [19–22]. SacI restriction sites were individually introduced into LAG1 and LAC1 sequences encoding the N- or the C-termini, and into each of the hydrophilic loops predicted by hydropathy analysis (Figure 1B), and the Suc2A cassette was inserted into every SacI site. Because the authentic expression levels of Lag1p and Lac1p are very low, we expressed these constructs under GAL10 promoter.
Analysis of the Lag1–Suc2A and Lac1–Suc2A fusion proteins suggests eight TMDs in Lag1p and Lac1p
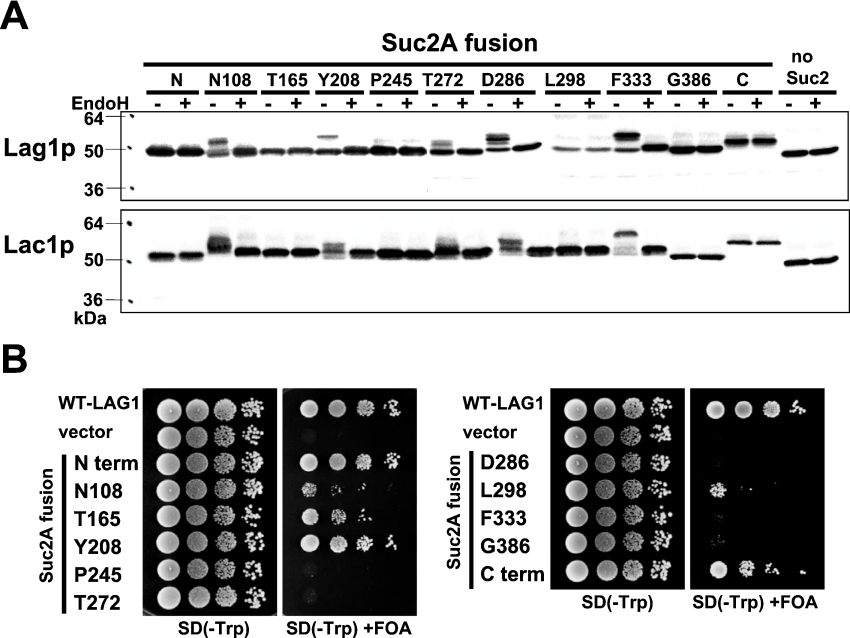
We examined the mobility of Lag1–Suc2A and Lac1–Suc2A fusion proteins treated with or without endo H by immunoblot analysis (Figure 2A). The insertion of Suc2A cassette after amino acid residue Asn108, Tyr208, Thr272, Asp286 or Phe333 resulted in a molecular mass difference between endo H-treated and mock-treated samples, demonstrating that in these proteins the Suc2A cassette had been glycosylated. This result suggests that these regions were exposed on the luminal side of the ER. On the other hand, the mobilities of Lag1–Suc2A and Lac1–Suc2A proteins in which the Suc2A cassette was inserted into the N-terminus, Thr165, Phe245, Leu298, Gly386, or the C-terminus were not altered after endo H treatment, which means that these chimaeric proteins were not glycosylated. These results suggest that these regions are oriented to the cytoplasmic side of the ER. These results also show that yeast Lag1p and Lac1p are not normally glycosylated, in contrast with what has been claimed for one of its mammalian homologues LASS6 [23]. Taken together, our results suggest that Lag1p and Lac1p contain eight TMDs.
Figure 2. Topology determination by insertion of glycosylation sites.
(A) The glycosylation status of the topological reporter Lag1–Suc2A and Lac1–Suc2A fusion proteins. RH382 cells were transformed with plasmids 1777–1798 to express the Lag1–Suc2A and Lac1–Suc2A fusion proteins. The numbers and amino acids represent the amino acid residues of Lag1p and Lac1p before which the 53-amino-acid cassette was inserted. Total cell lysates were prepared from the transformants, treated with or without endo H for 90 min at 37 °C, and resolved on 12% (w/v) polyacrylamide gels. The Lag1 and Lac1 proteins were detected by immunoblotting using anti-FLAG antibodies. (B) Functional complementation of lag1Δ lac1Δ cells by Lag1–Suc2A fusion proteins. lag1Δ lac1Δ double deletion mutant cells transformed with pRS416-LAG1 (LAG1 on uracil-based plasmid) (RH6602) were transformed with pRS414-LAG1-SUC2AN (LAG1-SUC2A fusion constructs on tryptophan-based plasmid) (plasmids 1799–1809). The transformants were diluted to a final concentration of 1×107 cells/ml, and 10 μl of 10-fold dilutions were spotted on SD (−Trp)- and SD (−Trp)-containing 5-FOA plates. The plates were incubated at 30 °C for 3 days. The deletion mutants transformed with wild-type LAG1 (plasmid 1751) and vector without insert (plasmid 1750) were used as controls.
To test whether the Lag1–Suc2A fusion proteins can functionally replace Lag1p, these chimaeric constructs were expressed under Lag1p's own promoter in lag1Δlac1Δ cells and we examined their ability to complement the growth defect of the cells (Figure 2B). lag1Δ lac1Δ cells have a severe growth defect. If the Lag1–Suc2A cassette fusion proteins can replace wild-type Lag1p, the transformants will grow on 5-FOA (5-fluoro-orotic acid) plates because they are able to grow without the URA3 plasmid carrying wild-type LAG1. In contrast, if the fusion protein does not complement the loss of wild-type Lag1p, cells cannot be cured of the wild-type LAG1 plasmid and they will not grow on 5-FOA plates due to the accumulation of toxic fluorodeoxyuridine that is formed from 5-FOA in the cells. Serial dilution analysis demonstrated that three of 11 fusion proteins (N-terminus, Tyr208 and C-terminus) complemented the growth defect of lag1Δ lac1Δ cells and were therefore assumed to be catalytically active and to have been inserted in their native topology. The fusion proteins in which Suc2A cassette was inserted into Asn108, Thr165 and Leu298 in Lag1p were partially functional. However, when the Suc2A cassette was inserted into Pro245, Thr272, Asp286, Phe333 and Gly386, the chimaeric proteins lost their function.
An alternative method for assigning topology of Lag1p and Lac1p
Since the Lag1–Suc2A chimaeric constructs in which the Suc2A cassette was inserted into and around Lag motif lost their function in vivo, we confirmed the topology around the Lag motif by using another method. Tandem factor-Xa (fXa)-protease-cleavage sites (IEGRIEGR) were inserted at some predicted loops of Lag1p and Lac1p, especially around the Lag motif. A tandem recognition sequence was inserted to increase the probability of cleavage. Their accessibility to fXa protease, which cleaves proteins containing the IEGR tetrapeptide on the C-terminal side of the arginine residue, in microsomes was assessed.
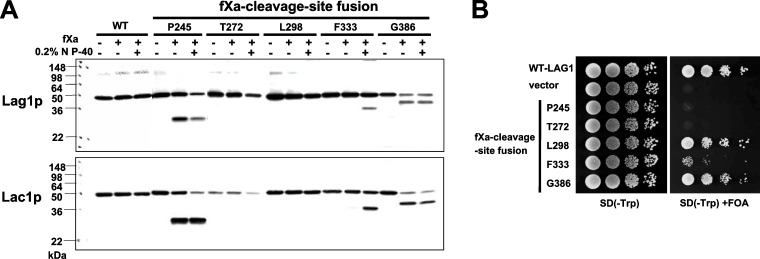
Lag1–fXa and Lac1–fXa cleavage site fusion proteins were expressed in cells, microsomes were prepared, and the sensitivity to fXa protease cleavage was assayed in the absence or presence of 0.2% NP40 (Figure 3A). When fXa-cleavage site was inserted into Pro245 and Gly386 of Lag1p and Lac1p, the chimaeric proteins were equally accessible to the fXa protease in the presence or absence of detergent, confirming the cytoplasmic location of these positions in the protein. In contrast, when the fXa-cleavage sites were inserted into Phe333 of Lag1p and Lac1p, the chimaeric proteins were cleaved only after treatment of microsomes with NP40, providing additional evidence that these residues lie in the lumen of the ER. However, when fXa-cleavage sites were inserted into Thr272 and Leu298 of Lag1p and Lac1p, the fusion proteins were refractory to cleavage, even in the presence of detergent.
Figure 3. Topology determination by insertion of protease cleavage sites.
(A) fXa protease cleavage of Lag1–fXa and Lac1–fXa fusion proteins. RH382 cells were transformed with plasmids 1810–1819 to express the Lag1–fXa and Lac1–fXa fusion proteins. The numbers and amino acids represent the amino acid residues of Lag1p and Lac1p before which the fXa protease cleavage site was inserted. The microsomes were mock-digested or digested with fXa protease in the absence or presence of NP40 on ice for 2 h, and resolved on 13.5% polyacrylamide gels. The Lag1 and Lac1 proteins were detected by immunoblotting using anti-FLAG antibodies. (B) Functional complementation of lag1Δ lac1Δ cells by Lag1–fXa cleavage site fusion proteins. lag1Δ lac1Δ double deletion mutant cells transformed with pRS416-LAG1 (LAG1 on uracil-based plasmid) (RH6602) were transformed with pRS414-LAG1-fXa4-8 (LAG1-fXa fusion constructs on tryptophan-based plasmid) (plasmids 1820–1824). The transformants were tested for complementation as in Figure 2(B).
We examined whether the fXa-cleavage site fusion constructs could complement the growth defect of lag1Δ lac1Δ cells (Figure 3B). When the fXa-cleavage sites were introduced into Leu298 and Gly386, these chimaeric proteins maintained their function almost completely. When the fXa-cleavage site was inserted at the position Phe333, the enzyme retained partial function, and the constructs at positions Pro245 and Thr272 lost their activity.
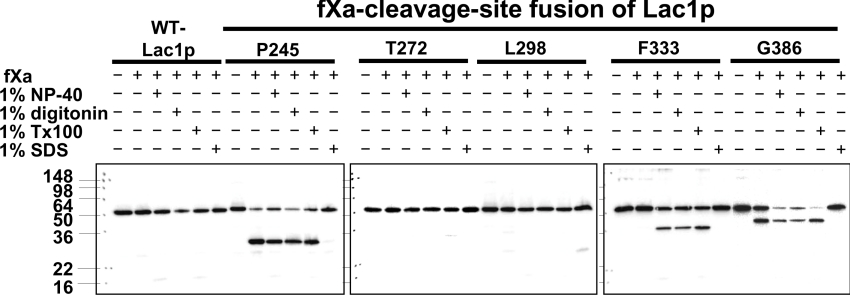
When the fXa-cleavage site was inserted into Thr272 and Leu298 of Lag1p and Lac1p, the chimaeric proteins were not cleaved even in the presence of 0.2% NP40. There are two possibilities; one is that the region is tightly packed or structured so that the site is not accessible to fXa protease. The other possibility is that the interaction with Lip1p might prevent the accession to the fXa protease. To address the latter possibility, we used different types of detergents. It has been reported that the Lip1p is co-immunoprecipitated with Lag1p and Lac1p upon 1% digitonin solubilization, and that the interactions between Lag1p or Lac1p and Lip1p are lost when ceramide synthase is solubilized using 1% Triton X-100 [10]. We used 1% NP40, 1% digitonin, 1% Triton X-100 and 1% SDS for membrane solubilization (Figure 4). We confirmed that the chimaeric proteins where fXa-cleavage sites were inserted into Pro245 and Gly386 of Lac1p were equally accessible to the fXa protease in the presence or absence of detergent, but when the cleavage sites were inserted into Phe333 of Lac1p, cleavage occurred only after treatment of microsomes with 1% NP40, 1% digitonin and 1% Triton X-100. In the case of 1% SDS, none of the chimaeric proteins were cleaved, suggesting that the fXa protease lost its activity under this condition. In other words, fXa protease maintained its activity in 1% NP40, 1% digitonin and 1% Triton X-100. When fXa-cleavage sites were inserted at Thr272 and Leu298, the chimaeric proteins were not cleaved even if 1% Triton X-100 was added to the reaction. These results indicate that the fXa-cleavage sites inserted into Thr272 and Leu298 cannot be cleaved, probably because of the structure of the region around Thr272 and Leu298 of Lag1p and Lac1p, and not because it interacts with Lip1p.
Figure 4. Effects of detergent on protease cleavage.
fXa protease cleavage of Lag1–fXa and Lac1–fXa fusion proteins with various detergent treatments. The microsomes from the transformants used in Figure 3 were prepared and mock-digested or digested with fXa protease in the absence or presence of 1% NP40, 1% digitonin, 1% Triton X-100 and 1% SDS on ice for 24 h, and resolved on 13.5% polyacrylamide gels. The Lag1 and Lac1 proteins were detected by immunoblotting using anti-FLAG antibodies.
Histidine and aspartic acid residues in the Lag1p motif are essential for the function of Lag1p in vivo
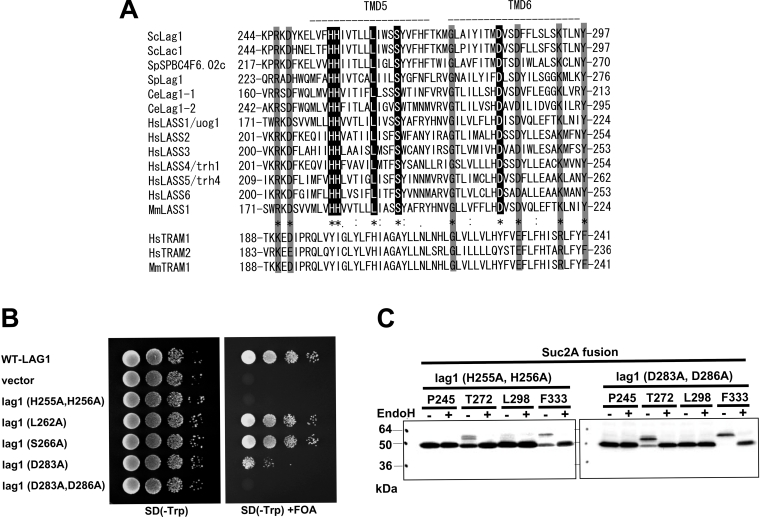
Lag1p and Lac1p are known to show significant sequence similarity to TRAM (translocating chain-associating membrane protein), a mammalian protein thought to be involved in protein translocation across the ER membrane. However, no translocation defects were detected in lag1Δ lac1Δ yeast cells [24]. Furthermore, human TRAM does not complement the growth defect of a lag1Δ lac1Δ double mutant, in contrast with another human gene called HsLAG1 (Homo sapiens LAG1) [11]. These results suggest that TRAM and Lag1p do not functionally overlap despite their sequence similarity. By comparing the whole-length amino acid sequences of Lag1 homologues from various organisms and TRAMs, we noticed that some amino acids, corresponding to His255, His256, Leu262, Ser266 and Asp283 of Lag1p and Lac1p, are conserved only among the Lag1 family, but not in TRAM proteins (Figure 5A). To examine the physiological importance of these amino acids, we mutated these residues to alanine residues and tested whether the mutants complemented the lag1Δ lac1Δ double mutant (Figure 5B). L262A and S266A mutations did not affect the function of Lag1p. On the other hand, when His255 and His256 were mutated to alanine, the complementation activity was completely lost. The D283A mutant partially complemented the growth defect of lag1Δ lac1Δ. When both Asp283 and Asp286 were converted into alanine, the mutant gene lost complementation activity completely. These results indicate that the histidine and aspartic acid residues in the Lag motif are critical for function of Lag1p in vivo. We tested whether these mutations have an effect on the topology of Lag1p. H255A/H256A and D283A/D286A mutations were introduced into some of the LAG1-SUC2A chimaeric constructs used in Figure 2(A), and the mobility of the fusion proteins was examined as described in Figure 2(A) (Figure 5C). Importantly, these mutations did not change the membrane topology of Lag1p, suggesting that the enzyme is assembled normally into the membrane.
Figure 5. Alignment of the Lag motif.
(A) Alignment of amino acid sequences of the Lag motif of Lag1 homologues and TRAM. The CLUSTALW algorithm was used to create the sequence alignment. The ‘*’ indicates positions that have a single, fully conserved residue among Lag1 homologues. The ‘:’ and ‘.’ indicate that the stronger- and weaker-score groups are fully conserved among Lag1 homologues respectively. Sc, S. cerevisiae; Sp, S. pombe; Ce, Caenorhabditis elegans; Hs, H. sapiens; Mm, Mus musculus. Note that some amino acid residues showed similarity in both Lag1 homologues and TRAM (shaded box) homologues, but others are conserved only among Lag1 homologues (black box). (B) Mutation analysis of the conserved amino acids among Lag1 homologues but not in TRAM. lag1Δ lac1Δ double deletion mutant cells transformed with pRS416-LAG1 (LAG1 on uracil-based plasmid) (RH6602) were transformed with pRS414-lag1(H255A/H256A), pRS414-lag1(L262A), pRS414-lag1(S266A), pRS414-lag1(D283A) and pRS414-lag1(D283A/D286A) (plasmids 1825–1829). The transformants were tested for complementation as in Figure 2(B). (C) H255A/H256A and D283A/D286A mutations were introduced into the LAG1-SUC2A chimaeric constructs used in Figure 2(B), resulting in plasmids 1853–1860. Protein expression and detection were performed as described in Figure 2(A).
DISCUSSION
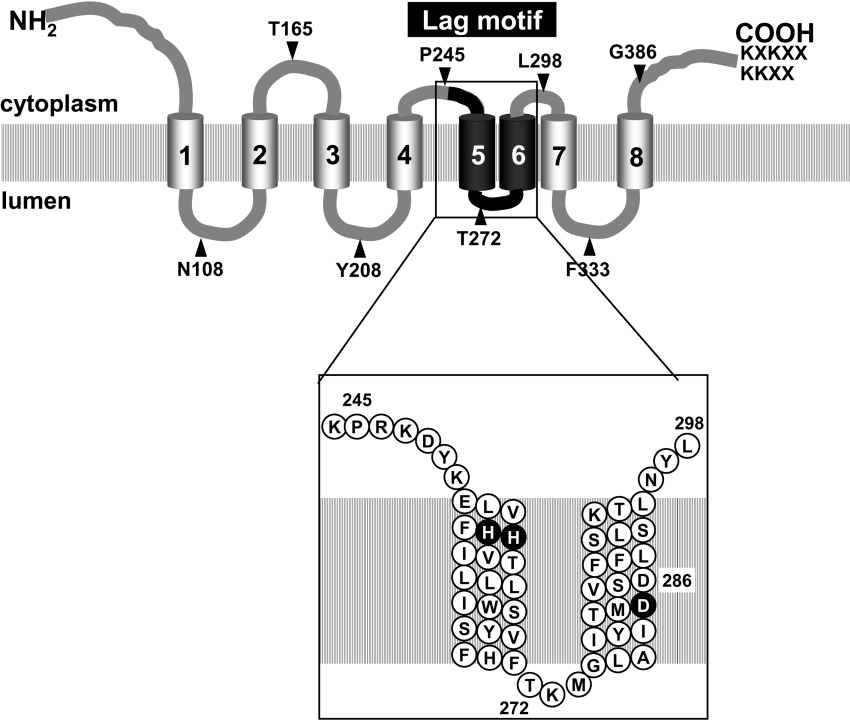
The present study reports a detailed topology mapping of Lag1p and Lac1p, the subunits of ceramide synthase. Our results suggest the presence of eight TMDs in Lag1p and Lac1p, as shown in schematic form in Figure 6. Lag1p and Lac1p show high homology throughout their length and exhibit striking similarities in hydrophobicity profiles. Thus it seems reasonable to assume that the membrane topology of Lag1p and Lac1p are the same.
Figure 6. Model for the topology of Lag1p and Lac1p.
Lag1p and Lac1p are integral membrane proteins with eight TMDs. The N-terminus and C-terminus are located on the cytoplasmic side of the ER and have dilysine-containing ER retrieval signals on their C-terminus. The positions of the Suc2A topological-reporter cassette and fXa-protease-cleavage site insertions are indicated by arrowheads. The numbers refer to the amino acid at which the insertions were fused. The region of the Lag motif, which is indicated in black, is expanded in a box. Most of the Lag motif is predicted to be embedded in the membrane. His255, His256 and Asp283 are conserved only among Lag1 homologues, not in TRAM, and are essential for the function of Lag1p in vivo (black circles).
Based on the results of hydrophobicity profiles, we postulated that there are eight transmembrane regions in Lag1p and Lac1p, and examined their topology using two separate methods, insertion of an Suc2A glycosylation cassette, which is recognized in the lumen of the ER and fXa-cleavage sites, which can be accessed from the cytoplasmic side of the ER by a specific protease. The Suc2A cassettes that were inserted at the N-terminus, at positions Thr165, Pro245, Leu298, Gly386 or at the C-terminus of Lag1p and Lac1p were not glycosylated, indicating that these sites are exposed to the cytoplasmic side of the ER. On the other hand, the Suc2A cassettes that were inserted at positions Asn108, Tyr208, Thr272 or Phe333 were glycosylated, suggesting that these regions are localized in the lumen of the ER. Curiously, the insertion of the Suc2A cassette at the C-termini of Lag1p and Lac1p made the mobility of the fusion protein in SDS/PAGE slower than the other constructs (Figure 2A). We do not know the reason for this behaviour, but one possibility is that since the dilysine ER-retrieval signals should be abrogated in these constructs, the chimaeric proteins might be mislocalized and further modified, but this modification does not seem to be related to N-linked glycosylation because the protein is not endo H-sensitive.
We confirmed the topology around the Lag motif by using the fXa-cleavage sites. The chimaeric proteins in which the fXa-cleavage sites were inserted at positions Pro245 and Gly386 were equally cleaved with or without detergent, again suggesting that these sites are localized on the cytoplasmic side of the ER. On the other hand, the chimaeric protein where the fXa-cleavage site was inserted at Phe333 was cleaved only when detergent was added to the reaction mixture, indicating that the site is exposed to the luminal side of the ER. However, in contrast with the results obtained by fusion proteins with Suc2A cassette, we could not obtain the clear topological information by using the chimaeric constructs in which the fXa-cleavage sites were inserted at the position Thr272 and Leu298. Since the chimaeric proteins could not be cleaved even if 1% Triton X-100 was added to the reaction mixture, a condition where the interaction with Lip1p should not be maintained [10], we speculate that the region around the Lag motif is not accessible to fXa protease cleavage because of the structure of this region. Since N-glycosylation can occur co-translationally and thus prior to complete folding, the fusion constructs in which the glycosylation cassettes were inserted at the position Thr272 could be glycosylated, even though the regions will be tightly packed after their translocation and folding. Similar behaviour has been reported for Sec61p and Lcb1p; not every site where an fXa cleavage site was inserted was protease-sensitive [20,25]. We propose that the region around the Lag motif is highly structured and that most of the residues are embedded in the membrane.
A variety of programmes designed to identify potential transmembrane regions suggest the presence of four to eight membrane-spanning domains in Lag1p and Lac1p. Most of the programs used to predict potential membrane-spanning domains identified TMDs 1, 2, 3, 4, 5, 7 and 8 of Lag1p and Lac1p (Figure 1B and Table 2). In contrast, TMD 6 of Lag1p and TMD 6 of Lac1p were predicted as TMDs by some programs, but not by others. The TMD 6 was most likely eliminated by some programs due to the fact that it is very close to TMDs 5 and 7. Analysis of the glycosylation state of Lag1–Suc2A and Lac1–Suc2A fusion proteins revealed that Thr272 is on the luminal side and Leu298 is on the cytoplasmic side of the ER membrane, suggesting that the α-helix of TMD 6 is located in the membrane. We also confirmed the existence of TMDs 5 and 6 by using chimaeric constructs that were fused to full-length SUC2 after every predicted loop of LAG1 and LAC1 (N. Kageyama-Yahara and H. Riezman, unpublished work). Full-length Suc2p was fused just after Pro245, Thr272 or Leu298 of Lag1p and Lac1p. These constructs were expressed in cells, treated with or without endo H, and their mobility was examined by Western-blot analysis (results not shown). From these results, Thr272 was located on the opposite side of the membrane from Pro245 and Leu298, suggesting again that TMDs 5 and 6 transverse the ER membrane. One might wonder how the insertion of the Suc2A cassette at Asp286 could give a glycosylated protein, as this residue is predicted to be inside the lipid bilayer in our model. However, its glycosylation is consistent with our model. This construct could be glycosylated because the last ten residues of the Suc2A cassette, PQRCVAIVTY, are sufficiently similar in hydrophobicity to substitute for the ten residues preceding Asp286, LAIYITMDVS. This would leave a luminal loop preceding transmembrane helix 6 of approx. 46 residues in this construct, which is sufficient for recognition by the glycosylation machinery.
Taking together the results obtained from the Suc2A- and fXa-fusion proteins, we conclude that the N-terminus, the loops corresponding to amino acid residues Thr165, Pro245, Leu298 and Gly386, and the C-terminus are on the cytoplasmic side, whereas the loops corresponding to amino acid residues Asn108, Tyr208, Thr272 and Phe333 are on the luminal side of the ER membrane. There is no discordance between the two methods of analysis, and the results of both are most consistent with the presence of eight TMDs in Lag1p and Lac1p (Figure 6). At least one of the constructs confirming the topology at the two termini and positions Asn108, Thr165, Tyr208, Leu298, Phe333 and Gly386 showed complementation activity which allows us to be highly confident in our assignments of TMDs 1, 2, 3, 7 and 8, because this ensures that the protein is inserted into the membrane with its normal topology. Taking into account that the number of TMDs must be an even number to have both termini exposed to the cytoplasm, we are also highly confident in our assignment of TMD 4. On the other hand, and not surprisingly, insertions near or in the conserved Lag motif, Pro245, Thr272 and Asp286 always inactivated the complementing activity of our constructs. Our results are consistent with the presence of two additional TMDs, TMDs 5 and 6, but since our constructs are inactive these results can only be interpreted as suggestive. An alternative topology that would, however, not be consistent with our mapping results would be that both of these hydrophobic regions form a tightly folded structure on the cytoplasmic side of the ER membrane.
The eight transmembrane helices and their orientation in Lag1p and Lac1p are similar to the topological model proposed for TRAM. TRAM shows a significant homology with Lag1p (24% identical and 42% similar over 202 amino acids) [11]. TRAM has been proposed to be stimulatory or required for the process of membrane translocation of secretory proteins [26–28]. Since TRAM is likely to enter into direct contact with translocating nascent chains and is present in ER membranes in amounts at least equivalent to membrane-bound ribosomes, it seems to be a component of a translocation complex. TRAM may form or contribute to a hydrophilic environment through or along which polypeptides can be transported [26]. Due to the predicted structural similarity and our results, we would predict that the required histidine and aspartic acid residues in the Lag motif are embedded in the lipid bilayer (Figure 6), suggesting that Lag1p and Lac1p might form a hydrophilic ‘channel’ in the membrane.
In spite of the similarity in amino acid sequences and structures, human TRAM does not complement the yeast LAG1 gene and the TRAM protein does not contain a complete Lag motif [11]. Furthermore, no translocation defects were detected in lag1Δlac1Δ yeast cells [24]. These results suggest that their functions are not conserved, even though they seem to have some structural homology. A multiple sequence alignment of Lag1 family and TRAM family shows that some amino acids are conserved only in Lag1 family but not in the TRAM family. From point mutational analysis, we showed that histidine residues at positions 255 and 256 and aspartic acid residues at the positions 283 and 286 are critical for the function of Lag1p in vivo. These amino acids are in the Lag motif, which could potentially be the active site, and the predictions from our topology results are that they are embedded in the membrane (Figure 6). Since histidine and aspartic acid residues are charged amino acids, they might be important for the interaction with sphinganine or fatty acyl-CoA, the substrates of ceramide synthase. It would be very interesting to determine if the ceramide synthase forms a channel-like structure. If so, this could explain how it might be able to accept sphinganine from either side of the ER membrane, which would be consistent with data on the topology of two enzymes that can create this substrate, the 3-ketosphinganine dehydrogenase [13] and sphingosine 1-phosphate phosphatase [14]. It might also begin to help to explain how the enzyme can perform a reaction with the hydrophilic ends of sphinganine and fatty acyl-CoA and at the same time be able to recognize the length of the hydrocarbon chain of the fatty acid. Future experiments will hopefully lead to a high-resolution structure of the ceramide–synthase complex and direct tests of the presence of a hydrophilic ‘channel’ in the enzyme.
Acknowledgments
This work was supported by the Uehara Memorial Foundation (N.K.-Y.), the Nakatomi Foundation (N.K.-Y.), the Swiss National Science Foundation (H.R.) and the Human Frontiers Science Program Organization (H.R.). We are grateful to Beatrice Vallée (Center for Molecular Biophysics, CNRS, Orleans, France) and members of the Riezman laboratory for helpful discussions.
References
- 1.Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry D. K., Hannun Y. A. The role of ceramide in cell signaling. Biochim. Biophys. Acta. 1998;1436:233–243. doi: 10.1016/s0005-2760(98)00145-3. [DOI] [PubMed] [Google Scholar]
- 3.Hannun Y. A., Luberto C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- 4.Hannun Y. A., Obeid L. M. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 5.Acharya U., Patel S., Koundakjian E., Nagashima K., Han X., Acharya J. K. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science. 2003;299:1740–1743. doi: 10.1126/science.1080549. [DOI] [PubMed] [Google Scholar]
- 6.Morell P., Radin N. S. Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A's by brain microsomes. J. Biol. Chem. 1970;245:342–350. [PubMed] [Google Scholar]
- 7.Akanuma H., Kishimoto Y. Synthesis of ceramides and cerebrosides containing both alpha-hydroxy and nonhydroxy fatty acids from lignoceroyl-CoA by rat brain microsomes. J. Biol. Chem. 1979;254:1050–1060. [PubMed] [Google Scholar]
- 8.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallee B., Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J. C., Kirchman P. A., Zagulski M., Hunt J., Jazwinski S. M. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 12.Mandon E., Ehses I., Rother J., van Echten G., Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- 13.Kihara A., Igarashi Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 2004;279:49243–49250. doi: 10.1074/jbc.M405915200. [DOI] [PubMed] [Google Scholar]
- 14.Kihara A., Sano T., Iwaki S., Igarashi Y. Transmembrane topology of sphingoid long-chain base-1-phosphate phosphatase, Lcb3p. Genes Cells. 2003;8:525–535. doi: 10.1046/j.1365-2443.2003.00653.x. [DOI] [PubMed] [Google Scholar]
- 15.Funakoshi T., Yasuda S., Fukasawa M., Nishijima M., Hanada K. Reconstitution of ATP- and cytosol-dependent transport of de novo synthesized ceramide to the site of sphingomyelin synthesis in semi-intact cells. J. Biol. Chem. 2000;275:29938–29945. doi: 10.1074/jbc.M004470200. [DOI] [PubMed] [Google Scholar]
- 16.Funato K., Riezman H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 2001;155:949–959. doi: 10.1083/jcb.200105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine T. P., Wiggins C. A. R., Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschberg K., Rodger J., Futerman A. H. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 1993;290:751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilstring C. F., Ljungdahl P. O. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J. Biol. Chem. 2000;275:31488–31495. doi: 10.1074/jbc.M005047200. [DOI] [PubMed] [Google Scholar]
- 20.Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 2004;279:53707–53716. doi: 10.1074/jbc.M410014200. [DOI] [PubMed] [Google Scholar]
- 21.Kota J., Ljungdahl P. O. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 2005;168:79–88. doi: 10.1083/jcb.200408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H., von Heijne G., Nilsson I. Membrane topology of the STT3 subunit of the oligosaccharyl transferase complex. J. Biol. Chem. 2005;280:20261–20267. doi: 10.1074/jbc.M412213200. [DOI] [PubMed] [Google Scholar]
- 23.Mizutani Y., Kihara A., Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barz W. P., Walter P. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 1999;10:1043–1059. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson B. M., Critchley A. J., Stirling C. J. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J. Biol. Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- 26.Gorlich D., Hartmann E., Prehn S., Rapoport T. A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature (London) 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich D., Rapoport T. A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 28.Voigt S., Jungnickel B., Hartmann E., Rapoport T. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 31.Jones D., Taylor W., Thornton J. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 32.Tusnady G. E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann K., Stoffel W. TMbase – a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]