Abstract
The mechanism of hyaluronan biosynthesis in vertebrates had been proposed to occur at the reducing end of growing chains. This mechanism was questioned because a recombinant synthase appeared to add new monosaccharides to the non-reducing end. I reinvestigated this problem with membranes from the eukaryotic B6 cell line. The membranes were incubated with UDP-[3H]GlcNAc and UDP-[14C]GlcA to yield differentially labelled reducing terminal and non-reducing terminal domains. Digestion of the product with a mixture of the exoglycosidases β-glucuronidase and β-N-acetylglucosaminidase truncated the hyaluronan chain strictly from the non-reducing end. The change in 3H/14C ratio of the remaining hyaluronan fraction, during the course of exoglycosidase digestion, confirmed the original results that the native eukaryotic synthase extended hyaluronan at the reducing end. This mechanism demands that the UDP-hyaluronan terminus is bound to the active site within the synthase and should compete with the substrates for binding. Accordingly, increasing substrate concentrations enhanced hyaluronan release from the synthase. A model is proposed that explains the direction of chain elongation at the reducing end by the native synthase and at the non-reducing end by the recombinant synthase based on a loss of binding affinity of the synthase towards the growing UDP-hyaluronan chain.
Keywords: chain elongation, eukaryotic cell line, hyaluronan biosynthesis, mechanism, membrane, recombinant synthase
INTRODUCTION
Hyaluronan is synthesized, without any primer requirement, by alternate addition of D-glucuronic acid and N-acetyl-D-glucosamine units from the corresponding UDP-sugar donors (UDP-GlcA and UDP-GlcNAc) [1,2]. The growing chains are exported from streptococci and from eukaryotic cells by ABC transporters (ATP-binding-cassette transporters). Transport can be inhibited by inhibitors of multidrug resistance transporters [3–6]. Hyaluronan synthases have been tentatively classified based on primary structure, molecular mass, predicted membrane topology and enzymological characteristics [7]. Class I thus includes the streptococcal, the vertebrate and the viral enzymes, whereas Class II contains a single member, the Pasteurella multocida enzyme.
In 1983, I proposed that hyaluronan chains are elongated at the reducing end, because the digestion kinetics of pulse–chase-labelled chains with exoglycosidases immediately reduced the incorporated radioactivity that was introduced into the chains during the first pulse reaction [2]. These experiments were confirmed by Asplund et al. [8] with a human glioma cell line and by Tlapak-Simmons et al. [9] with streptococci. Both of these native enzyme systems failed to further extend preformed, exogenously added hyaluronan oligosaccharides. In contrast, the structurally unrelated synthase from P. multocida synthesizes hyaluronan at the non-reducing end [10]. Uncertainty about the direction of hyaluronan synthesis in eukaryotic cells arose, because a recombinant hyaluronan synthase from Xenopus laevis expressed in yeast added new sugar residues to the non-reducing end [11]. In addition, a recombinant synthase was able to elongate hyaluronan oligosaccharides, and this should not occur if synthesis proceeded at the reducing end [12]. Because of these contradictory results, I decided to reinvestigate the directionality of hyaluronan chain elongation catalysed by native eukaryotic hyaluronan synthase applying the double labelling method of Bodevin-Authelet et al. [11]. The results confirmed that the native eukaryotic synthase added new sugars at the reducing end.
The results also shed light on the factors that determine chain length. I previously demonstrated that hyaluronan is liberated from the synthase by dissociation [13]. If nascent hyaluronan is degraded, the synthase elongates it to a size that is determined by thermodynamic factors [14,15]. It is an unexplained observation that the three eukaryotic hyaluronan synthases produce hyaluronan with different chain lengths [16]. My results suggest that other determinants for chain length are the binding affinity of the synthase to the reducing hyaluronan chain terminus and the concentrations of the nucleotide sugar precursors that compete with the growing hyaluronan chain for the active site in the enzyme.
EXPERIMENTAL
Materials
Radiolabelled UDP-[14C]GlcA [specific radioactivity 0.196 Ci/mmol (2.40×1011 c.p.m./mmol)] was obtained from Amersham Biosciences, and UDP-[3H]GlcNAc (specific radioactivity 38.5 Ci/mmol) from PerkinElmer Life Sciences. For preparation of the substrate solution, 50 μCi of UDP-[3H]GlcNAc was mixed with 132 μg of unlabelled UDP-GlcNAc and diluted to 20 ml for the substrate solution to achieve a final concentration of 10 μM and a specific radioactivity of 0.1 Ci/mmol (4.74×1010 c.p.m./mmol). Additional chemicals were from Sigma–Aldrich Chemical.
Isolation of plasma membranes
B6 cells, a hybrid cell line derived from mouse mammary carcinoma, and a Chinese hamster lung cell line which produced large amounts of hyaluronan [17,18] were grown in suspension culture in Dulbecco's modified Eagle's medium supplemented with streptomycin/penicillin (100 units of each/ml), kanamycin (100 units/ml) and 10% (v/v) foetal calf serum. A suspension culture was grown to a density of 2×105/ml in four culture flasks (180 cm2 surface area). Hyaluronan synthesis was stimulated, 4 h before cell harvest, by supplementation of additional foetal calf serum (5%). The cells were sedimented at 1500 g for 5 min, washed with PBS and suspended in 30 ml of ice-cold PBS [3]. The cells were transferred into a Parr-bomb, exposed to a nitrogen pressure of 900 lbf/in2 (1 lbf/in2=6.9 kPa) for 15 min and disrupted by nitrogen cavitation [19]. Nuclei were sedimented at 2000 g for 10 min and the particulate fraction was obtained by centrifugation at 20000 g for 20 min.
Hyaluronan synthesis and isolation
For synthesis of [3H]hyaluronan, the membrane fraction was suspended in 1 ml of prewarmed substrate for hyaluronan synthesis that contained 8 μM UDP-GlcA, 10 μM UDP-[3H]GlcNAc, 4 mM dithiothreitol and 20 mM MgCl2 in 50 mM Tris/malonate (pH 7.0) and incubated again at 37 °C for 30 min. The membranes were sedimented by centrifugation for 5 min at 14000 g. An aliquot (1/3) was solubilized in 1% SDS and heated at 90 °C for 5 min. Undissolved material was sedimented by centrifugation for 5 min at 14000 g and the supernatant was applied on to a gel filtration Sephacryl S-1000 column (1.4 cm×64 cm) with 10 mM NH4HCO3 as eluent at a flow rate of 14 ml/h. Fractions of 2 ml were collected and the radioactivity of 100 μl was determined.
For synthesis of dual-labelled hyaluronan, the rest of the membrane fraction (2/3) was sedimented and suspended in 1 ml of prewarmed substrate for hyaluronan synthesis that contained 8 μM UDP-[14C]GlcA, 150 μM UDP-GlcNAc, 4 mM dithiothreitol and 20 mM MgCl2 in 50 mM Tris/malonate (pH 7.0) and incubated at 37 °C for 15 min. The membrane fraction was again sedimented, solubilized in SDS and applied on to a gel filtration Sephacryl S-1000 column as described above.
Enzymatic degradation of hyaluronan
The dual-labelled hyaluronan fractions from the Sephacryl S-1000 column (fractions 25–35) were collected, freeze-dried to dryness and dissolved in 4 ml of 0.1 M sodium acetate (pH 5.2). The enzymes β-N-acetylglucosaminidase (0.8 unit) and β-glucuronidase (10000 units) were added and incubated in four aliquots in dialysis bags at 37 °C and simultaneously dialysed against 5 litres of 10 mM sodium acetate (pH 5.2) for time periods as indicated in Table 1. After different time periods, the radioactivity of the remaining [3H/14C]hyaluronan in the dialysis bags was determined.
Table 1. Digestion of dual-labelled hyaluronan with exoglycosidases.
| Digestion period (h) | 3H (c.p.m.) | 14C (c.p.m.) | 3H/14C ratio |
|---|---|---|---|
| Experiment 1 | |||
| 0 | 7162 | 14216 | 0.50 |
| 3 | 4281 | 10157 | 0.42 |
| 9 | 2076 | 6498 | 0.32 |
| 27 | 1606 | 5780 | 0.28 |
| Experiment 2 | |||
| 0 | 3725 | 10560 | 0.35 |
| 3 | 2799 | 9512 | 0.29 |
| 8 | 1434 | 5264 | 0.27 |
| 24 | 1461 | 5616 | 0.26 |
RESULTS
Synthesis of dual-labelled hyaluronan
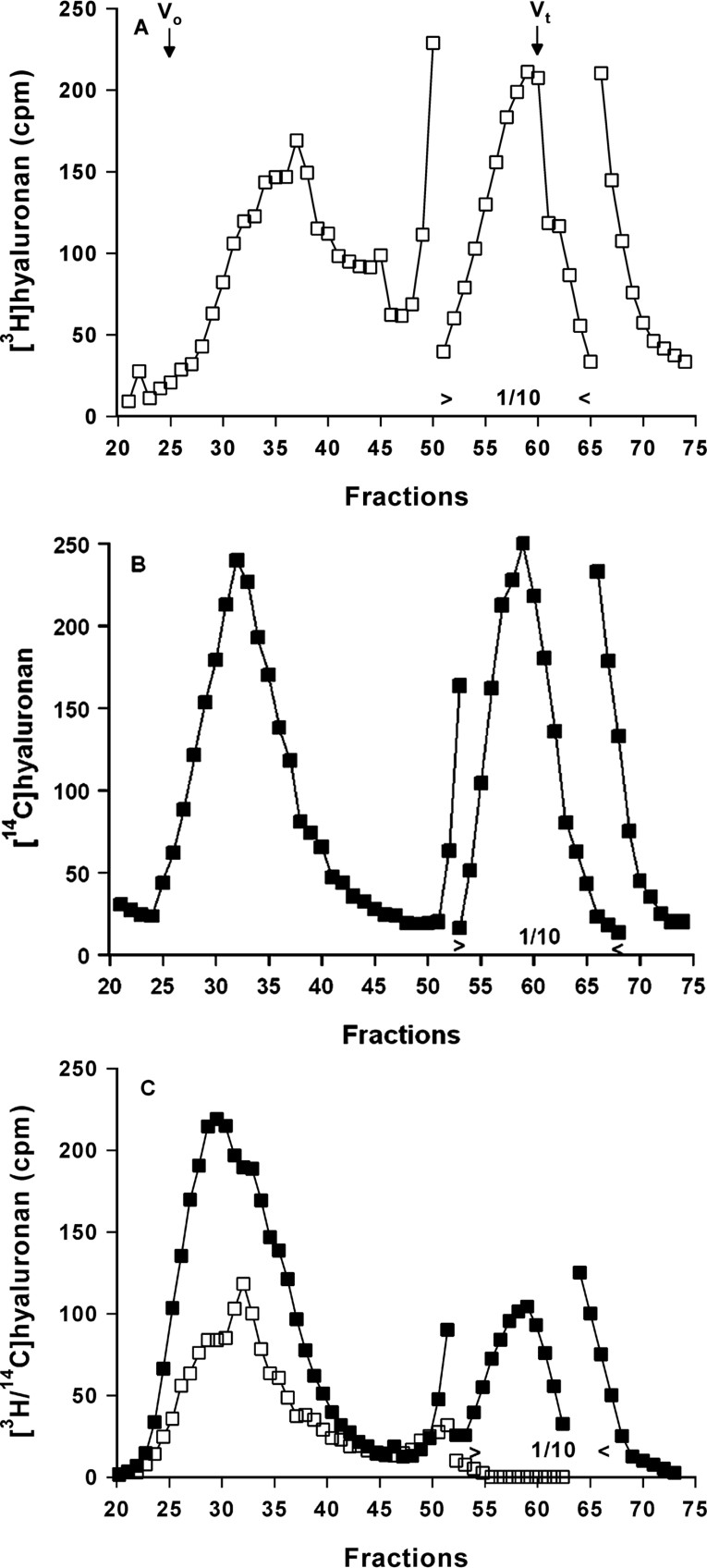
The eukaryotic hyaluronan synthase has a rather high KM value for UDP-GlcNAc (∼60 μM) and thus high substrate concentrations are required to obtain sufficient chain elongation. On the other hand, large amounts of UDP-[3H]GlcNAc with high specific radioactivity impede the purification of [3H/14C]hyaluronan. Therefore a series of preliminary experiments were performed to evaluate the optimal experimental conditions to produce dual-labelled hyaluronan chains by incubation of membranes from the eukaryotic B6 cell line with different substrate concentrations, specific radioactivities and synthesis periods. A compromise was found that produced both a sufficient chain elongation rate and a measurable [3H]hyaluronan radioactivity. Hyaluronan was first labelled by incubation of membranes with UDP-[3H]GlcNAc at a concentration of 10 μM and unlabelled 8 μM UDP-GlcA for 30 min at 37 °C. The [3H]hyaluronan was analysed by gel filtration using Sephacryl S-1000. Figure 1(A) shows that most of the [3H]hyaluronan was included in the middle of the column eluate. In a second experiment, the membranes were incubated with a substrate solution containing 8 μM UDP-[14C]GlcA and unlabelled 150 μM UDP-GlcNAc for 15 min and subjected again to gel filtration. Figure 1(B) shows that this labelling condition yielded a size of hyaluronan that was also included in the column eluate. In the third experiment, the two incubation conditions were utilized sequentially and the size of [3H/14C]hyaluronan was again analysed by gel filtration. Figure 1(C) shows that [3H]hyaluronan was further elongated by [14C]hyaluronan. For the subsequent exoglycosidase digestion, the [3H/14C]hyaluronan-containing fractions were isolated.
Figure 1. Synthesis of dual-labelled hyaluronan.
(A) A membrane fraction of B6 cells was incubated with UDP-[3H]GlcNAc and unlabelled UDP-GlcA for 30 min at 37 °C. The membranes were solubilized in 1% SDS and subjected to gel filtration on a Sephacryl S-1000 column (1.4 cm×64 cm). Fractions of 2 ml were collected and analysed for 3H (□) radioactivity. The scale of the fractions 51–65 containing the substrate was reduced by a factor of 10. (B) The membranes were incubated with UDP-[14C]GlcA and unlabelled UDP-GlcNAc for 15 min at 37 °C and analysed as in (A) for 14C (■) radioactivity. (C) The membranes were first incubated with UDP-[3H]GlcNAc and unlabelled UDP-GlcA for 30 min at 37 °C and then with UDP-[14C]GlcA and unlabelled UDP-GlcNAc for 15 min and analysed as in (A) for 3H (□) and 14C (■) radioactivity. Fractions 25–35 were combined for subsequent digestion.
Exoglycosidase digestion of dual-labelled hyaluronan
The dual-labelled hyaluronan of high molecular mass was subjected to graded exoglycosidase (β-N-acetylglucosaminidase and β-glucuronidase) digestion as described in the Experimental section. The digestion was performed with simultaneous dialysis. Aliquots in the dialysis bags were withdrawn at various time periods to determine the residual radioactivity.
The results are shown in Table 1. The radioactivity of the dual-labelled hyaluronan decreased continuously. The ratio of 3H/14C decreased during digestion. This result was verified by two independent experiments. It indicated that the [3H]hyaluronan introduced first into the growing chain was digested faster. Consequently, most of the hyaluronan chains must have been elongated at the reducing end.
Effect of substrate concentration on hyaluronan release from the synthase
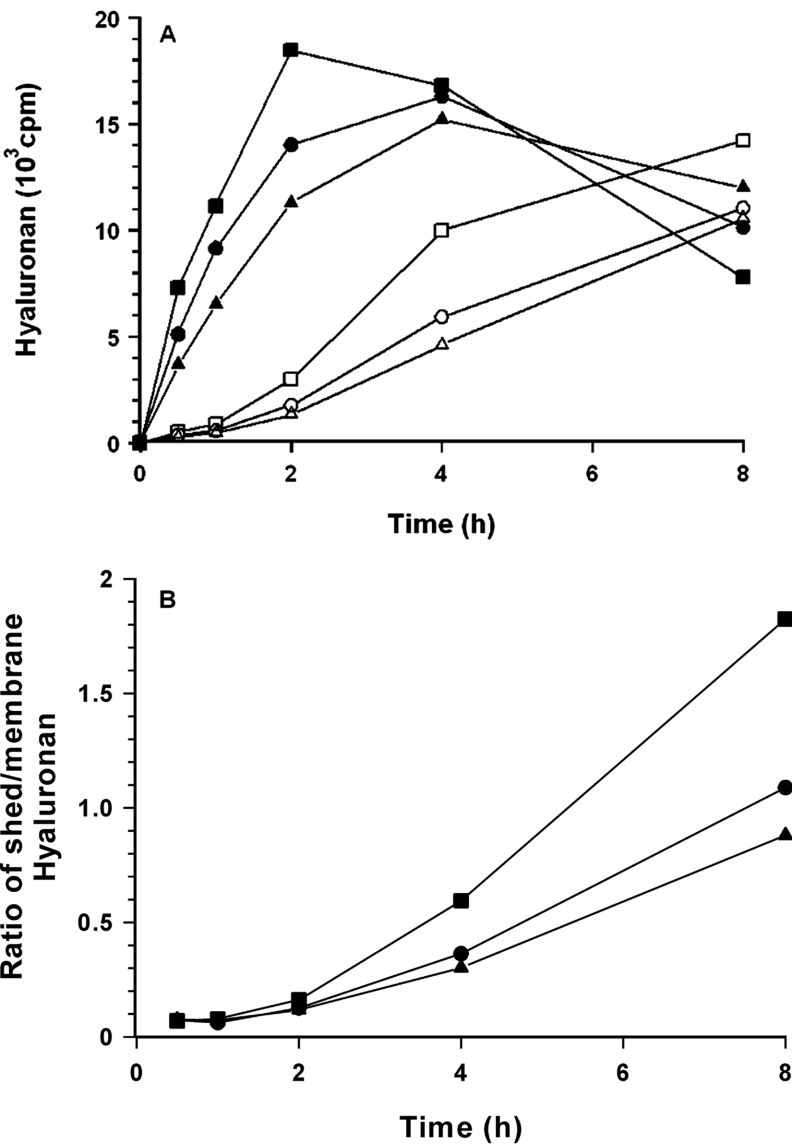
If the terminal UDP-hyaluronan is bound to the synthase during elongation at the sites where the substrates also bind, the concentration of the substrates should influence not only the rate of elongation, but also the rate of chain release. Therefore membranes from B6 cells were incubated with different concentrations of substrates and the release of hyaluronan from the membranes into the soluble fractions was determined. Figure 2(A) shows that hyaluronan was first incorporated into the membrane fraction and then released. The proportion of soluble hyaluronan increased with the substrate concentrations (Figure 2B). Similar results were obtained when only the concentration of substrate was altered (results not shown). Thus the substrates appeared to compete with UDP-hyaluronan for binding to the synthase.
Figure 2. Release of hyaluronan from the membrane-bound synthase.
Hyaluronan was synthesized by B6 membranes in the presence of a substrate containing 8 μM UDP-[14C]GlcA and unlabelled 166 μM UDP-GlcNAc (■, □) or a substrate diluted 1:2 (●, ○) or 1:3 (▲, △) with a buffer (4 mM dithiothreitol and 20 mM MgCl2 in 50 mM Tris-malonate). At the times indicated, the membranes were sedimented for 3 min at 14000 g, dissolved in 1% SDS and applied to descending paper chromatography (solid symbols). The supernatants (open symbols) were applied directly [1]. The origins containing hyaluronan were excised and the radioactivities were determined (A). From these data, the ratios of released hyaluronan and membrane-bound hyaluronan were calculated (B).
DISCUSSION
It would be desirable to obtain a homogeneous preparation of double labelled hyaluronan for experimental proof of the direction of hyaluronan chain elongation by exoglycosidase digestion. However, I have shown previously that continuous new chain initiation occurred during hyaluronan synthesis [1], and thus conclusions can only be derived from enriched hyaluronan preparations in our study on the native enzyme as well as on the recombinant enzyme by Bodevin-Authelet et al. [11]. Therefore the double labelled hyaluronan preparation used for exoglycosidase digestion was heterogeneous and its size overlapped with the single labelled hyaluronan. But most of the first labelled [3H]hyaluronan increased in molecular mass during the second incubation and co-eluted with the second 14C-labelled hyaluronan, indicating that it had been further elongated to some extent. This heterogeneity is reflected in the simultaneous release of both labels during exoglycosidase digestion, and conclusions are only based on differences of release rates. In contrast with the study of Bodevin-Authelet et al. [11] on recombinant hyaluronan synthase, I found that hyaluronan was elongated at the reducing end by the native synthase, confirming the original mechanism [1,2].
It is surprising that a recombinant eukaryotic hyaluronan synthase expressed in yeast synthesizes hyaluronan at the non-reducing end [11], because the structural homology between streptococcal and vertebrate synthases should reflect similar mechanisms, as opposed to the synthase from Pasteurella. This discrepancy calls for an explanation. It can be found in a detailed consideration of the experimental conditions and the probable mechanism of enzymatic reactions.
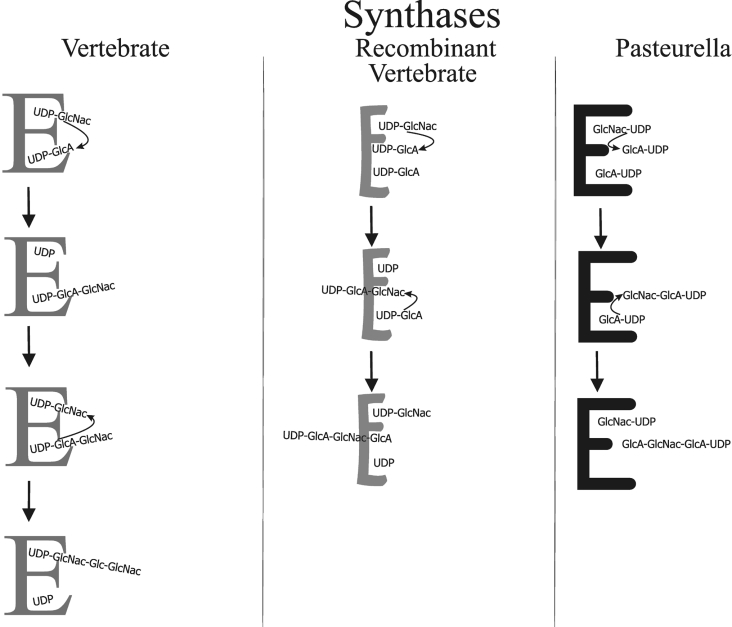
It is obvious that the two substrates UDP-GlcA and UDP-GlcNAc must bind to two active sites within the enzyme. Since no other molecule is required for chain initiation and elongation, the growing UDP-hyaluronan shuttles from one substrate to the other during elongation, as illustrated in Figure 3. Polymerization at the reducing end is therefore dependent on a high affinity of the growing UDP-hyaluronan for both active sites. It is likely that the binding affinities of UDP-hyaluronan to these sites are proportional to those for the two substrates. The binding affinity of UDP-GlcA and UDP-GlcNAc is reflected by the KM values which have been measured for both the native and the recombinant synthases. Table 2 demonstrates that the recombinant synthase has dramatically reduced affinities for both substrates. Even detergent solubilization appeared to decrease binding affinity. It is thus likely that UDP-hyaluronan that binds to the same sites has also reduced affinity for the synthase. An additional component that holds the growing hyaluronan chain close to the synthase in the native membrane is the hyaluronan exporter, because it has been demonstrated that hyaluronan export and synthesis are closely associated in eukaryotic plasma membranes [5] and that growing hyaluronan chains exert feedback inhibition on the synthase [14,15]. In addition, the exporter is also absent from the recombinant enzyme preparations.
Figure 3. Transfer reactions for hyaluronan synthesis at the reducing and non-reducing ends.
Left-hand column, mechanism for the native synthase. After dissaccharide formation, high-affinity binding of UDP-hyaluronan to the active sites of the native enzyme E isolated from vertebrate cells ensures that elongation proceeds at the reducing end. Middle column, mechanism for the recombinant synthase. Reduced binding affinity to the recombinant enzyme E leads to transfer of monosaccharides to soluble substrates at the non-reducing end. Right-hand column, mechanism for the synthase from Pasteurella.
Table 2. Comparison of published Michaelis–Menten constants foxr native and recombinant eukaryotic hyaluronan synthases.
| Km for UDP-GlcA (μM) | Km for UDP-GlcNAc (μM) | Vmax for UDP-GlcA (nmol·h−1·mg−1) | Vmax for UDP-GlcNAc (nmol·h−1·mg−1) | Reference | |
|---|---|---|---|---|---|
| Native chick fibroblasts | 20 | 50 | 10 | 13 | [22] |
| Native human fibroblasts | 3 | 21 | 70 | 70 | [23] |
| Native human fibroblasts | 1 | 12 | 6 | 7 | [24] |
| Native mouse fibroblasts | 10.8 | 58.4 | 7 | 1 | [25] |
| Detergent-solubilized native mouse oligodendroglioma | 50 | 100 | [26] | ||
| Recombinant Xenopus hyaluronan synthase DG42 | 60 | 235 | [27] | ||
| Recombinant mouse hyaluronan synthase HAS1 | 60 | 950 | 2772 | 3090 | [28] |
The possible scenario for synthesis at the non-reducing end by the recombinant synthase is illustrated in Figure 3. At the high substrate concentrations that were used for the assessment of growth direction of the recombinant synthase (150 μM UDP-GlcNAc and 50 μM UDP-GlcA) [11], the active sites will be constantly occupied by substrates. Any initially formed UDP-hyaluronan will be ousted from the synthase. However, the enzymatic activities for the transferase reactions remain the same or are even increased, as indicated by the increased Vmax values. Thus the synthase tries to find targets for transfer of monosaccharides from the nucleotide sugars. The only abundant targets are soluble nucleotide sugars. It is thus likely that the synthase transfers the monosaccharides to soluble substrate, instead of transferring bound UDP-hyaluronan. In fact, Bodevin-Authelet et al. [11] only analysed soluble hyaluronan after precipitation of membrane proteins with trichloroacetic acid and, unfortunately, did not indicate whether there existed any membrane-bound hyaluronan [11]. Such a trichloroacetic acid precipitation has previously been shown to precipitate most of the newly synthesized hyaluronan [20], especially after the short periods of chain growth that were used for determination of direction of elongation.
The chemical reaction is virtually identical with the initial step of polymerization that results in the first disaccharide unit for elongation at the reducing end. But now elongation proceeds at the non-reducing end. The only difference is the transferred moiety: UDP-hyaluronan to the nucleotide sugars for the synthesis at the reducing end and the nucleotide sugars to hyaluronan for the synthesis at the non-reducing end. This may also explain the failure of Bodevin-Authelet et al. [11] to chase a pulse-labelled hyaluronan chain. In contrast, a chase was demonstrated previously for the natural eukaryotic synthase [21] and again in the present paper (Figure 1).
In accord with hyaluronan chain elongation at the reducing end were results suggesting that nucleotide sugars should compete with UDP-hyaluronan for binding to the enzyme, and high concentrations increased shedding and reduced the chain length. Thus intracellular concentrations of the nucleotide sugars and the affinity of the growing UDP-hyaluronan chain for the enzyme determine the dissociation rate.
Acknowledgments
I thank Dr R. Pohlmann (Münster, Germany) and Dr R. Stern (San Francisco, CA, U.S.A.) for a critical review of this paper, and U. Rasmussen and R. Schulz for excellent technical assistance.
References
- 1.Prehm P. Synthesis of hyaluronate in differentiated teratocarcinoma cells: characterization of the synthase. Biochem. J. 1983;211:181–189. doi: 10.1042/bj2110181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prehm P. Synthesis of hyaluronate in differentiated teratocarcinoma cells: mechanism of chain growth. Biochem. J. 1983;211:191–198. doi: 10.1042/bj2110191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prehm P. Hyaluronate is synthesized at plasma membranes. Biochem. J. 1984;220:597–600. doi: 10.1042/bj2200597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouskova G., Spellerberg B., Prehm P. Hyaluronan release from Streptococcus pyogenes: export by an ABC transporter. Glycobiology. 2004;14:931–938. doi: 10.1093/glycob/cwh115. [DOI] [PubMed] [Google Scholar]
- 5.Prehm P., Schumacher U. Inhibition of hyaluronan export from human fibroblasts by inhibitors of multidrug resistance transporters. Biochem. Pharmacol. 2004;68:1401–1410. doi: 10.1016/j.bcp.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Prehm P. Inhibitors of hyaluronan export prevent proteoglycan loss from osteoarthritic cartilage. J. Rheumatol. 2005;32:690–696. [PubMed] [Google Scholar]
- 7.DeAngelis P. L. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pathogens, and algal viruses. Cell. Mol. Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asplund T., Brinck J., Suzuki M., Briskin M. J., Heldin P. Characterization of hyaluronan synthase from a human glioma cell line. Biochim. Biophys. Acta. 1998;1380:377–388. doi: 10.1016/s0304-4165(98)00010-5. [DOI] [PubMed] [Google Scholar]
- 9.Tlapak-Simmons V. L., Baron C. A., Gotschall R., Haque D., Canfield W. M., Weigel P. H. Hyaluronan biosynthesis by class I streptococcal hyaluronan synthases occurs at the reducing end. J. Biol. Chem. 2005;280:13012–13018. doi: 10.1074/jbc.M409788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeAngelis P. L. Molecular directionality of polysaccharide polymerization by the Pasteurella multocida hyaluronan synthase. J. Biol. Chem. 1999;274:26557–26562. doi: 10.1074/jbc.274.37.26557. [DOI] [PubMed] [Google Scholar]
- 11.Bodevin-Authelet S., Kusche-Gullberg M., Pummill P. E., DeAngelis P. L., Lindahl U. Biosynthesis of hyaluronan: direction of chain elongation. J. Biol. Chem. 2005;280:8813–8818. doi: 10.1074/jbc.M412803200. [DOI] [PubMed] [Google Scholar]
- 12.Hoshi H., Nakagawa H., Nishiguchi S., Iwata K., Niikura K., Monde K., Nishimura S. An engineered hyaluronan synthase: characterization for recombinant human hyaluronan synthase 2 Escherichia coli. J. Biol. Chem. 2004;279:2341–2349. doi: 10.1074/jbc.M305723200. [DOI] [PubMed] [Google Scholar]
- 13.Prehm P. Release of hyaluronate from eukaryotic cells. Biochem. J. 1990;267:185–189. doi: 10.1042/bj2670185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nickel V., Prehm S., Lansing M., Mausolf A., Podbielski A., Deutscher J., Prehm P. An ectoprotein kinase of group C streptococci binds hyaluronan and regulates capsule formation. J. Biol. Chem. 1998;273:23668–23673. doi: 10.1074/jbc.273.37.23668. [DOI] [PubMed] [Google Scholar]
- 15.Lüke H. J., Prehm P. Synthesis and shedding of hyaluronan from plasma membranes of human fibroblasts and metastatic and non-metastatic melanoma cells. Biochem. J. 1999;343:71–75. [PMC free article] [PubMed] [Google Scholar]
- 16.Itano N., Sawai T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J. Biol. Chem. 1999;274:25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 17.Koyama H., Ono T. Initiation of a differentiated function (hyaluronic acid synthesis) by hybrid formation in culture. Biochim. Biophys. Acta. 1970;217:477–487. doi: 10.1016/0005-2787(70)90545-9. [DOI] [PubMed] [Google Scholar]
- 18.Koyama H., Yatabe I., Ono T. Isolation and characterization of hybrids between mouse and Chinese hamster cell lines. Exp. Cell Res. 1970;62:455–463. doi: 10.1016/0014-4827(70)90577-x. [DOI] [PubMed] [Google Scholar]
- 19.Klempner M. S., Mikkelsen R. B., Corfman D. H., Andre S. J. Neutrophil plasma membranes. I. High-yield purification of human neutrophil plasma membrane vesicles by nitrogen cavitation and differential centrifugation. J. Cell Biol. 1980;86:21–28. doi: 10.1083/jcb.86.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugahara K., Schwartz N. B., Dorfman A. Biosynthesis of hyaluronic acid by Streptococcus. J. Biol. Chem. 1979;254:6252–6261. [PubMed] [Google Scholar]
- 21.Prehm P. Inhibition of hyaluronate synthesis. Biochem. J. 1985;225:699–705. doi: 10.1042/bj2250699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimoto N., Temin H. M., Strominger J. L. Studies of carcinogenesis by avian sarcoma viruses. II. Virus-induced increase in hyaluronic acid synthetase in chicken fibroblasts. J. Biol. Chem. 1966;241:2052–2057. [PubMed] [Google Scholar]
- 23.Appel A., Horwitz A. L., Dorfman A. Cell-free synthesis of hyaluronic acid in Marfan syndrome. J. Biol. Chem. 1979;254:12199–12203. [PubMed] [Google Scholar]
- 24.Mian N. Analysis of cell-growth-phase-related variations in hyaluronate synthase activity of isolated plasma-membrane fractions of cultured human skin fibroblasts. Biochem. J. 1986;237:333–342. doi: 10.1042/bj2370333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinowski N. M., Cysyk R. L., August E. M. A filter paper assay for hyaluronic acid synthetase: application to the enzyme from Swiss 3T3 fibroblasts. Biochem. Mol. Biol. Int. 1995;35:1123–1132. [PubMed] [Google Scholar]
- 26.Ng K. F., Schwartz N. B. Solubilization and partial purification of hyaluronate synthetase from oligodendroglioma cells. J. Biol. Chem. 1989;264:11776–11783. [PubMed] [Google Scholar]
- 27.Pummill P. E., Achyuthan A. M., DeAngelis P. L. Enzymological characterization of recombinant Xenopus DG42, a vertebrate hyaluronan synthase. J. Biol. Chem. 1998;273:4976–4981. doi: 10.1074/jbc.273.9.4976. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida M., Itano N., Yamada Y., Kimata K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J. Biol. Chem. 2000;275:497–506. doi: 10.1074/jbc.275.1.497. [DOI] [PubMed] [Google Scholar]