Abstract
The ESE (epithelium-specific Ets) subfamily of Ets transcription factors plays an important role in regulating gene expression in a variety of epithelial cell types. Although ESE proteins have been shown to bind to regulatory elements of some epithelial genes, the optimal DNA-binding sequence has not been experimentally ascertained for any member of the ESE subfamily of transcription factors. This has made the identification and validation of their targets difficult. We are studying ESE-2 (Elf5), which is highly expressed in epithelial cells of many tissues including skin keratinocytes. Here, we identify the preferred DNA-binding site of ESE-2 by performing CASTing (cyclic amplification and selection of targets) experiments. Our analysis shows that the optimal ESE-2 consensus motif consists of a GGA core and an AT-rich 5′- and 3′-flanking sequences. Mutational and competition experiments demonstrate that the flanking sequences that confer high DNA-binding affinity for ESE-2 show considerable differences from the known consensus DNA-binding sites of other Ets proteins, thus reinforcing the idea that the flanking sequences may impart recognition specificity for Ets proteins. In addition, we have identified a novel isoform of murine ESE-2, ESE-2L, that is generated by use of a hitherto unreported new exon and an alternate promoter. Interestingly, transient transfection assays with an optimal ESE-2 responsive reporter show that both ESE-2 and ESE-2L are weak transactivators. However, similar studies utilizing GAL4 chimaeras of ESE-2 demonstrate that while the DNA-binding ETS (E twenty-six) domain functions as a repressor, the PNT (pointed domain) of ESE-2 can act as a potent transcriptional activation domain. This novel transactivating property of PNT is also shared by ESE-3, another ESE family member. Identification of the ESE-2 consensus site and characterization of the transcriptional activation properties of ESE-2 shed new light on its potential as a regulator of target genes.
Keywords: cyclic amplification and selection of targets (CASTing), consensus DNA-binding sequence, epithelium-specific Ets (ESE), keratinocyte, pointed domain (PNT), transcription
Abbreviations: CASTing, cyclic amplification and selection of targets; DBD, DNA-binding domain; EMSA, electrophoretic mobility-shift assay; ETS domain, E twenty-six domain; ESE, epithelium-specific Ets; ESE-2L, the longer isoform of ESE-2; EST, expressed sequence tag; GST, glutathione S-transferase; HA, haemagglutinin; mK, mouse keratinocyte; PNT, pointed domain; RT, reverse transcriptase; tk, thymidine kinase
INTRODUCTION
The Ets transcription factors play a crucial role in transcriptional regulation of genes involved in a variety of developmental and cellular responses including tumorigenesis and differentiation [1,2]. The Ets family is quite large, consisting of at least 26 unique members in mouse, all of which contain an evolutionarily conserved DBD (DNA-binding domain) called the ETS (E twenty-six) domain. Ets proteins are expressed in a wide variety of tissues and organs and comprise some members that are ubiquitously expressed and others that display cell- and tissue-specific expression [3]. For example, Ets proteins such as PU.1 and Spi-B are expressed strongly in haematopoietic cell lineages and haematopoietic organs such as thymus and spleen. On the other hand, several Ets proteins such as Ets1 and Ets2 are widely expressed during development and differentiation of many tissues [4].
Within the Ets family are a subset of proteins that are specifically expressed in epithelial cells of various tissues and organs [5]. These ESE (epithelium-specific Ets) factors consist of ESE-1 (also known as Ert/Jen/Elf3/Esx), ESE-2 (also known as Elf5), ESE-3 (also known as EHF) and PDEF (also known as Pse) [6–10]. ESE-1, ESE-2 and ESE-3 can be clustered together as a distinct subgroup within the Ets family based on phylogenetic considerations of the sequence of their ETS domain. However, using the same criteria, PDEF is phylogenetically distant from the three other members but has been considered to be part of this group because of its epithelial-restricted expression pattern. All four members of the ESE family of proteins share not only sequence similarity in the conserved approx. 85-amino-acid ETS DBD, but also within the N-terminal PNT (pointed domain). Interestingly, the approx. 70% homology in the Ets DBD between ESE factors is relatively moderate compared with other Ets factors such as ELF1 and MEF that share close to 90% homology.
ESE-1 is broadly expressed in organs such as lung, stomach, kidney, colon and skin [11,12]. Particularly high expression of ESE-1 is detected in the small intestine, suggesting an important role for this protein in regulating the morphogenesis and differentiation of small intestinal epithelium. Indeed, this is supported by evidence from targeted inactivation of the ESE-1 gene in mice, which results in severe alterations of the epithelial architecture in the small intestine [13]. In addition, ESE-1 has also been implicated in proliferation and differentiation programmes in other sites such as the corneal epithelial cells and the mammary gland [14,15]. ESE-3 was originally isolated from early stage pituitary somatotroph tumours and is constitutively expressed in the bronchial and mucous gland epithelial cells of lung [16,17]. Other organs including prostate, pancreas, salivary gland and trachea also express ESE-3 [18]. PDEF has been shown to be expressed specifically in prostate glandular epithelial cells and has been shown to be important for prostate-specific antigen gene regulation [9].
ESE-2 displays a more restricted pattern of expression compared with ESE-1 and ESE-3. ESE-2 mRNA has been shown to be present in the tissues rich in glandular or secretory epithelial cells such as mammary gland, salivary gland, kidney and stomach [10,19]. One model system where the expression profile of both ESE-1 and ESE-2 has been examined carefully is skin keratinocytes. Both the genes are up-regulated during keratinocyte differentiation in cell culture, but ESE-2 is clearly induced at a later stage than ESE-1 [8,20]. ESE-2 is also expressed in the extraembryonic ectoderm lineage, which is essential for mammalian placental formation and the survival of the embryo in utero [21]. Embryos with a null mutation in the ESE-2 gene die very early during embryogenesis and exhibit specific defects in the formation of extraembryonic ectoderm [21,22]. Studies on heterozygous ESE-2 mice have shown that ESE-2 is also important for proliferation and differentiation of mammary alveolar epithelial cells during pregnancy and lactation [22].
Although expression analysis and genetic experiments have so far provided some information about the biological functions of ESE proteins, biochemical studies of these proteins have been lacking. Ets proteins regulate gene expression primarily by direct binding to specific regulatory regions, such as promoters and enhancers of target genes. The selectivity of an Ets transcription factor for a specific promoter depends to an extent on the nature of the Ets-binding site. Thus, though all DNA-binding ETS domains recognize a core motif of GGA, different Ets proteins exhibit a preference for different flanking sequences in order to bind differentially to specific DNA sites [23]. For instance, Ets-1 prefers to bind to a 5′-(A/G)CCGGA(A/T)G(T/C)-3′ consensus sequence, whereas PU.1 binds preferentially to 5′-(A/t)(A/t)(A/t)(G/A)(G/A)(G/A)GAA(A/G/C)(C/T)(T/G/A)-3′ [24–26]. Most Ets proteins respond to various signalling pathways, bind to their cognate sites and act as either transcriptional activators or repressors. This function of ETS protein is often mediated by interaction with other transcription factors or through specific domains that recruit co-activators or co-repressors.
Understanding the biochemical properties of Ets proteins is an important first step in deciphering their role in regulation of gene expression. To gain insights into the biochemical properties of the ESE subfamily, here we have performed extensive studies on ESE-2. First, we performed CASTing (cyclic amplification and selection of targets) experiments to determine the optimum DNA-binding sequence of ESE-2. Our studies show that optimum DNA binding by ESE-2 requires a GGA core and an AT-rich 5′- and 3′-flanking sequence. The flanking sequences that confer high DNA-binding affinity for ESE-2 are important and show considerable differences from the known consensus DNA-binding sites of other Ets proteins. Secondly, we have identified a novel isoform of ESE-2, ESE-2L, that is generated by alternate promoter usage and contains an additional 31 N-terminal amino acids. Finally, using reporter assays in keratinocytes, we have dissected the transactivation properties of ESE-2 and ESE-2L. These studies have led to the identification of the domains of ESE-2 that are likely to be critical in mediating the capacity of ESE-2 to transactivate differentiation-specific genes in keratinocytes and other epithelial cell types.
EXPERIMENTAL
CASTing
A 79-bp oligonucleotide containing 25 random nucleotides in the centre flanked by sequences complementary to primers A and B was synthesized (IDT Technologies). The sequences of the oligonucleotides are as follows: 79-base oligonucleotide, 5′-GT-CGCTCGAGCGGTATGACGAGATCTA(N)25TAGATCTGCG-TCACTAGTCTAGACTAG-3′; primer A, 5′-GTCGCTCGAGCGGTATGACG-3′; primer B, 5′-CTAGTCTAGACTAGTGAC-GC-3′. A random sequence library of double-stranded radiolabelled oligonucleotides was prepared by annealing the 79-bp oligonucleotide to 5-fold molar excess of primer B followed by extension with Klenow enzyme. EMSAs (electrophoretic mobility-shift assays) were performed by adding 100 ng of recombinant GST (glutathione S-transferase)–ESE-2 protein to radiolabelled DNA in DNA binding buffer [5% (v/v) glycerol, 10 mM Hepes (pH 7.9), 75 mM KCl, 1 mM dithiothreitol, 2.5 mM MgCl2 and 1 mM EDTA] containing 0.5 μg of poly(dA-dT)·poly(dA-dT) and 5 μg of BSA. The reaction was incubated at room temperature (21–25 °C) for 30 min and subsequently the DNA–protein complexes were resolved by electrophoresis. The complexes formed specifically in the presence of recombinant GST–ESE-2 proteins were detected by autoradiography, excised from gels and eluted overnight at 37 °C in DNA-elution buffer containing 0.3 M NaCl, 1 mM EDTA and 0.1% SDS. The eluted DNA was extracted once in phenol/chloroform and then precipitated with ethanol. Purified DNA was subjected to re-amplification by PCR in the presence of [α-32P]dCTP. The amplified radiolabelled DNA was purified using G-50 Nick columns (Amersham) and was used in subsequent EMSA experiments. After four cycles of CASTing, the final amplified DNA was cloned directly using a pCR2.1-TOPO TA Cloning kit (Invitrogen). Nucleotide sequences of 66 independent clones were determined. The degenerate portion of the sequences was compiled and analysed for shared sequence patterns by visual inspection and by weblogo software (http://weblogo.berkeley.edu).
Preparation of GST fusion proteins
Protein expression constructs for GST–ESE-1, GST–ESE-2, GST–PDEF, GST–ETS-1 and GST–PU.1 were generated by cloning the appropriate PCR-amplified cDNA fragments in-frame downstream of GST into the pGEX-5X-1 vector (Amersham). Expression and purification of GST fusion proteins were carried out as described previously [27]. The quality and quantity of the proteins were verified by SDS/PAGE followed by Coomassie Brilliant Blue staining.
EMSAs with GST–ETS proteins
EMSAs were performed with 100–300 ng of recombinant GST fusion proteins and end-labelled double-stranded oligonucleotides. Complementary oligonucleotides (25–35 bases) were synthesized (IDT Technologies), annealed, and 2 pmol of double-stranded oligonucleotides was used for radioactive labelling with [α-32P]dCTP by Klenow enzyme. The labelled probes were purified by using G-50 Nick columns. Binding reactions were performed at room temperature for 30 min in 20 μl of DNA-binding buffer with labelled probes and recombinant GST fusion proteins in the presence of 0.5 μg of poly(dA-dT)·poly(dA-dT) added as a non-specific DNA competitor. The protein–DNA complexes were resolved by gel electrophoresis and visualized by autoradiography.
Expression vectors and luciferase reporter gene constructs
The plasmids used in the present study were generated by standard molecular cloning procedures and details can be obtained from S.S. upon request. The ESE expression plasmids, HA (haemagglutinin)-ESE-2, HA-ESE-2L, HA-ESE-2(DBD) and HA-ESE-2N-(35–253), were generated by cloning appropriate cDNA fragments of ESE-2 in the multiple cloning site of pCMV-HA vector (Clontech). The HA-VP16-ESE-2(DBD) plasmid was created by fusing a 78-amino-acid segment of the VP16 activation domain in frame with the ESE-2(DBD). For the GAL4 experiments, first the cDNA corresponding to the DBD of yeast GAL4 protein was cloned into the pCMV-HA mammalian expression vector so that all proteins could be monitored by anti-HA antibodies. The GAL4 DBD–ESE-2 chimaeras were constructed by cloning the full length or a series of 5′- or 3′-deletions of the mouse ESE-2 cDNA in frame with the DBD of yeast GAL4 protein into a pCMV-HA mammalian expression vector (Clontech) for the expression of GAL4 DBD–ESE-2 fusion proteins in transiently transfected cells. The LTK construct containing the minimal tk (thymidine kinase) promoter upstream of the luciferase reporter gene in pGL3-basic vector has been previously described and was used to generate (ESE-2)3LTK [28]. The ESE-2 responsive unit in the plasmid (ESE-2)3LTK comprises three copies of the sequence AATGAGGAAGTAATTC, which was cloned into the KpnI and SacI sites of the pGL3TK vector. The GAL4-dependent luciferase reporter plasmid, pFR-Luc, contains five copies of the GAL4-binding site upstream of a minimal promoter and the firefly luciferase gene and was obtained from a commercial source (Stratagene). All constructs were verified by DNA sequencing to ensure proper reading frame and absence of any PCR-generated mutation.
Cell culture, transient transfection and reporter gene assays
A spontaneously arising immortalized mK (mouse keratinocyte) cell line was grown and maintained in a low Ca2+ medium comprised of a 3:1 mixture of Ham's F12 and Dulbecco's modified Eagle's medium supplemented with 15% (v/v) chelated foetal bovine serum. Plasmids used for transfection assays were prepared using plasmid Midi kit from Qiagen. Transient transfections of all GAL4 DBD–ESE-2 fusion constructs were carried out with FuGENE 6™ transfection reagent (Roche) according to the manufacturer's instructions. Briefly, mK cells were seeded in 6-well plates and transfected when they reached 40–50% confluence. For each well, the plasmids (1 μg) encoding the HA-tagged ESE-2 full-length or truncated proteins or GAL4 DBD–ESE-2 chimaeras were co-transfected along with either 1 μg of (ESE-2)3LTK or pFR-Luc reporter plasmid. CMV-LacZ plasmid (0.5 μg) was used to serve as an internal control for adjusting variations in the transfection efficiency. mK cells were induced to differentiate by increasing the Ca2+ concentration in the medium to 1.2 mM (from 0.05 mM) 12 h after transfection and cells were allowed to undergo differentiation for an additional 24 h.
For reporter gene assays, transfected cells were harvested 36 h after transfection, washed with PBS, and lysed in Reporter lysis reagent (Promega). The cell extracts were assayed for luciferase activity using the Luciferase Assay system (Promega) and, for β-galactosidase activity, using the Galacton Plus kit (Applied Biosystems) according to the manufacturers’ instructions. The luciferase activity obtained was then normalized to the β-galactosidase activity, and the average values were determined. All transfections were performed in duplicate, and results of at least three independent experiments were calculated as the means±S.D. values for relative luciferase activity of each construct. The expression levels for all GAL4 DBD fusion proteins were verified by Western blotting of lysates from transfected cells using an antibody against the HA tag (Roche).
Identification of ESE-2L and RT (reverse transcriptase)–PCR analysis
A novel isoform of ESE-2, ESE-2L, was identified by an EST (expressed sequence tag) database search with sequences similar to the ESE-2 cDNA. One EST (NCBI database accession no. AI603932) was identified that showed sequence match at the 3′-end with a published ESE-2 sequence. A BLAST search of the mouse genomic sequence with the ESE-2L cDNA was performed at the National Center for Biotechnology Information website to derive the genomic organization. Total RNA was isolated from skin of adult mice by using TRIzol® reagent (Invitrogen) in accordance with the manufacturer's instructions. To generate cDNA, 2 μg of total RNA was reverse-transcribed using oligo(dT) primer and SuperScript II RT (Invitrogen). A 315-bp fragment of mouse ESE-2 was PCR-amplified using specific primers 5′-GACCGAGATCTCTGATCTGTTCAGCAATGAAG-3′ (sense) and 5′-CCGCGGTACCTCAGGTCTCTTCAGCATCATTG-3′ (antisense), and a 518-bp fragment of mouse ESE-2L isoform was amplified using specific primers 5′-CCTCTGGCTGATAGCTGGCCATTGTGG-3′ (sense) and 5′-GCCGCTGATGTTGAAGTGACAGAAGG-3′ (antisense). As a control, a 214-bp β-actin was amplified using specific primers 5′-ACCAACTGGGACGATATGGAGAAGA-3′ (sense) and 5′-TACGACCAGAGGCATACAGGGACAA-3′ (antisense). PCR amplification was performed with JumpStart Taq polymerase (Sigma).
RESULTS AND DISCUSSION
Identification of the ESE-2 consensus DNA-binding sequence
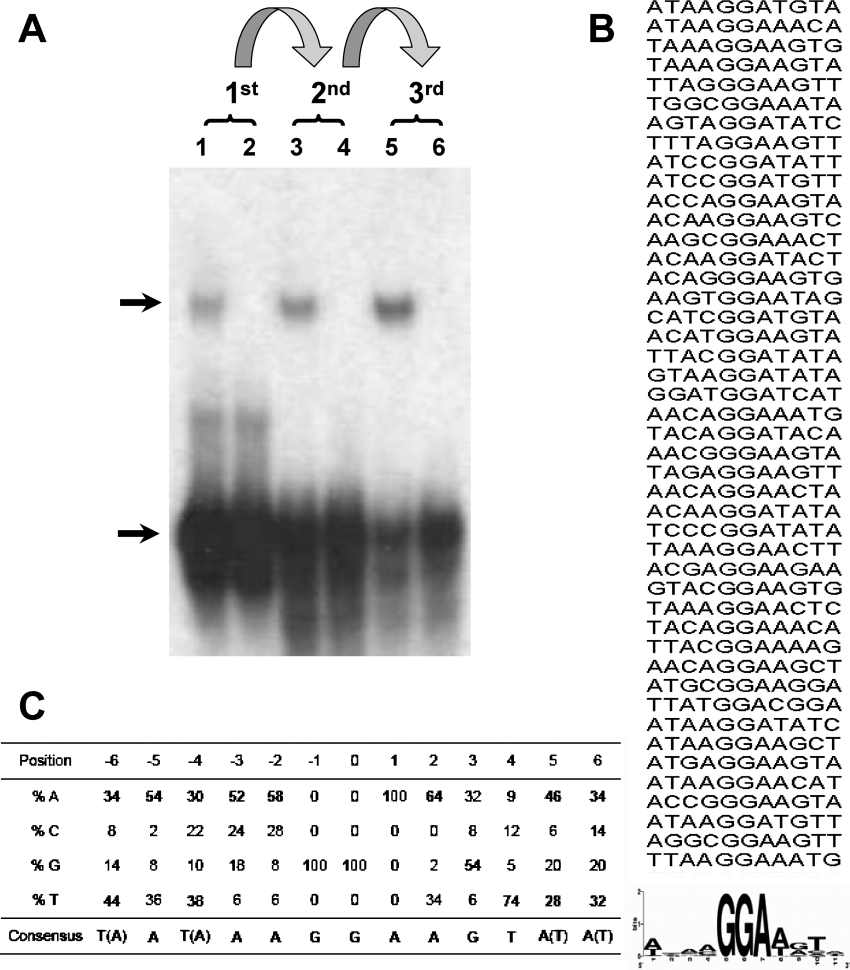
Although all characterized ETS transcription factors bind to sequences that contain the core GGA trinucleotide motif, it is thought that binding specificity of individual ETS proteins to their cognate targets is achieved in part through the sequences that flank this core region. The ESE proteins constitute a distinctive niche subgroup that exhibits high levels of sequence similarity and selective expression in epithelial cells, yet so far the DNA sequence requirements for any member of this subfamily have not been determined experimentally. We are studying ESE-2, a transcription factor that is highly expressed in epithelial cells of various types including skin keratinocytes. As a first step towards understanding the biochemical properties of ESE-2, we performed the selection of ESE-2-binding sequences using the CASTing method [29]. For this purpose, recombinant GST–ESE-2 was incubated at room temperature for 30 min with a pool of doublestranded oligonucleotides containing 25 nt of random core sequences in the centre flanked by primer sequences. The DNA–protein complexes were then separated by gel-shift assays and the protein-bound DNA was subsequently recovered from the gel and subjected to 15–20 cycles of PCR amplification. As illustrated in Figure 1(A), with each round of such selection, an enrichment of specific binding sites was observed as judged by an increased efficiency of DNA–ESE-2 complex formation. After four such rounds of selection, the final PCR products were cloned into PCR2.1 Topo vector and 66 different clones were sequenced. The sequences obtained were then analysed by aligning them as shown in Figure 1(B). Strikingly, all selected clones contained at least one copy of the GGA (or its complementary TCC) core motif. For the sake of clarity, we have numbered the residues with the central guanine being designated as position ‘0’. While positions −1 and +1 contain the invariant guanine and adenine residues respectively, the rest of the positions showed some degree of variability. Nevertheless, analysis of the sequences suggests that the predominant residues that flank the GGA core motifs consist of either an adenine or thymine residue, except for position 3, where a guanine is preferred (Figure 1C). Taken together, our experiments define the optimal ESE-2 DNA-binding consensus motif as (T/a)A(T/a)AAGGAAGT(A/t)(A/t), consisting of a GGA core and an AT-rich 5′- and 3′-flanking sequences. A previous study had derived a consensus site for ESE-2 (ANCAGGAAGTAN) that is slightly different from the one we report here in this paper [8]. It should be noted that the previous data were based on a study with a limited number of promoter sequences that contain selected Ets-binding sites found in various epithelium-specific promoters. Interestingly, the 13-base consensus sequence of ESE-2 identified by our CASTing studies closely matches with the binding sites present within intron 1 of cytokeratin 18, one of the recently identified ESE-2 target genes [30]. The availability of the consensus binding sequence for ESE-2 will be useful in testing its biochemical properties and in identification and validation of potential ESE-2 target genes.
Figure 1. CASTing for the ESE-2 consensus DNA-binding site.
(A) EMSA with 79-base oligonucleotides containing PCR-amplifiable sequences flanking 25-bp random nucleotides and purified recombinant GST–ESE-2 proteins. The DNA recovered from this first cycle of selection was then amplified using PCR, mixed with fresh GST–ESE-2 proteins, and then subjected to additional cycles of selection. Lanes 1, 3 and 5 show the DNA–protein complexes from first cycle, second cycle and third cycle respectively. Lanes 2, 4 and 6 show DNA alone used in each cycle. Upper arrow and lower arrow indicate the positions of ESE-2–DNA complex and random oligonucleotides respectively. (B) Alignment of the individual sequences. Shown in the upper panel is the alignment of sequences by the position of GGA. The consensus sequence is shown in the lower panel as a WebLogo [36]. (C) Binding site preference for ESE-2. The residues are based on the invariant GGA motif numbered, with the central guanine residue being designated as position ‘0’. For each position, the frequency of each nucleotide is indicated as a percentage. The ESE-2 consensus DNA-binding sequence obtained is derived from the following criteria: any nucleotide that is preferred over 50% is selected; if no nucleotide is preferred for more than 50%, then the two nucleotides that are most preferred are selected.
Analysis of selected ESE-2-binding sequences
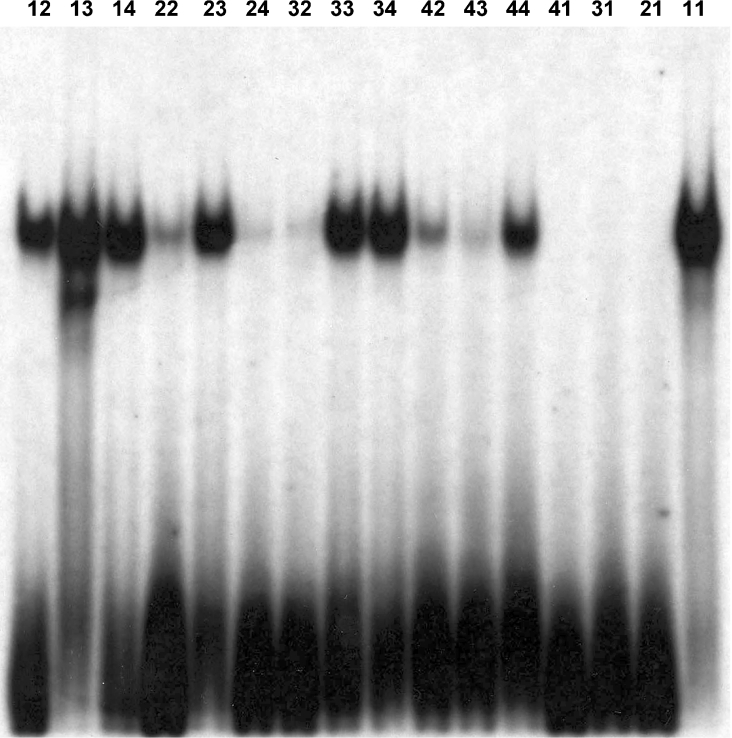
The fact that the ESE-2 consensus site consisted of a core GGA sequence and a preference for A/T-rich flanking sequences prompted us to examine experimentally the sequence requirements in more detail. To facilitate these studies, we divided the compiled sequences into four groups, each group representing different levels of match with the optimum consensus sequence (Table 1). Thus Group 1 consists of four sequences (11–14) that matched both on the 5′ and 3′ of the core GGA sequence. Group 2 (21–24) and Group 3 (31–34) consist of sequences that match with the consensus site only on the 5′- or the 3′-side of the core GGA motif respectively. In each case, the mismatch sequences of the 5′- and 3′-side consist of nucleotides that were selected at the lowest frequency for each position (Figure 1C). Finally, Group 4 (41–44) contains sequences that contain only the core GGA motif but show the most divergent 5′- and 3′-flanking sequences based on our CASTing analysis. The 11-, 21-, 31- and 41-sequences (Table 1, italics) represent artificial sequences that were designed to exemplify each group and were not derived from CASTing, whereas the remaining sequences were picked from the 66 clones that were sequenced. We next generated 16 oligonucleotides that contained these sequences, labelled them to the same specific activity and tested them in EMSAs with GST–ESE-2. EMSA experiments with these oligonucleotides suggested that while all four sequences from Group 1 bound to ESE-2 very well, sequences from Groups 2 and 3 showed moderate binding, whereas sequences from Group 4 showed the weakest binding (Figure 2). What was striking in our finding was that the oligonucleotides corresponding to sequences 21 and 31, which completely deviated from the consensus AT-rich 3′- and 5′-flanking sequences respectively, showed absolutely no binding to ESE-2. This suggested an important role of flanking sequences in governing the binding of ESE-2 to DNA. However, other oligonucleotides from Groups 2 and 3 (numbers 23, 33 and 34) showed reasonable binding, suggesting that moderate deviations from the consensus sequences are potentially tolerated.
Table 1. The selected DNA-binding sites for ESE-2 can be classified into four groups.
The sequences of selected clones were classified into four groups as follows: Group 1 contains consensus flanking sequences on both sides of the GGA motif. Groups 2 and 3 contain consensus flanking sequences only on the 5′- and 3′-sides respectively. Group 4 contains no consensus flanking sequence at either end. The underlined stretches of nucleotides GATCTA and TAG indicate the additional sequences that were added to increase the length of the oligonucleotides for EMSA. The relative binding affinity was determined by estimation of ESE-2–DNA complex and was scaled from ++++ (strong) to +/− (weak). Boldface indicates the core motif of the consensus sequence.
| Group | Oligonucleotide | Sequence | Binding affinity |
|---|---|---|---|
| Group 1 | 11 | GATCTATATAAGGAAGTAATATTAG | ++++ |
| 12 | GATCTATTAAAGGAAGTGTTATTAG | ++ | |
| 13 | GATCTAAATGAGGAAGTAATTCTAG | ++++ | |
| 14 | GATCTAATAGAGGAAGTTGTCATAG | +++ | |
| Group 2 | 21 | GATCTATATAAGGACTGCCCGGTAG | − |
| 22 | GATCTATATAAGGAACATTATTTAG | + | |
| 23 | GATCTAAATAAGGAAACACGCTTAG | +++ | |
| 24 | GATCTATACAAGGATACTGAATTAG | +/− | |
| Group 3 | 31 | GATCTACCGTTGGAAGTAATATTAG | − |
| 32 | GATCTATACATGGAAGTAAATTTAG | +/− | |
| 33 | GATCTAAAACGGGAAGTACTAGTAG | +++ | |
| 34 | GATCTACACCGGGAAGTATAACTAG | +++ | |
| Group 4 | 41 | GATCTACCGTTGGACTGCCCGGTAG | − |
| 42 | GATCTAGTTACGGAAAAGTAATTAG | + | |
| 43 | GATCTAATTACGGATATAGAGGTAG | +/− | |
| 44 | GATCTAATCCCGGATATATAGATAG | ++ |
Figure 2. Binding specificity of ESE-2 to individual oligonucleotides containing different degrees of match with the consensus sequence.
ESE-2-specific binding is demonstrated by EMSAs. Sequences of each oligonucleotide used in these experiments are listed in Table 1.
In addition to testing the direct binding of the various oligonucleotides to ESE-2, we also confirmed the affinity of these sequences to ESE-2 by performing competition experiments. For this purpose, radiolabelled oligonucleotides corresponding to sequence 11 were incubated with GST–ESE-2, and 50-, 200- and 400-fold excess of unlabelled oligonucleotides corresponding to the four groups was used to compete with the binding of ESE-2. Similar to the data obtained from direct binding, sequences 21, 31 and 41 did not compete for the binding at all, excess amounts of sequences 22, 24, 32, 42 and 43 slightly competed, whereas sequences 11, 12, 13, 14, 23, 33, 34 and 44 exhibited strong competition ability (results not shown). Not surprisingly, the strong DNA-binding affinity of ESE-2 was found in most sequences of Group 1 that contained the ideal consensus flanking sequences on both sides of the GGA motif. These relative binding affinities of the different sequences are summarized in Table 1. These results also indicate that sequence 13 obtained from the CASTing experiments can bind to ESE-2 very well and can be considered a good candidate for an ideal ESE-2 DNA-binding motif. Collectively, our EMSA studies indicate that not only the GGA motif but also flanking sequences on both sides play an important role in dictating the DNA-binding affinity of ESE-2.
Comparison of the DNA-binding specificity of ESE-2 with other Ets-domain proteins
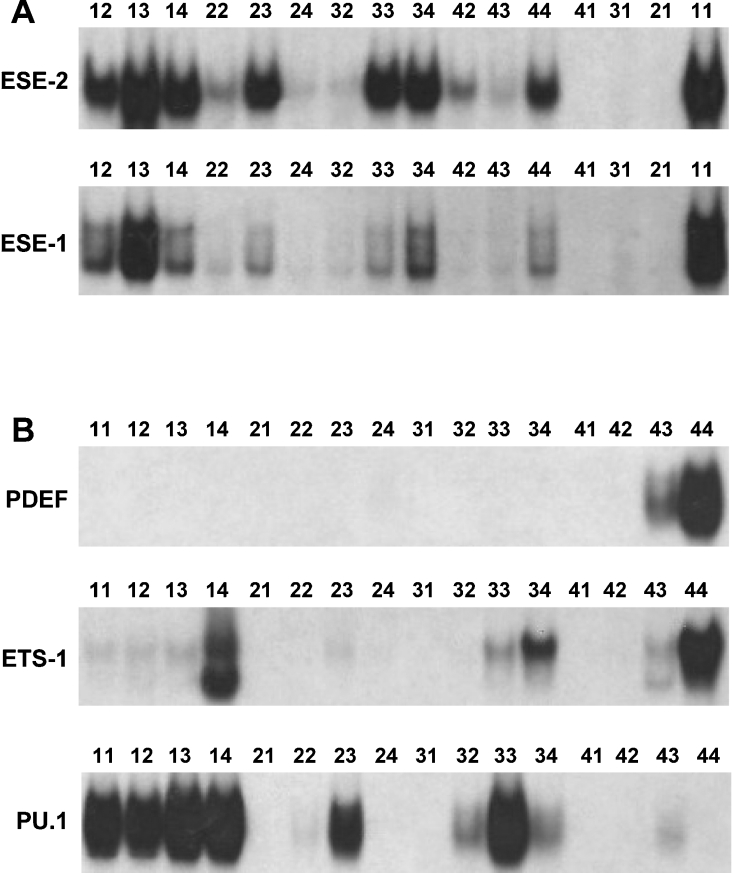
Selection studies have identified consensus DNA-binding sites centred upon the GGA motif with the variable flanking nucleotides for many Ets proteins. What is evident from these studies is that there is a great deal of heterogeneity of DNA-binding sites for ETS proteins, which may reflect a difference in their DNA-binding properties. To further analyse this experimentally, we compared the DNA-binding properties of ESE-2 and several other Ets-domain proteins by performing EMSA with recombinant proteins. We utilized the 16 oligonucleotides that represented the four different classes of ESE-2-binding sites that were derived from the CASTing experiments described above. For the first experiment, we chose ESE-1, which is phylogenetically close to ESE-2 and shares a high level of sequence similarity in the ETS DNA-binding region. As shown in Figure 3(A), GST–ESE-1 showed different degrees of binding activities with the 16 oligonucleotides. The relative binding affinities to all oligonucleotides were similar but not identical between ESE-2 and ESE-1. This is in agreement with the reported ESE-1 consensus DNA-binding site, which is virtually identical with sequences of Group 1 of the ESE-2-binding sequences. However, oligonucleotides 23 and 33 showed significantly better binding to ESE-2 than ESE-1. This suggests that there could be subtle qualitative and/or quantitative differences in DNA binding of ESE-2 and ESE-1, which may have important implications for target-gene specificity in vivo.
Figure 3. EMSA with ESE-2 and other Ets transcription factors with different Ets-binding sites.
Binding activities of ESE-2 were compared with ESE-1 (A) and other Ets factors (B). Numbers on top of each lane refer to the specific oligonucleotides used in this experiment and the sequences can be found in Table 1. Each oligonucleotide was labelled to the same specific activity and was used in EMSA with recombinant ESE-2, ESE-1, PDEF, ETS-1 or PU.1 proteins. Only the DNA–protein complexes are shown.
To extend these observations further, we generated GST fusion proteins of three additional ETS proteins, ETS-1, PU.1 and PDEF. As shown in Figure 3(B), PU.1 efficiently bound to most of the sites that have been selected by ESE-2 and exhibited a similar pattern, in particular showing strong binding to sites of Group 1. In contrast, ETS-1 showed a significantly different pattern of binding, with very weak binding to Group 1 sequences. These results are in agreement with the established consensus sequence for ETS-1, which shows differences in the flanking sequence requirements when compared with ESE-2. Finally, as expected from the fact that ESE-2 and PDEF share very low sequence homology in their ETS domains, EMSA studies with PDEF showed the most striking results, with virtually no DNA binding to any of the oligonucleotides except 43 and 44. It has been reported that PDEF uniquely prefers binding to a GGAT core instead of a GGAA [9]. Indeed, a recent crystal structure of PDEF provides evidence that two amino acid residues that are unique to PDEF dictate its preference for GGAT sequences [31]. Our results provide further experimental support that the DNA-binding sites for PDEF are distinct from other Ets-domain proteins, showing an apparent preference for GGAT versus GGAA in the core of the binding site (thus binding only to oligonucleotides 43 and 44). It should be emphasized that, although the consensus DNA-binding sites for any given Ets-domain proteins may represent only subtle differences, the binding specificities of individual sequences may vary considerably. Whether these differences are also important in the specificity of Ets proteins for their target genes in vivo is a question that can be addressed in the future when the targets for each Ets protein have been unequivocally established. In addition, it is likely that co-operative protein–protein interactions between Ets proteins and accessory transcription factors at adjacent DNA sites may influence the DNA binding in vivo [32].
Isolation of a novel isoform of mouse ESE-2
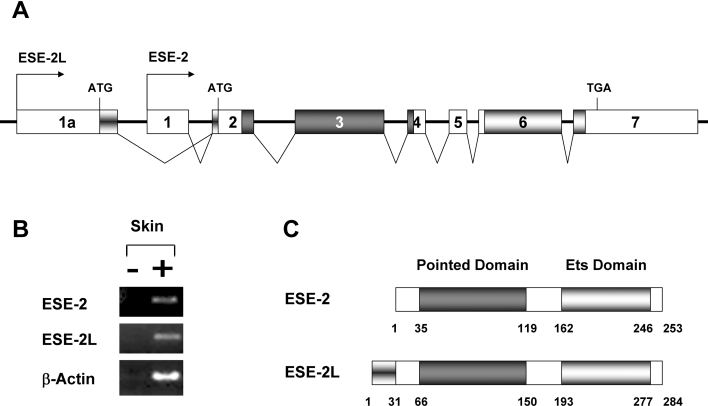
The human ESE-2 gene has been shown to have two alternatively spliced isoforms (hESE-2a and hESE-2b) that differ in their 5′-ends [8]. As a result, the coding region of hESE-2b begins at the second methionine of hESE-2a and lacks the first ten amino acids of hESE-2a. Interestingly, the human ESE-2a and ESE-2b isoforms show differential expression patterns: the hESE-2a is expressed in kidney epithelium, whereas the hESE-2b is more strongly expressed in the prostate glands, salivary glands and skin keratinocytes. To identify whether similar isoforms of ESE-2 are also expressed in mouse skin, we performed RT–PCR and searched the mouse EST database for sequences that match ESE-2 cDNA. We were able to detect the expression of the mouse homologue of the hESE-2b isoform but not the hESE-2a isoform (results not shown). Interestingly, one EST from skin tissue was identified, which matched part of the published mouse ESE-2 sequence, but in addition contained an extra sequence that extended the 5′-region of ESE-2. Analysis of the mouse genomic DNA sequences suggests that there is an additional upstream exon (exon 1a) for ESE-2 with its own promoter, which when utilized should give rise to a longer isoform of ESE-2 (Figure 4A). We performed RT–PCR analysis on mouse skin cDNA using primers specific for ESE-2 as well as the longer isoform of ESE-2 (ESE-2L). RT–PCR analysis revealed that the expression of both ESE-2 and ESE-2L was detected in skin tissue, suggesting that it is a bona fide isoform (Figure 4B). This inclusion of the new exon extended the coding region of ESE-2 by an additional 31 amino acids at the N-terminus (Figure 4C). The sequences of these amino acids do not match any other protein in the database and may confer on ESE-2 unique functional properties such as transactivation or interaction with specific partner proteins.
Figure 4. The mouse ESE-2 locus gives rise to a longer ESE-2 isoform (ESE-2L).
(A) Genomic structure of the mouse ESE-2 gene. Exons are shown as boxes and are numbered 1–7 and 1a. Exon 1, the specific exon 1 of ESE-2; exon 1a, the specific exon 1 of ESE-2L; exons 2–7, common exons for both ESE-2 and ESE-2L. The exons and introns are not drawn to scale. The arrows indicate the transcriptional start sites of ESE-2 and ESE-2L. The putative start codon (ATG) and stop codon (TGA) are indicated. Dark grey boxes and light grey boxes indicate the pointed and the Ets domain respectively. (B) Expression of both isoforms of ESE-2 in skin. RT–PCR analysis of reverse-transcribed total RNA from skin was performed using primers specific for mouse ESE-2 (top panel) or ESE-2L (middle panel). As a control, β-actin (bottom panel) was amplified. PCR was performed with RNA samples processed with (+) or without (−) RT. (C) Schematic representation of the two ESE-2 proteins. The PNT and the Ets domain are depicted. The numbers indicate the positions of amino acids.
Transcriptional activity of ESE-2
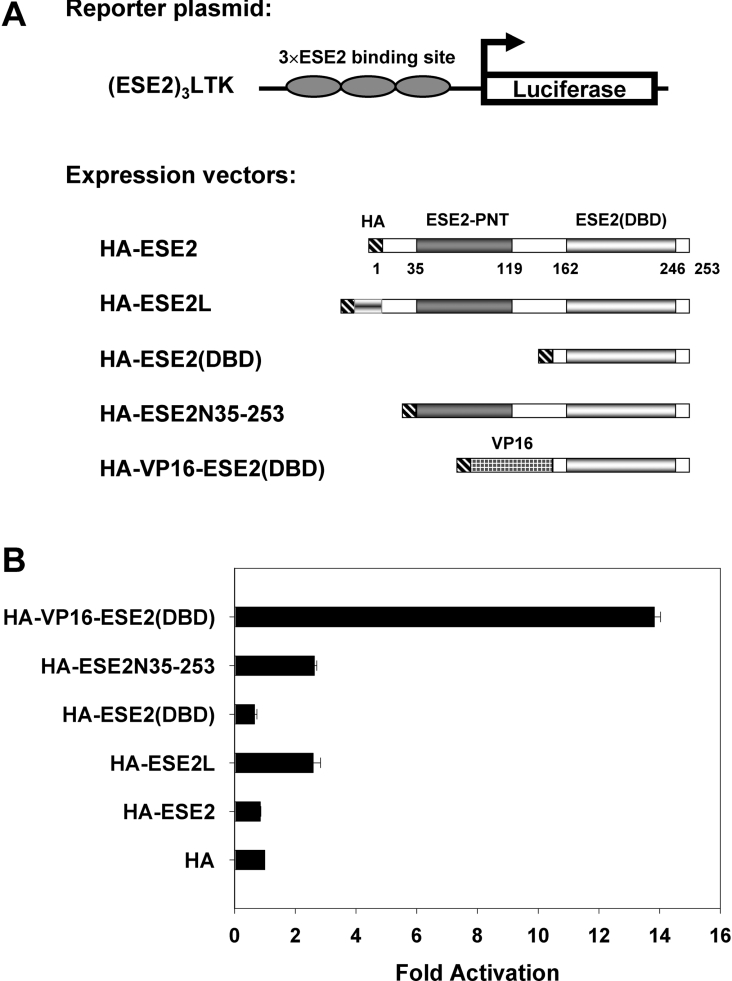
Having established the optimum DNA-binding site and identified a novel isoform of ESE-2, our next goal was to validate the binding of the ESE-2 to its optimum binding site in a cellular context and to examine the transcriptional activity of ESE-2 protein and the novel isoform ESE-2L in transient transfection experiments. For this purpose, three copies of an optimum ESE-2 DNA-binding site were cloned upstream of the luciferase gene in front of the minimal tk promoter to generate (ESE-2)3LTK reporter plasmid (Figure 5A). We chose the sequence 13 (AATGAGGAAGTAATTC), which as described above possesses strong binding affinity to ESE-2 (boldface indicates conserved core sequence). We also generated plasmids that can express HA epitope-tagged full-length ESE-2, ESE-2L and truncated versions of ESE-2 (Figure 5A). The (ESE-2)3LTK reporter construct was co-transfected into mK cells along with either control expression plasmid pCMVHA or plasmids that expressed the various isoforms of ESE-2. A CMV-LacZ plasmid was co-transfected to serve as an internal control for transfection efficiency. Interestingly, ESE-2 did not activate reporter gene expression, whereas ESE-2L showed a moderate 3-fold activation (Figure 5B). This is in agreement with previous published results [9,30]. Similarly, a deletion construct expressing a truncated protein containing only the DNA-binding ETS domain also had no effect on ESE-2 responsive reporter gene expression. It has been reported that full-length human ESE-2 contains a negative regulatory domain at the N-terminus, which may influence DNA binding and activation. For this reason, we also tested in co-transfection assays the effects of a plasmid that expresses a truncated ESE-2 protein, ESE-2N-(35–253), lacking the first 34 amino acids at the N-terminus. As shown in Figure 5(B), expression of ESE-2N-(35–253) increased transcription 3-fold, suggesting that amino acids 1–34 may potentially have a repressive effect on full-length ESE-2. Taken together, these results suggest that under our experimental conditions, full-length ESE-2 shows no transcriptional activity, whereas ESE-2L containing 31 additional amino acids in its N-terminus is a moderate transcriptional activator. Western-blot analysis showed that all proteins were stably expressed when transfected (Supplementary Figure S1A at http://www.BiochemJ.org/bj/398/bj3980497add.htm).
Figure 5. Transcriptional activity of ESE-2 as measured by luciferase assays with an optimum ESE-2 responsive reporter.
(A) A schematic representation of the constructs. (ESE-2)3LTK, a luciferase reporter construct containing three copies of ESE-2-binding sites linked to a minimal tk promoter; HA–ESE-2, a construct containing full-length ESE-2 cDNA with HA tag at the 5′-end; HA–ESE-2L, a construct containing the alternatively spliced ESE-2 isoform with HA tag at the 5′-end; HA–ESE-2(DBD), a construct containing ESE-2(DBD); HA–ESE-2N-(35–253), a truncated construct lacking the first 34 amino acids of ESE-2 at the N-terminus; HA–VP16-ESE-2(DBD), a fusion construct containing the coding sequence for the VP16 transcriptional activation domain fused in frame with the coding sequence of ESE-2(DBD). (B) Transcriptional reporter gene assay for the consensus DNA-binding site of ESE-2 is shown. CMV-LacZ plasmid was used as an internal control for the efficiency of transfection, and the reporter gene expression was normalized to the β-galactosidase activity and presented as values relative to the HA control. Results are shown as means±S.D.
One possibility for the lack of transcriptional activity is that the sequence of ESE-2-binding site chosen for the (ESE-2)3LTK construct may not bind well to ESE-2 in vivo. To test this, we generated a construct that expresses a chimaeric protein consisting of the DBD of ESE-2 fused to the acidic activation domain of the VP16. Co-transfection of the (ESE-2)3LTK reporter construct into mK cells with the VP16-ESE-2(DBD) construct resulted in a strong transcriptional activity (14-fold higher than the empty vector), as measured by luciferase values. This result suggests that the ESE-binding site chosen for the (ESE-2)3LTK construct is indeed a suitable binding site for ESE-2 in vivo. The poor transcriptional potency of ESE-2 proteins may be an inherent property of the proteins or a limitation of the transient transfection assays. Indeed, one caveat to the transient transfection experiments is the fact that the mK cells express many ETS proteins, which may compete with ESE-2 in binding to the Ets site present in the (ESE-2)3LTK reporter construct. This notion is supported by the fact that the (ESE-2)3LTK reporter has relatively higher basal expression than the LTK reporter even in the absence of ESE-2 expression vector (results not shown). Hence, under these conditions, overexpression of ESE-2 may not activate transcription above and beyond these elevated basal levels. Other studies on natural promoters such as the parotid gland-specific promoter, PSA (prostate-specific antigen) promoter, and WAP (whey acidic protein) promoter have shown that ESE-2 can activate or repress transcription in a context-dependent manner, although the effects were weak [8,10]. As expected, these promoters are complex, and often contain additional elements such as AP-1 (activator protein-1)-binding sites that may act synergistically with ESE-2 to activate transcription.
Identification of a transcriptional activation domain within ESE-2
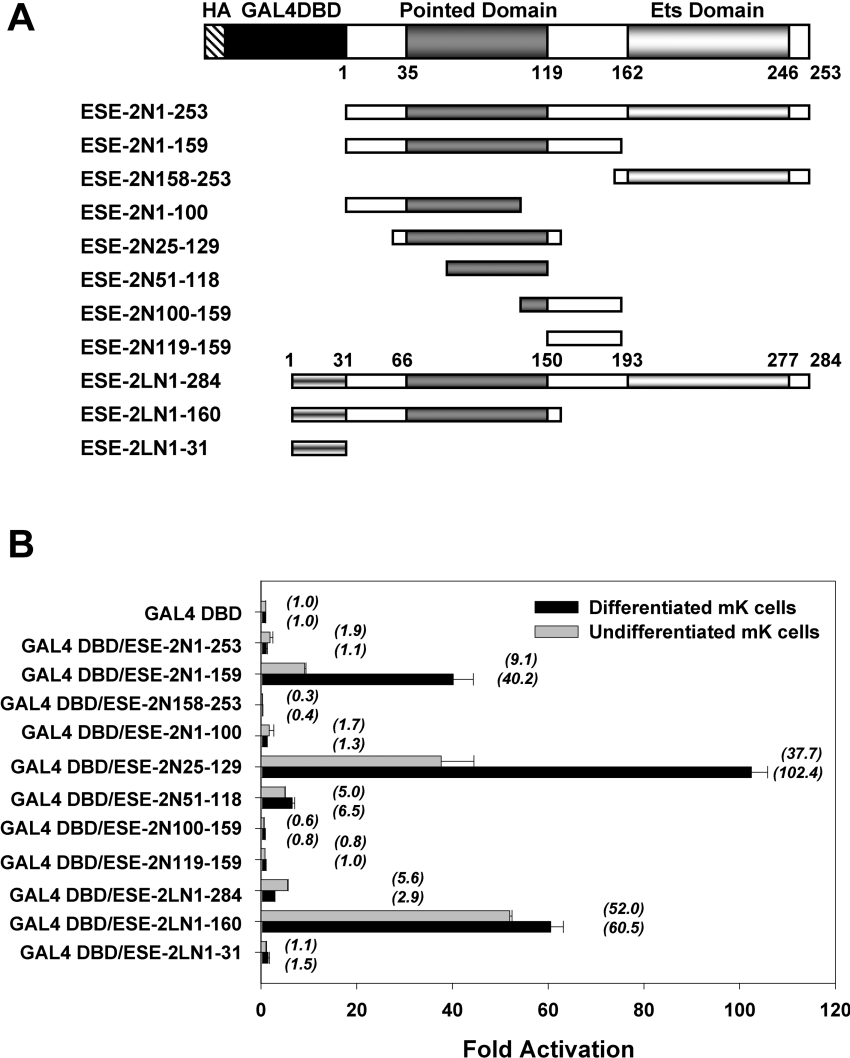
Besides the role for the Ets domain in binding to target genes, very little is known about other functional domains of ESE-2, in particular whether there are segments of the protein involved in transcriptional activation and/or repression. To identify and characterize the potential activation or repression properties of ESE-2 independent of its DNA-binding activity, we prepared constructs that would express chimaeric proteins containing various segments of ESE-2 fused to the heterologous DBD of yeast GAL4 transcription factor (GAL4 DBD) (Figure 6A). Next, we performed transient transfection experiments using a luciferase reporter gene linked to a minimal promoter and five copies of GAL4 binding sites, and determined the transcriptional activities of GAL4 DBD–ESE-2 fusion proteins. These experiments were performed in mKs grown under two separate conditions. One set of transfections was performed in keratinocytes growing in proliferative state, where they express low levels of ESE-2. Additionally, transfection studies were also performed in keratinocytes, which were induced to differentiate by changing the Ca2+ levels in the growth media, thus resulting in higher levels of ESE-2. Each of the chimaeric GAL4 fusion protein was expressed at relatively similar levels as judged by Western-blot analysis (Supplementary Figure S1B at http://www.BiochemJ.org/bj/398/bj3980497add.htm).
Figure 6. Transcriptional activation and repression potential of GAL4 DBD–ESE-2 fusion proteins.
Transcriptional activation was measured by ESE-2-induced expression of a luciferase reporter gene in transiently transfected mK cells. (A) Schematic illustration of full-length and various truncated ESE-2 and ESE-2L constructs. The HA tag, GAL4 DBD, PNT and Ets domains are depicted. Horizontal bars indicate the regions of ESE-2 that were fused to GAL4 DBD at their N-termini. Numbers specify positions of the terminal amino acids in the truncated ESE-2 constructs. (B) Transcriptional activities determined by GAL4 DBD–ESE-2 fusion proteins. mK cells were transiently transfected with expression vectors for the indicated GAL4 DBD–ESE-2 fusion proteins along with the pFR-Luc reporter plasmid containing GAL4 responsive elements upstream of a luciferase reporter gene. CMV-LacZ was used as a control for the efficiency of transfection. The normalized value of fold activation for each GAL4 DBD–ESE-2 fusion protein in mK cells, relative to the activity of the GAL4 DBD alone, which was set to 1, is presented as the mean±S.D. for at least three separate experiments performed in duplicate. Black bars and shaded bars represent the activities determined in calcium-induced differentiated mK cells and undifferentiated mK cells respectively. Parentheses denote the numerical mean values.
The GAL4 DBD–ESE-2N-(1–253) chimaera containing the full-length ESE-2 was almost transcriptionally inert since it showed a weak 2-fold activity compared with GAL4 DBD (Figure 6B). This is not surprising since GAL4 chimaeras that contain full-length transcription factors with their own DBD are often poor transcriptional activators. The mechanism for this phenomenon is not well understood; however, one possibility is that the presence of a second DBD leads to targeting/sequestration of the chimaeric protein to the multitude of the potential binding sites present in genomic DNA rather than the GAL4 sites of the reporter construct. The transfection assay showed that the GAL4 DBD–ESE-2N-(158–253) fusion protein significantly reduced the reporter gene expression in both undifferentiated and differentiated mK cells, indicating that the ESE-2 Ets domain had a significant transcriptional repression activity. To avoid the transrepression by the ETS domain, ESE-2 fragments lacking the Ets domain were fused to the GAL4 DBD to allow identification of regions that were responsible for transcriptional activation. As shown, the transcriptional activities of GAL4 DBD–ESE-2N-(1–159) were 9- and 40-fold higher than the activity of GAL4 DBD alone in undifferentiated and differentiated mK cells respectively (Figure 6B). Interestingly, the transcriptional activity of this domain (N1–159) is further enhanced by small N-terminal and C-terminal truncation. Indeed, co-transfection of the GAL4 DBD–ESE-2N-(25–129) fusion construct induced an approx. 100-fold increase in luciferase activity in differentiated mK cells compared with that seen after co-transfection of the GAL4 DBD alone. These results suggest that a major transactivation domain of ESE-2 is located between residues 25 and 129, corresponding to the PNT and that this domain is more active in differentiated keratinocytes. We also tested additional constructs that contained only a partial PNT, such as ESE-2N-(1–100), ESE-2N-(51–118), ESE-2N-(100–159) and ESE-2N-(119–159). Other than the GAL4 DBD–ESE-2N-(51–118) construct, which showed weak luciferase activity, none of the other PNT deleted constructs showed any transcriptional activity. These results suggest that the entire PNT of ESE-2 is needed for optimum activation since this activity is completely lost even with minor N- or C-terminal deletions.
Finally, we also tested the ESE-2L isoform for its activity in similar assays. The GAL4 DBD–ESE-2LN-(1–284) chimaera containing the longer isoform of ESE-2 showed moderate transcriptional activity since it showed a 6-fold activity compared with GAL4 DBD in mK cells. This is similar to the effect we observed in previous experiments with ESE-2 responsive promoter, where ESE-2L showed 3-fold higher activation potential compared with ESE-2. Surprisingly, the activity of this isoform was lower in differentiated than proliferating keratinocytes, an observation that is interesting but difficult to explain without additional studies. Also, GAL4 DBD–ESE-2LN-(1–160) showed strong activity since it contained the PNT. However, the additional 31 amino acids that are exclusive to the ESE-2L isoform failed to show any transcriptional activity in this assay, suggesting that its unique function might not be linked to transcriptional properties. Neither the transcriptional activation potential of the ESE-2 PNT nor the transcriptional repression potential of the ETS domain was restricted to mK cells. When similar experiments were performed in NIH 3T3 fibroblasts, both the transactivation potential and the transrepression potential were observed (results not shown), although the activation was not as dramatic as that seen in mK cells. Taken together, our results show that the Ets domain of ESE-2 acts as a transcriptional repressor, whereas the PNT of ESE-2 can function as a transcriptional activator.
It is possible that the overexpression of a potent transactivator paradoxically reduces the expression of a co-transfected reporter plasmid in a transient transfection assay, with the resulting decline in reporter activity thought to be due to sequestration of general transcription factors. This process is called ‘squelching’ and this phenomenon has been observed with many transcription factors [33]. To determine if squelching was affecting the GAL4 chimaeric constructs, we repeated the experiments using different amounts (0.125–2.0 μg) of two representative expression plasmids. Analysis of luciferase activities revealed that the reporter gene was inhibited to a similar level by both low-level expression of ESE-2N-(100–159) as well as high-level expression, indicating that the inhibition was not due to transcriptional squelching (Supplementary Figure S2 at http://www.BiochemJ.org/bj/398/bj3980497add.htm). In addition, the activation of luciferase gene expression by GAL4 DBD–ESE-2N-(1–159) was increased in a dose-dependent manner. This suggested that the effects that we saw with the different constructs were not dependent on levels of proteins and that the results obtained were unlikely to be due to the squelching effect.
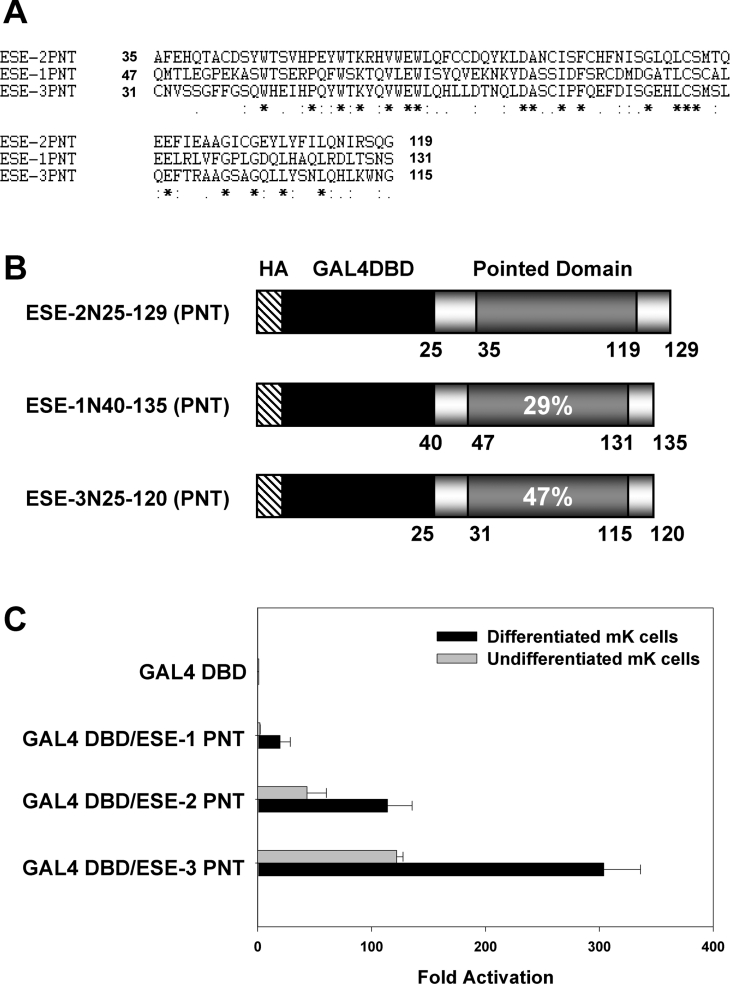
Transactivation potential of the PNTs in ESE family members
The fact that the transactivation domain of ESE-2 was located within the N-terminal PNT is an interesting finding and to our knowledge has not been reported for any other PNT. Indeed, to date, PNTs of other Ets proteins have been implicated in protein–protein interactions, oligomerization and even transcriptional repression [2]. Activation domains are typically characterized by the nature of the amino acid composition (i.e. acidic, glutamine-rich), although a growing number of activation domains do not fit any known pattern, suggesting that it is not the mere sequence but the overall structure that confers the transactivation properties [34]. The PNTs of ESE family members (ESE-1, ESE-2 and ESE-3) show moderate sequence similarity at the amino acid level (29% between ESE-2 and ESE-1 and 47% between ESE-2 and ESE-3) (Figures 7A and 7B). The novel finding that the ESE-2 PNT is a strong activator prompted us to test whether this property was unique to ESE-2. For this purpose, we generated GAL4 DBD fusion constructs containing the PNTs of mouse ESE-1 and ESE-3 to assess their transactivation potential. As before, mK cells were transiently transfected with GAL4 DBD–ESE-1 PNT, GAL4 DBD–ESE-2 PNT and GAL4 DBD–ESE-3 PNT, together with a pFR-Luc reporter plasmid. Equal expression levels of all three chimaeric proteins was observed, as shown by Western-blot analysis (results not shown). Both ESE-2 PNT and ESE-3 PNT exhibited high levels of transactivation potential in both undifferentiated and differentiated mK cells (Figure 7C). In particular, ESE-3 PNT induced a 300-fold increase in the luciferase activity in differentiated keratinocytes. On the other hand, PNT of ESE-1 showed no transactivation activity in undifferentiated mK cells and very low levels in differentiated keratinocytes. Our results for ESE-1 are in agreement with a previous study, where it was shown that exon 4 of ESE-1 encoding a unique 31 amino acid sequence had strong activation potency but not the PNT [35]. These results suggest that the PNT of both ESE-2 and ESE-3 may be important for their transcriptional activity and may provide a potential docking site for co-activators and other members of the transcriptional machinery. It is interesting to note that the PNTs of ESE-2 and ESE-3 contain a number of acidic amino acids, which may play an important role in this process. Our results suggest that a subset of the PNTs may possess a unique signature of amino acids and/or structural features that confer on them a transcriptional activation potential. There is also an intriguing possibility that this activation potential may also be influenced by the cell type and cellular environment. Future studies with additional PNTs should shed light on the role of these domains in regulating gene expression. Finally, our identification and characterization of the functional domains of ESE-2 will provide a better understanding of the mechanisms by which ESE-2 controls the expression of its target genes.
Figure 7. Transcriptional activities of the PNTs of ESE family members.
(A) Alignment and comparison of amino acid sequences of the PNTs of ESE-1 (NCBI database accession no. NM_007921) and ESE-3 (NCBI database accession no. NM_007914) with those of ESE-2 (NCBI database accession no. NM_010125). ClustalW program was used for the multiple alignments. Asterisks indicate amino acid residues that are identical in all sequences. Symbols ‘:’ and ‘.’ denote conserved substitutions and semi-conserved substitutions respectively. (B) Schematic illustration of the PNTs of ESE-2, ESE-1 and ESE-3 constructs, fused to GAL4 DBD at their N-termini. The HA tag, GAL4 DBD and PNT are depicted. The numbers indicate the positions of amino acids of each ESE protein. Percentage identity of each PNT with ESE-2 is shown. (C) Transactivation activities determined for the PNTs of ESE-1, ESE-2 and ESE-3. mK cells were transiently transfected with expression vectors containing the PNTs of ESE family members as indicated. The relative, normalized, luciferase activity (±S.D.) is presented in the form of a bar graph. Black bars and shaded bars represent the activities determined in calcium-induced differentiated mK cells and undifferentiated mK cells respectively.
Online data
Acknowledgments
We are especially grateful to Dr Lee Ann Garrett-Sinha (Department of Biochemistry, State University of New York, Buffalo, NY, U.S.A.) for helpful discussions and advice. We also thank Irene Kulik for excellent technical assistance in preparing DNA for transfection experiments and other members of our laboratory for useful comments on this study. This work was supported by a grant R01GM069417 from NIH (National Institutes of Health; Bethesda, MD, U.S.A.) (S.S.).
References
- 1.Sementchenko V. I., Watson D. K. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 2.Sharrocks A. D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 3.Hollenhorst P. C., Jones D. A., Graves B. J. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maroulakou I. G., Bowe D. B. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- 5.Feldman R. J., Sementchenko V. I., Watson D. K. The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Res. 2003;23:2125–2131. [PubMed] [Google Scholar]
- 6.Choi S. G., Yi Y., Kim Y. S., Kato M., Chang J., Chung H. W., Hahm K. B., Yang H. K., Rhee H. H., Bang Y. J., Kim S. J. A novel ets-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J. Biol. Chem. 1998;273:110–117. doi: 10.1074/jbc.273.1.110. [DOI] [PubMed] [Google Scholar]
- 7.Oettgen P., Alani R. M., Barcinski M. A., Brown L., Akbarali Y., Boltax J., Kunsch C., Munger K., Libermann T. A. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol. Cell. Biol. 1997;17:4419–4433. doi: 10.1128/mcb.17.8.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oettgen P., Kas K., Dube A., Gu X., Grall F., Thamrongsak U., Akbarali Y., Finger E., Boltax J., Endress G., et al. Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J. Biol. Chem. 1999;274:29439–29452. doi: 10.1074/jbc.274.41.29439. [DOI] [PubMed] [Google Scholar]
- 9.Oettgen P., Finger E., Sun Z., Akbarali Y., Thamrongsak U., Boltax J., Grall F., Dube A., Weiss A., Brown L., et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J. Biol. Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J., Ng A. Y., Tymms M. J., Jermiin L. S., Seth A. K., Thomas R. S., Kola I. A novel transcription factor, ELF5, belongs to the ELF subfamily of ETS genes and maps to human chromosome 11p13-15, a region subject to LOH and rearrangement in human carcinoma cell lines. Oncogene. 1998;17:2719–2732. doi: 10.1038/sj.onc.1202198. [DOI] [PubMed] [Google Scholar]
- 11.Tymms M. J., Ng A. Y., Thomas R. S., Schutte B. C., Zhou J., Eyre H. J., Sutherland G. R., Seth A., Rosenberg M., Papas T., et al. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15:2449–2462. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 12.Andreoli J. M., Jang S. I., Chung E., Coticchia C. M., Steinert P. M., Markova N. G. The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res. 1997;25:4287–4295. doi: 10.1093/nar/25.21.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng A. Y., Waring P., Ristevski S., Wang C., Wilson T., Pritchard M., Hertzog P., Kola I. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida N., Yoshida S., Araie M., Handa H., Nabeshima Y. Ets family transcription factor ESE-1 is expressed in corneal epithelial cells and is involved in their differentiation. Mech. Dev. 2000;97:27–34. doi: 10.1016/s0925-4773(00)00419-6. [DOI] [PubMed] [Google Scholar]
- 15.Neve R., Chang C. H., Scott G. K., Wong A., Friis R. R., Hynes N. E., Benz C. C. The epithelium-specific ets transcription factor ESX is associated with mammary gland development and involution. FASEB J. 1998;12:1541–1550. doi: 10.1096/fasebj.12.14.1541. [DOI] [PubMed] [Google Scholar]
- 16.Bochert M. A., Kleinbaum L. A., Sun L. Y., Burton F. H. Molecular cloning and expression of Ehf, a new member of the ets transcription factor/oncoprotein gene family. Biochem. Biophys. Res. Commun. 1998;246:176–181. doi: 10.1006/bbrc.1998.8560. [DOI] [PubMed] [Google Scholar]
- 17.Silverman E. S., Baron R. M., Palmer L. J., Le L., Hallock A., Subramaniam V., Riese R. J., McKenna M. D., Gu X., Libermann T. A., et al. Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am. J. Respir. Cell Mol. Biol. 2002;27:697–704. doi: 10.1165/rcmb.2002-0011OC. [DOI] [PubMed] [Google Scholar]
- 18.Kas K., Finger E., Grall F., Gu X., Akbarali Y., Boltax J., Weiss A., Oettgen P., Kapeller R., Libermann T. A. ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J. Biol. Chem. 2000;275:2986–2998. doi: 10.1074/jbc.275.4.2986. [DOI] [PubMed] [Google Scholar]
- 19.Lapinskas E. J., Palmer J., Ricardo S., Hertzog P. J., Hammacher A., Pritchard M. A. A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem. Cell Biol. 2004;122:521–526. doi: 10.1007/s00418-004-0713-x. [DOI] [PubMed] [Google Scholar]
- 20.Tummala R., Sinha S. Differentiation-specific transcriptional regulation of the ESE-2 gene by a novel keratinocyte-restricted factor. J. Cell Biochem. 2006;97:766–781. doi: 10.1002/jcb.20685. [DOI] [PubMed] [Google Scholar]
- 21.Donnison M., Beaton A., Davey H. W., Broadhurst R., L'Huillier P., Pfeffer P. L. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–2308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J., Chehab R., Tkalcevic J., Naylor M. J., Harris J., Wilson T. J., Tsao S., Tellis I., Zavarsek S., Xu D., et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves B. J., Petersen J. M. Specificity within the ets family of transcription factors. Adv. Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 24.Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 25.Ray-Gallet D., Mao C., Tavitian A., Moreau-Gachelin F. DNA binding specificities of Spi-1/PU.1 and Spi-B transcription factors and identification of a Spi-1/Spi-B binding site in the c-fes/c-fps promoter. Oncogene. 1995;11:303–313. [PubMed] [Google Scholar]
- 26.Fisher R. J., Mavrothalassitis G., Kondoh A., Papas T. S. High-affinity DNA–protein interactions of the cellular ETS1 protein: the determination of the ETS binding motif. Oncogene. 1991;6:2249–2254. [PubMed] [Google Scholar]
- 27.Tummala R., Romano R. A., Fuchs E., Sinha S. Molecular cloning and characterization of AP-2 epsilon, a fifth member of the AP-2 family. Gene. 2003;321:93–102. doi: 10.1016/s0378-1119(03)00840-0. [DOI] [PubMed] [Google Scholar]
- 28.Sinha S., Degenstein L., Copenhaver C., Fuchs E. Defining the regulatory factors required for epidermal gene expression. Mol. Cell. Biol. 2000;20:2543–2555. doi: 10.1128/mcb.20.7.2543-2555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright W. E., Funk W. D. CASTing for multicomponent DNA-binding complexes. Trends Biochem. Sci. 1993;18:77–80. doi: 10.1016/0968-0004(93)90156-h. [DOI] [PubMed] [Google Scholar]
- 30.Yaniw D., Hu J. Epithelium-specific ets transcription factor 2 upregulates cytokeratin 18 expression in pulmonary epithelial cells through an interaction with cytokeratin 18 intron 1. Cell Res. 2005;15:423–429. doi: 10.1038/sj.cr.7290310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Feng L., Said M., Balderman S., Fayazi Z., Liu Y., Ghosh D., Gulick A. M. Analysis of the 2.0 A crystal structure of the protein–DNA complex of the human PDEF Ets domain bound to the prostate specific antigen regulatory site. Biochemistry. 2005;44:7095–7106. doi: 10.1021/bi047352t. [DOI] [PubMed] [Google Scholar]
- 32.Wei P., Taniguchi S., Sakai Y., Imamura M., Inoguchi T., Nawata H., Oda S., Nakabeppu Y., Nishimura J., Ikuyama S. Expression of adipose differentiation-related protein (ADRP) is conjointly regulated by PU.1 and AP-1 in macrophages. J. Biochem. (Tokyo) 2005;138:399–412. doi: 10.1093/jb/mvi136. [DOI] [PubMed] [Google Scholar]
- 33.Cahill M. A., Ernst W. H., Janknecht R., Nordheim A. Regulatory squelching. FEBS Lett. 1994;344:105–108. doi: 10.1016/0014-5793(94)00320-3. [DOI] [PubMed] [Google Scholar]
- 34.Triezenberg S. J. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 35.Chang C. H., Scott G. K., Baldwin M. A., Benz C. C. Exon 4-encoded acidic domain in the epithelium-restricted Ets factor, ESX, confers potent transactivating capacity and binds to TATA-binding protein (TBP) Oncogene. 1999;18:3682–3695. doi: 10.1038/sj.onc.1202674. [DOI] [PubMed] [Google Scholar]
- 36.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.