Abstract
The Gpbar1 [G-protein-coupled BA (bile acid) receptor 1] is a recently identified cell-surface receptor that can bind and is activated by BAs, but its physiological role is unclear. Using targeted deletion of the Gpbar1 gene in mice, we show that the gene plays a critical role in the maintenance of bile lipid homoeostasis. Mice lacking Gpbar1 expression were viable, developed normally and did not show significant difference in the levels of cholesterol, BAs or any other bile constituents. However, they did not form cholesterol gallstones when fed a cholic acid-containing high-fat diet, and liver-specific gene expression indicated that Gpbar1-deficient mice have altered feedback regulation of BA synthesis. These results suggest that Gpbar1 plays a critical role in the formation of gallstones, possibly via a regulatory mechanism involving the cholesterol 7α-hydroxylase pathway.
Keywords: bile acid, cholesterol, gall-bladder, gallstone, gene ablation, G-protein-coupled bile acid receptor 1 (Gpbar1)
Abbreviations: BA, bile acid; CA, cholic acid; CBC, complete blood count; CDCA, chenodeoxycholic acid; CGD, cholesterol gallstone disease; CSI, cholesterol saturation index; Cyp27a1, sterol 27-hydroxylase; Cyp7a1, cholesterol 7α-hydroxylase; D2, type 2 iodothyroninedeiodinase; ES cell, embryonic stem cell; FAM, 6-carboxyfluorescein; FGF, fibroblast growth factor; FGFR4, FGF receptor 4; FTF, α-fetoprotein transcription factor; FXR, farnesoid X receptor; Gpbar1, G-protein-coupled BA receptor 1; GPCR, G-protein-coupled receptor; LXR, liver X receptor α; NSS, normal sheep serum; PXR, pregnane X receptor; SHP, small heterodimer partner; TEA, triethanolamine; TG, triacylglycerol
INTRODUCTION
Bile acids (BAs) are cholesterol derivatives essential for the absorption of dietary lipids and fat-soluble vitamins [1–3]. BA biosynthesis represents a major pathway of cholesterol catabolism and excretion. The primary BAs, CA (cholic acid) and CDCA (chenodeoxycholic acid), are produced in the liver from cholesterol in multiple synthetic steps, initiated either by Cyp7a1 (cholesterol 7α-hydroxylase) of the classic/neutral pathway or by mitochondrial Cyp27a1 (sterol 27-hydroxylase) of the alternative/acidic pathway [1,2,4]. Most BAs are conjugated post-synthetically with taurine or glycine, excreted into bile and stored in the gall-bladder. After each meal, the gall-bladder contracts to expel bile into the intestine where BAs serve as detergents to facilitate the absorption of lipids. Upon completion of their function, BAs are absorbed from the intestine and returned to the liver, where they inhibit their own synthesis [5–7]. This feedback regulatory loop prevents the accumulation of high levels of BAs that can damage cell membranes, impair liver function and cause cholestasis and cirrhosis.
BA homoeostasis is regulated by several nuclear receptors, two of which, FXR (farnesoid X receptor) and PXR (pregnane X receptor), have been shown to bind BA [8–13]. Recently, a new cell-surface receptor has been cloned and shown to serve as a Gs-coupled receptor for BAs [14,15]. This novel BA receptor, named Gpbar1 (G-protein-coupled BA receptor 1; aka M-BAR/BG37 and TGR5), is located on chromosome 1qC3 and is only distantly related to other known GPCRs (G-protein-coupled receptors), the closest relative being hGPCR19 (human GPCR19) [15]. Cells stably transfected with human Gpbar1 are activated by a number of BAs, including lithocholic acid, deoxycholic acid, CDCA and CA [14,15]. The rank order of potency for these BAs is distinct from that for the nuclear BA receptors FXR, PXR and VDR (vitamin D receptor) [12,16], leading to the proposal that Gpbar1 is a novel membrane type BA receptor, distinct from the known BA nuclear receptors [14,15].
Although previous studies have revealed the ability of Gpbar1 to bind BAs, they did not elaborate on the role of this receptor in BA physiology [14,15]. We studied Gpbar1 gene expression in multiple mouse tissues and found that it is transcribed almost exclusively in the gall-bladder. To gain insight into its physiological function, we generated mice with targeted deletion of the Gpbar1 gene. In this paper, we show that mice lacking Gpbar1 expression are resistant to CGD (cholesterol gallstone disease) when fed a lithogenic diet. Gpbar1−/− (Gpbar1-null) mice on this diet also show alterations of several key cholesterol metabolism genes, including Cyp7a1 and Cyp27a1, pointing to potential mechanisms for the gallstone resistance.
EXPERIMENTAL
Construction of the Gpbar1 targeting vector and generation of Gpbar1 knockout mice
A DNA vector designed to remove the entire coding region of the Gpbar1 gene was constructed using sequencing information from GenBank® accession number AC098570. DNA fragments corresponding to the 5′ and 3′ regions of the Gpbar1 locus were subcloned into a vector at either end of the neomycin resistance gene (neo). This targeting vector was linearized using the restriction enzyme NotI, electroporated into 129S3/SvImJ-derived ES cells (embryonic stem cells), and colonies resistant to G418 were picked and expanded for DNA analysis. DNAs from these colonies were screened for targeted Gpbar1 gene by a PCR-based strategy using one primer (5′-agaggaagagctagcaagcaccacctg-3′) corresponding to a region upstream of the Gpbar1 DNA and another primer (5′-cgccccgactgcatctgcgtgtt-3′) corresponding to the neomycin resistance gene (neo) cassette. Out of 759 129S3/SvImJ ES cell clones analysed, three yielded the 1386 bp diagnostic PCR amplification product. The predicted structure of the targeted Gpbar1 locus in the PCR-positive cells was confirmed by Southern-blot analysis using probes that hybridize outside of and adjacent to the construct arms. Cells from several correctly targeted ES cell lines were injected into C57BL/6 blastocysts to generate chimaeric mice. Gpbar1 heterozygous (Gpbar1+/−) offspring were identified by a PCR-based screening strategy using three oligonucleotide primers in a multiplex reaction corresponding to the region of homology, the neo gene, and the deleted region of the Gpbar1 gene. These primers were designed to detect both wild-type (363 bp) and targeted (503 bp) alleles. Oligonucleotide sequences were as follows: Gpbar1 deleted region forward 5′-ctacgctagcgacagcacattatcactgaggctttg-3′, Gpbar1 arm of homology reverse 5′-tggccagttactgtcctctcttg-3′, neo 5-cgccccgactgcatctgcgtgtt-3′. Gpbar1+/− mice were then interbred to generate Gpbar1−/− mice. Disruption of Gpbar1 expression was confirmed by real-time quantitative PCR analysis.
Animals and diets
For the experiments described in this paper, we used Gpbar1−/− mice generated in a hybrid (129S3/SvImJ×C57BL/6) background. Mice were bred in house and age- and gender-matched animals were used between 6 and 12 weeks of age. All mice were housed in an SPF (specific pathogen-free) environment and all experiments with animals were conducted according to the Schering-Plough Animal Care and Use Committee guidelines for animal care.
Diets were prepared by Research Diets. Mice were maintained on normal chow, containing <0.02% cholesterol, until 8–10 weeks of age. Then wild-type and knockout littermates were fed a lithogenic diet (RD D12109), containing 1.2% (w/v) cholesterol, 0.5% CA and 15% total fat. After feeding the lithogenic diet for 8 weeks, cholecystectomy was performed and gall-bladder bile was aspirated completely and used for analyses.
Collection and microscopic analysis of gall-bladder bile specimens and gallstones
After 8 weeks on the lithogenic diet, the Gpbar1+/+ mice (n=33) and Gpbar−/− littermates (n=30) were fasted for 2 h but allowed free access to water. Before killing, animals were weighed. After cholecystectomy, gall-bladder was opened at the fundus, and bile was examined under polarized light microscopy for the presence of mucin gel, liquid crystals, cholesterol crystals and true stones, according to published criteria [17].
Plasma, bile and hepatic lipid measurements
Total and lipoprotein cholesterol plasma levels were determined by a modification of the cholesterol oxidase method of Allain et al. [18] using kit reagents (Wako Pure Chemical Industries, Osaka, Japan). TG (triacylglycerol) concentrations were determined by a modification of the lipase–glycerol phosphate oxidase method (GPO-Trinder; Sigma, St. Louis, MO, U.S.A.). Plasma lipoprotein profiles were determined by FPLC with a Pharmacia Superose 6 column. FPLC fraction cholesterol levels were determined using the Wako Cholesterol CII enzymatic colorimetric method (Wako Chemicals U.S.A., Richmond, VA, U.S.A.). The accumulation of hepatic free cholesterol and cholesteryl esters was used as a surrogate marker of cumulative cholesterol absorption and its inhibition in mice [19]. Hepatic free cholesterol and cholesteryl esters were extracted using a method described by Folch et al. [20]. Lipid extracts were dried under nitrogen into HPLC sample vials, resuspended in hexane/propan-2-ol, and assayed for cholesteryl ester and free cholesterol concentrations chromatographically as previously described [21].
Histological analysis
Mice were killed and various tissues were collected, immersion fixed in 10% (v/v) neutral-buffered formalin, paraffin-embedded, and 5–6 μm sections were stained by standard haematoxylin and eosin methods. All tissues were examined by light microscopy and graded for severity by one investigator. Slides were judged against concurrent controls for the severity of the findings.
Blood analysis
Gpbar1−/− and Gpbar1+/+ littermate mice were fasted for 2 h and anaesthetized, and blood was collected from the abdominal aorta. Blood chemistry and CBC (complete blood count) analyses were performed by AniLytics (Gaithersburg, MD, U.S.A.).
mRNA expression analysis
mRNA from various tissues was extracted utilizing the Ultraspec RNA isolation kit from Biotecx (Houston, TX, U.S.A.) following the manufacturer's instructions. cDNA was generated by reverse transcription using random hexamers (Promega, Madison, WI, U.S.A.) and oligo-dT primers (Life Technologies, Gaithersburg, MD, U.S.A.). Real-time quantitative PCR analysis was performed on an ABI 7700 sequence-detection instrument (TaqMan) following the manufacturer's instructions. For TaqMan analysis, 25 ng of cDNA was used together with primers at 0.9 μM final concentration and an FAM (6-carboxyfluorescein)-labelled diagnostic probe at a final concentration of 0.25 μM. Gpbar1 primers and probe sequences were as follows: forward primer 5′-cctttccctgcttgccaat-3′, reverse primer 5′-ccggagtggctgcaaca-3′, probe 5′-6-FAM-tgctgctggtgcatg-MGB-3′ (where MGB is minor groove binder). Primer sequences for the genes involved in the synthesis and metabolism of BAs, cholesterol, phospholipids and fatty acids are shown in Supplementary Table S1 (http://www.BiochemJ.org/bj/398/bj3980423add.htm). Ribosomal RNA primers and probe (PE Applied Biosystems, Foster City, CA, U.S.A.) were used as an internal control. Quantitative PCR conditions were as follows: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Plasmid containing the Gpbar1 gene was used as a standard, ranging from 1 ng to 1 fg. Data were analysed using Sequence Detection Systems software version 1.7.
In situ hybridization
RNA probes specific for mouse Gpbar1 were generated as follows: a plasmid containing the full-length ORF (open reading frame) of mouse Gpbar1 was used as a template to PCR-amplify a set of approx. 250 bp fragments for in vitro transcription. Sequences of the gene-specific primers containing T3 and T7 promoter sequences at the 5′-end were as follows: forward primer 5′-aattaaccctcactaaagggtcctgcctccttctccacttgac-3′, reverse primer 5′-gtaatacgactcactatagggcaggccataaacttccaggtagagg-3′. These PCR products were gel-purified and used for in vitro transcription and digoxigenin labelling according to the manufacturer's instructions (Digoxigenin RNA Labelling kit; Roche Molecular Biochemicals). Gall-bladder, liver and small intestine from Gpbar1+/+ and Gpbar1−/− mice were cryo-sectioned at 10 μm, thaw-mounted on to microscope slides and fixed with 4% (w/v) paraformaldehyde in 1× PBS for 20 min at room temperature (22 °C). The sections were then washed in 1× PBS, dehydrated through graded alcohols [75, 95 and 100% (v/v) ethanol] and stored at −80 °C until further processing. Prior to the in situ procedure, they were left at room temperature until thawed, and then rehydrated through 100, 95 and 75% ethanol and 1× PBS. The sections were treated with 0.2 M HCl in 1× PBS for 10 min at room temperature, washed in 1× PBS, and digested with 2 μg/ml proteinase K for 30 min at room temperature. The reaction was stopped with 4% paraformaldehyde and the sections were washed with 1× PBS. They were washed in 0.1 M TEA (triethanolamine) buffer and treated with 0.1 M TEA and 0.25% acetic anhydride for 2×5 min at room temperature, further washed in 1× PBS, and then dehydrated with graded alcohols as described above. Prehybridization was carried out for 2 h at 55 °C, in a buffer containing 50% (v/v) formamide, 2.5× Denhardt's solution (0.02% Ficoll 400, 0.02% polyvinylpyrrolidone and 0.02% BSA), 0.6 M NaCl, 10 mM Tris/HCl, 1 mM EDTA, 0.1% SDS and 10 mM dithiothreitol. The buffer was filtered and 0.25 ml of 100 mg/ml yeast tRNA was added per 50 ml of buffer. The tissues were hybridized overnight at 55 °C with hybridization buffer containing 1 ng/μl of the probe that was denatured at 50 °C and then put on ice prior to adding it to the buffer. On the following day, the sections were washed as follows: 4× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) for 2×15 min at room temperature, 2× SSC for 2×15 min at 37 °C, RNase treatment for 30 min at 37 °C, and finally 2× SSC and 50% formamide at 60 °C for 2×30 min. The antibody reaction for digoxigenin consisted of washing in buffer 1 (100 mM Tris/HCl, pH 7.5, and 150 mM NaCl) from the labelling kit for 5 min, blocking with 2% (v/v) NSS (normal sheep serum) in buffer 1 containing 0.1% Triton X-100 for 2 h at room temperature, and incubation in the anti-digoxigenin antibodies (1:500 in buffer 1 containing 1.0% NSS and 0.1% Triton X-100) overnight at 4 °C. On the next day, the sections were washed with buffer 1 for 2×15 min, followed by a change to buffer 2 (100 mM Tris/HCl, pH 9.5, 100 mM NaCl and 50 mM MgCl), and developed in NBT (Nitro Blue Tetrazolium)/BCIP (5-bromo-4-chloroindol-3-yl phosphate) for 4 h.
Statistical analysis
GraphPad Prism version 4 was used for graphics (GraphPad Software, San Diego, CA, U.S.A.). Results shown are means±S.D. Statistical comparisons for significance between groups were performed using the Student's t test.
RESULTS
Gpbar1 is highly expressed in mouse gall-bladder
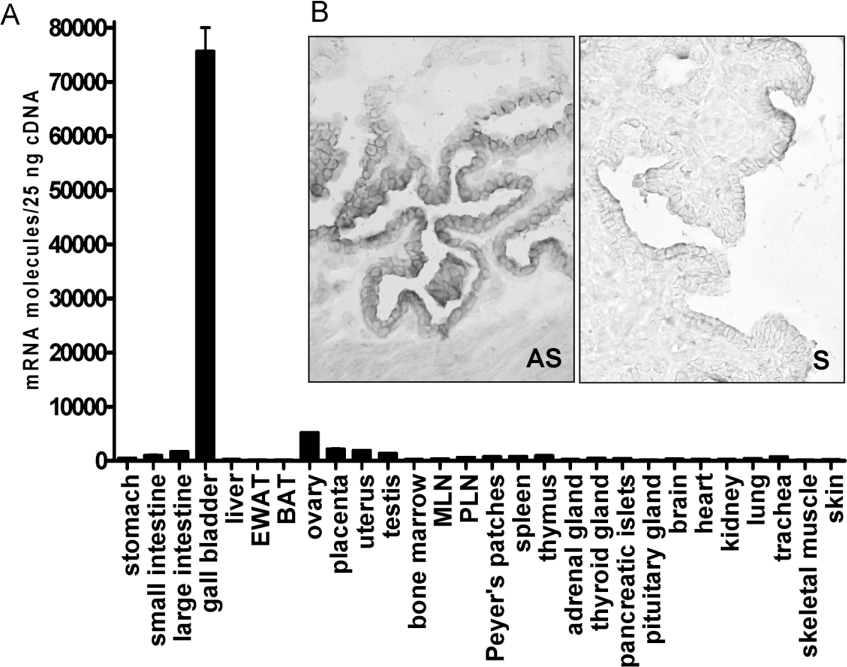
Real-time quantitative PCR analysis was carried out to assess the expression levels of Gpbar1 mRNA in 26 different mouse tissues. Although Gpbar1 is a BA-responsive receptor and BAs are stored in the gall-bladder, previous analyses have not examined its expression in this tissue. Gpbar1 was expressed predominantly in the gall-bladder where its mRNA level was 60–100 times greater than in all the other tissues analysed (Figure 1A). In situ hybridization histochemistry in gall-bladder revealed that hybridization signals for Gpbar1 antisense probe were restricted to epithelial cells. Hybridization of the sense probe for Gpbar1 resulted in only background labelling of the tissue sections (Figure 1B). Cellular expression of the Gpbar1 transcript in the other tissues examined, including intestine, was also limited to epithelial cells (results not shown).
Figure 1. Expression analysis of Gpbar1 gene.
(A) Real-time quantitative PCR analysis (TaqMan) was performed using C57BL/6 mouse tissues from three mice and in duplicates. EWAT, epididymal white adipose tissue; BAT, brown adipose tissue; MLN, mesenteric lymph nodes; PLN, peripheral lymph nodes. (B) In situ hybridization: gall-bladder. Magnification, ×40. AS, antisense probe; S, sense probe.
Targeted deletion of the Gpbar1 gene and generation of knockout mice
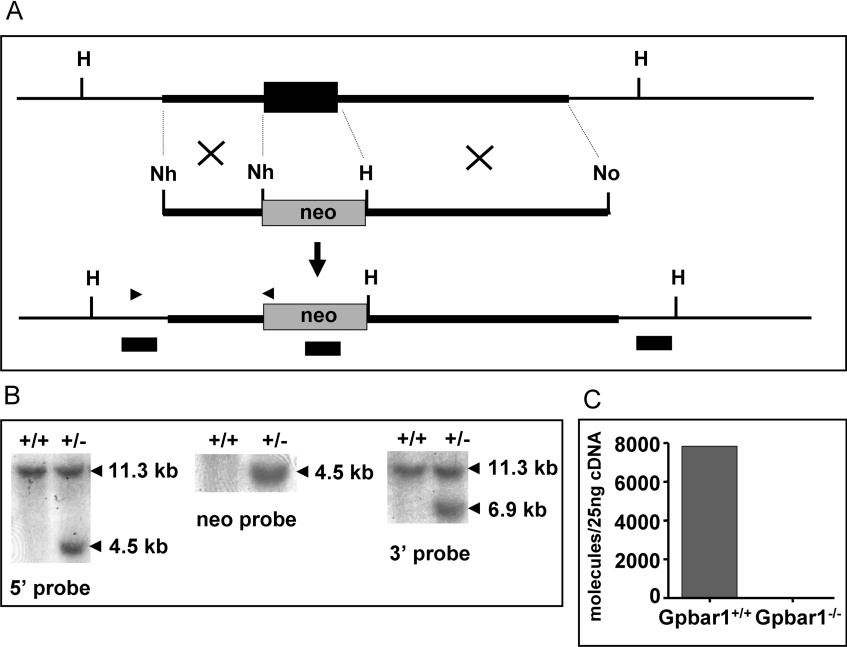
A gene-targeting vector was constructed using Gpbar1 genomic DNA (Figure 2A) and used to transfect ES cells. The vector was designed to allow for excision of the entire coding region of Gpbar1 after homologous recombination with its gene locus. Two independent 129S3/SvImJ-derived ES cell clones containing the targeted locus were identified by Southern-blot analysis (Figure 2B) and injected into C57BL/6 mouse blastocysts. Chimaeras obtained from these clones were bred with C57BL/6 mice to generate heterozygotes (Gpbar1+/−). PCR analysis of tail DNA from these mice confirmed germline transmission of the targeted allele. Further intercrossing of Gpbar1 heterozygous mice yielded wild-type (Gpbar1+/+), heterozygous (Gpbar1+/−) and homozygous (Gpbar1−/−) mice in the expected Mendelian ratio of 1:2:1, demonstrating that Gpbar1 expression is not essential for embryonic development. Real-time quantitative PCR analysis of mRNA prepared from the gall-bladders of Gpbar1+/+ and Gpbar1−/− mice confirmed that Gpbar1 expression was detected in the Gpbar1+/+ mouse, but not in any of the Gpbar1−/− mouse lines (Figure 2C).
Figure 2. Gene targeting of Gpbar1.
(A) Top: wild-type Gpbar1 locus; black rectangle represents the exon; transcription is from left to right. Middle: targeting vector; thick lines represent regions of homology to Gpbar1 and shaded rectangle represents neo cassette. The restriction enzyme sites used to subclone these regions are indicated; H, HindIII; No, NotI; Nh, NheI. Bottom: Gpbar1 targeted locus; the positions of the oligonucleotide primers used to screen targeted ES cells are indicated with black arrowheads. (B) DNA probes (black rectangles) from 5′ upstream and 3′ downstream homologous regions of Gpbar1 gene and neo were used to screen the ES cells. An 11.3 kb and a 4.5 kb HindIII fragment were expected for the wild-type Gpbar1 locus and for the targeted locus respectively when the 5′ probe was used. An 11.3 kb and a 6.9 kb HindIII fragment were expected for the wild-type Gpbar1 locus and for the targeted locus respectively when the 3′ probe was used. (C) Real-time quantitative PCR analysis of Gpbar1 RNA from the gall-bladders of wild-type and knockout mice. No Gpbar1 mRNA is detected in the gall-bladders of Gpbar1−/− mice.
Gpbar1−/− mice are viable and develop normally
The Gpbar1−/− mice appeared healthy and were fertile. To determine if any abnormalities would become apparent in older mice, Gpbar1−/− and Gpbar1+/+ littermate mice were followed until 20 months of age, but no abnormalities were detected upon gross examination.
CBC and chemistry tests were performed on Gpbar1−/− mice and Gpbar1+/+ littermates. These analyses did not reveal statistically significant differences between Gpbar1−/− mice and Gpbar1+/+ littermate mice with regard to CBC, cholesterol, BAs, TGs, HDL (high-density lipoprotein), LDL (low-density lipoprotein) or bilirubin (results not shown). Similarly, no differences were observed in the urine analysis, including glucose, bilirubin, ketone, specific gravity, blood, pH, protein, urobilinogen, nitrite and leucocytes (results not shown).
To determine whether the absence of Gpbar1 results in abnormalities of any major tissue, we examined formalin-fixed tissue sections of Gpbar1−/− and Gpbar1+/+ littermate mice by light microscopy. Routine histological analysis of all organs was unremarkable. No inflammation or developmental abnormalities were noted in the gall-bladder, liver, stomach, duodenum, jejunum, ileum, colon, lung, kidney, brain, heart, thymus, spleen, lymph nodes, pancreas, gonads or muscle (results not shown), suggesting that Gpbar1 is not required for the development of these organs.
Gpbar1−/− mice fed lithogenic diet do not develop gallstones
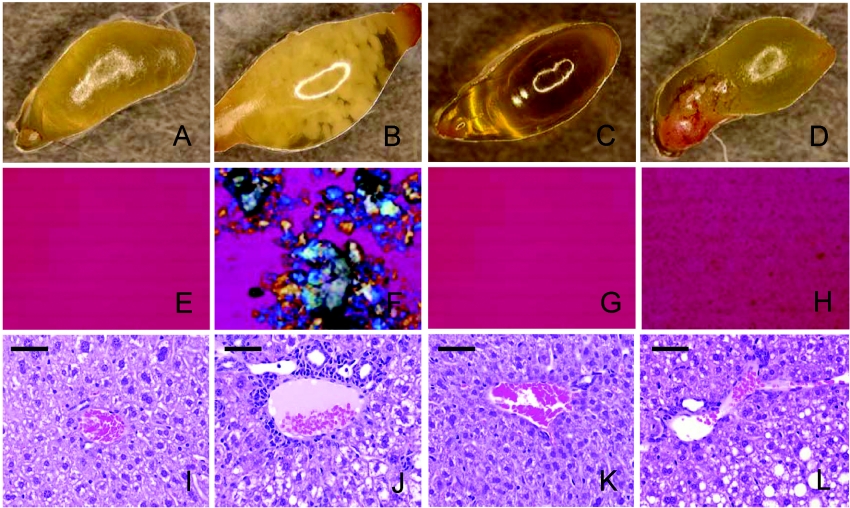
Given the high expression of Gpbar1 in the gall-bladder, the focus of the initial analysis was placed on this organ. Since cholelithiasis is the most common disease of the gall-bladder, we studied the susceptibility of Gpbar1−/− mice to induced cholesterol gallstone formation. Gpbar1−/− (n=30) and Gpbar1+/+ (n=33) littermates were fed a lithogenic diet for 8 weeks. BW (body weights) of both groups were similar at the start of the study, and animals from both groups gained similar weight on the lithogenic diet, with a tendency for Gpbar1−/− mice to be slightly lighter than their wild-type littermates at the end of the study (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/398/bj3980423add.htm). Upon completion of the treatment, animals were killed and their gall-bladders were removed. Bile was collected and examined microscopically for the presence of cholesterol crystals. White gallstones were clearly visible through the gall-bladder wall in 54% of the Gpbar1+/+ mice, but not in any of the Gpbar1−/− littermates (Figures 3A–3D). Histopathological analysis revealed that the gall-bladders of Gpbar1+/+ mice contained thick layers of mucus gel, interspersed with cholesterol monohydrate crystals, aggregated liquid crystals and real gallstones (Figure 3F). In contrast, gall-bladders of Gpbar1−/− littermate mice appeared either clear or formed small crystals (Figure 3H).
Figure 3. Biliary cholesterol crystallization and liver histopathology.
Gall-bladder from (A) Gpbar1+/+ mice on chow, (B) Gpbar1+/+ mice on lithogenic diet, (C) Gpbar1−/− mice on chow and (D) Gpbar1−/− mice on lithogenic diet. Polarizing light microscopy of gall-bladder bile from (E) Gpbar1+/+ mice on chow, (F) Gpbar1+/+ mice on lithogenic diet, (G) Gpbar1−/− mice on chow and (H) Gpbar1−/− mice on lithogenic diet. Histological images of liver sections in portal triad from (I) Gpbar1+/+ mice on chow, (J) Gpbar1+/+ mice on lithogenic diet, (K) Gpbar1−/− mice on chow and (L) Gpbar1−/− mice on lithogenic diet. Magnification, ×10 (A–D) and ×400 (E–H). (I–L) Scale bar, 50 μm.
Gpbar1+/+ and Gpbar1−/− animals administered the lithogenic diet had varying degrees of hepatocyte vacuolization. Gpbar1+/+ livers showed the expected bile duct hyperplasia, characterized by bile ducts that were prominent, mildly ectatic, and lined by hyperplastic epithelium characterized with occasional piling up and increased mitotic activity. However, when compared with their wild-type littermates, Gpbar1−/− mice showed a lower degree of bile duct hyperplasia (Figures 3I–3L).
Gpbar1−/− mice have lower bile CSI (cholesterol saturation index)
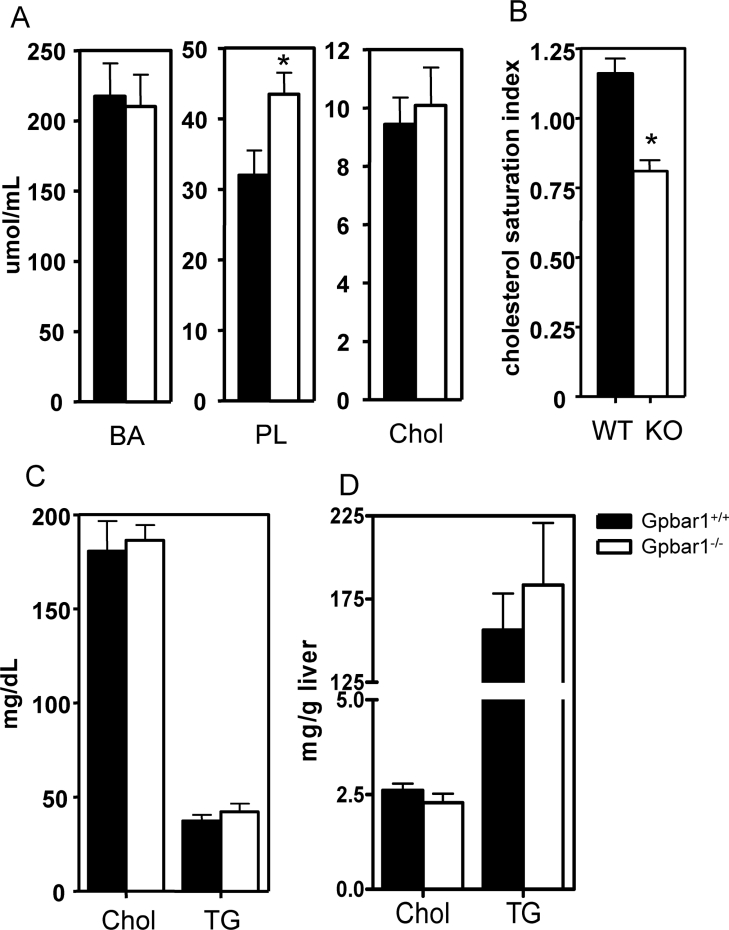
Bile was collected and grouped into pools to determine bile lipid composition and CSI calculated from the critical tables [22] (Figures 4A and 4B). Phospholipid levels were significantly higher in Gpbar1−/− mice fed a lithogenic diet (43.5±7.5 μmol/ml) compared with Gpbar1+/+ controls (32.0±9.3 μmol/ml), P=0.03 (Figure 4A). As a consequence of the altered lipid levels, the CSI values were significantly lower in Gpbar1−/− mice fed a lithogenic diet (CSI=0.81±0.08) compared with Gpbar1+/+ controls (CSI=1.16±0.12, P=0.03) (Figure 4B). Cholesterol and TG levels in blood and hepatic samples were not significantly different between Gpbar1−/− and Gpbar1+/+ littermates (Figures 4C and 4D).
Figure 4. Lipid profiles of mice fed on lithogenic diet.
(A) Lipid compositions of gall-bladder bile. (B) CSI values of gall-bladder bile. (C) Serum cholesterol and TG levels. (D) Hepatic cholesterol and TG levels. Individual values were measured in from six to eight mice per group on lithogenic diet. Error bars represent S.D. *P<0.05 when compared with wild-type samples. PL, phospholipids; Chol, cholesterol.
The negative feedback regulation of BA synthesis is altered in Gpbar1−/− mice
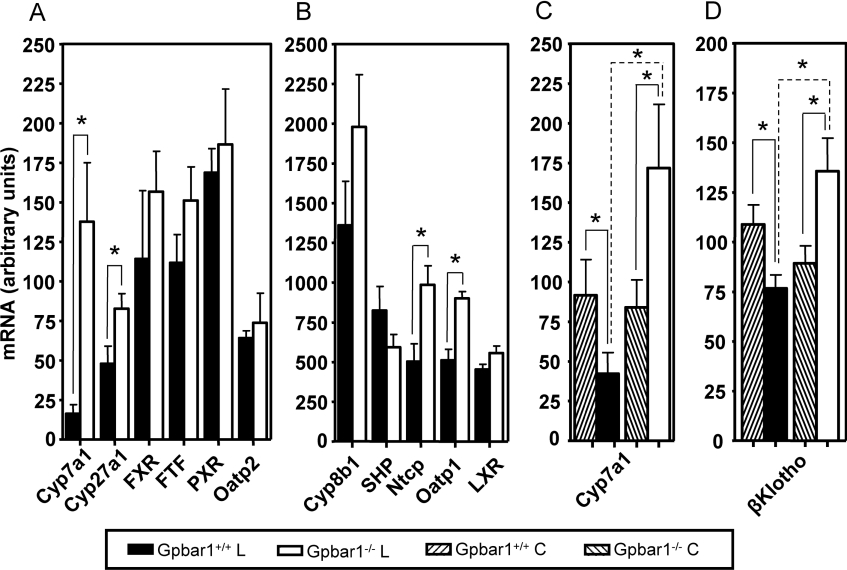
In order to investigate the molecular basis of the resistance of Gpbar1−/− mice to CA-induced gallstone formation, we performed quantitative PCR analysis of 22 genes involved in cholesterol homoeostasis on mRNA isolated from Gpbar1−/− and Gpbar1+/+ mouse livers and gall-bladders fed either chow or lithogenic diet. The changes in the expression levels of these genes in Gpbar1+/+ mice on chow and CA diet were in agreement with published results [23–26] (results not shown). When compared with their wild-type littermates, Gpbar1−/− mice on CA diet showed significantly higher hepatic expression levels in four genes involved in BA synthesis (Cyp7a1 and Cyp27a1), and in hepatocellular uptake [Ntcp1 (Na+-taurocholate-co-transporting protein 1) and Oatp1 (organic anion-transporting polypeptide 1)] (Figures 5A and 5B). Cyp7a1 is the rate-limiting enzyme in the elimination of cholesterol via conversion into BAs. As expected, upon feeding the lithogenic diet, Cyp7a1 mRNA expression levels were decreased in Gpbar1+/+ mice. In contrast, Cyp7a1 expression levels were significantly up-regulated in Gpbar1−/− littermate mice (Figure 5C), indicating that Gpbar1 is involved in the regulation of Cyp7a1 expression. These results are in agreement with previous observations that transgenic mice with constitutive liver-specific expression of Cyp7a1 are protected from gallstone formation [27].
Figure 5. Gene expression analysis.
Real-time quantitative PCR analysis was performed on mRNA from livers of Gpbar1−/− and Gpbar1+/+ littermate mice. (A, B) BA synthesis-, regulation- and uptake-related genes. Individual values were measured from six mice per group on lithogenic diet. (C) Cyp7a1 expression and (D) βKlotho expression. Individual values were measured from nine mice per group on chow (C) and lithogenic (L) diet. Error bars represent S.D. *P<0.05 when compared with wild-type samples.
The hepatic expression profile of an additional 11 genes known to be implicated in the synthesis and metabolism of cholesterol, phospholipids and fatty acids (Abca1, Hmgcr, Ldlr, Scarb1, Fasn, Lpl and Ptcp), and in canalicular bile secretion (Abcb11, Abcb4, Abcg5 and Abcg8), was not altered in Gpbar1−/− mice (see Supplementary Figures S2A and S2B at http://www.BiochemJ.org/bj/398/bj3980423add.htm).
At the same time, the expression profile of all 33 genes involved in the BA, cholesterol and phospholipids metabolism was not significantly affected in the gall-bladders of Gpbar1−/− mice (results not shown).
Recently, it has been shown that mice lacking the βKlotho gene are resistant to gallstone formation [28]. We analysed the mRNA expression levels of βKlotho in the livers of Gpbar1+/+ and Gpbar1−/− mice fed chow and lithogenic diet. Interestingly, βKlotho expression levels were decreased in Gpbar1+/+ mice on lithogenic diet, but significantly up-regulated in Gpbar1−/− littermate mice (Figure 5D).
DISCUSSION
CGD is one of the most common disorders of the GI (gastro-intestinal) tract, but its aetiology is poorly understood. The genetics of this disease is complex because biliary cholesterol homoeostasis is regulated by multiple genes [29]. Formation of cholesterol microcrystals from supersaturated bile is considered a critical step in gallstone formation. Cholesterol precipitates in bile when its concentration relative to BA and phospholipids becomes excessive [27,30]. Defects in cholesterol or BA metabolism are likely to be involved in the development of cholelithiasis. Gpbar1 is a recently cloned putative BA receptor gene shown to bind and internalize BAs. It is the first identified membrane-bound BA receptor that belongs to the GPCR family. However, the functional role of Gpbar1 in vivo is not known.
Our analysis of the Gpbar1 expression pattern in mice showed that the gene is transcribed at high levels in the gall-bladder, suggesting a functional role in this organ. Gpbar1 gene deletion in mice revealed that its product plays an important function in the formation of gallstones. Gpbar1−/− mice were viable, developed normally and had no physiological abnormalities when compared with their normal Gpbar1+/+ littermates. Further analysis of bile from Gpbar1−/− and Gpbar1+/+ animals did not show any significant differences in the content of cholesterol, BAs and phospholipids. However, when challenged with a CA-containing high-fat diet, Gpbar1−/− mice responded differently than their normal Gpbar1+/+ littermates. In mice with normal expression of Gpbar1 gene, 54% developed cholelithiasis with gallstones of cholesterol type. In contrast, mice with targeted disruption of the gene did not show a single occurrence of gallstones. This dramatic difference in response to lithogenic diet clearly indicates that gallstone formation during increased consumption of a high-fat CA diet requires a functional Gpbar1 gene.
Since the molecular mechanisms of gallstone formation are still poorly understood, it is difficult to explain the role of Gpbar1 in this process. Cholesterol is soluble in bile where it is incorporated in mixed micelles with bile salts and phospholipids. Cholesterol precipitation and crystallization from supersaturated bile are known to be affected by the ratio between bile salts and phospholipids [31,32]. In supersaturated bile, excess cholesterol is kept in vesicles, consisting of spherical bilayers of cholesterol and phospholipids. In the presence of excess phospholipids, resulting in a low bile salt/phospholipids ratio, excess cholesterol is solubilized in vesicles and crystal formation is slow. When bile salts are in excess, resulting in a high bile salt/phospholipid ratio, vesicles are absent and cholesterol crystals precipitate at a faster rate. In the biles of Gpbar1+/+ mice fed a lithogenic diet, the high bile salt/phospholipid ratio resulted in cholesterol crystallization and formation of gallstones. In contrast, Gpbar1−/− mice produced an excess of phospholipids in their gall-bladders, resulting in a low bile salt/phospholipids ratio unfavourable for gallstone formation. At this time, it is not clear how Gpbar1 activity could contribute to increased phospholipid levels.
In the present study, we have also shown that the loss of Gpbar1 function impairs the negative feedback regulation of BA synthesis, resulting in the failure of Gpbar1−/− mice to repress Cyp7a1 mRNA in response to CA feeding. Multiple regulatory pathways have been proposed for the inhibition of BA synthesis through the suppression of Cyp7a1 activity when BA concentrations increase by enterohepatic circulation [1,2,4,33,34]. In one of these receptor-mediated mechanisms, also referred to as the SHP (small heterodimer partner)-dependent mechanism, BA-activated FXR induces SHP, which in turn interacts with FTF (α-fetoprotein transcription factor) and inhibits Cyp7a1 gene transcription. It has been reported that mice with targeted deletion of FXR and SHP genes have increased Cyp7a1 expression [35–37]. In addition, several negative feedback regulatory pathways independent of SHP have been proposed. In one of these receptor-mediated pathways, Cyp7a1 activity is repressed by PXR [2]. In another proposed negative feedback suppression of BA synthesis, FGFR4 [FGF (fibroblast growth factor) receptor 4] and its ligand, FGF15, have been implicated as components of a gut–liver signalling pathway acting through hepatic SHP to control Cyp7a1 expression [38,39]. The lack of differences in the expression of the three key components of the SHP-dependent pathway (FXR, SHP and FTF) (Figures 5A and 5B), as well as in the expression levels of PXR (Figure 5A) and FGFR4 (results not shown) in the livers of Gpbar1+/+ and Gpbar1−/− mice fed a CA diet, does not support a role of Gpbar1 in these mechanisms. There were also no significant differences between Gpbar1−/− mice and their wild-type littermates in the expression levels of LXR (liver X receptor α), which is a known feedforward regulator of Cyp7a1 activity (Figure 5B). In addition, we did not observe any significant difference in the expression levels of the FXR target genes (Abcb11 and Abcb4) and of the genes controlled by LXR (Abcg5 and Abcg8) (see Supplementary Figure S2B at http://www.BiochemJ.org/bj/398/bj3980423add.htm).
Another recently proposed BA synthesis control pathway involves a yet unclear mechanism implicating βKlotho in Cyp7a1 regulation [28,33]. Since mice lacking the βKlotho gene are resistant to gallstone formation, we analysed the mRNA expression levels of βKlotho in the livers of Gpbar1+/+ and Gpbar1−/− mice fed chow and lithogenic diet. Interestingly, βKlotho expression levels were decreased in Gpbar1+/+ mice fed on a lithogenic diet, but significantly up-regulated in Gpbar1−/− littermate mice (Figure 5D). At this point, it is not clear how Gpbar1 regulates the expression of Cyp7a1 and whether it acts through one of the established regulatory pathways. Further studies of Cyp7a1 regulation in Gpbar1−/− mice may help to elucidate the molecular mechanisms of the negative feedback control.
We have shown that the hepatic expression levels of the Cyp7a1, Cyp27a1, Ntcp1 and Oatp1 genes are significantly higher in Gpbar1−/− mice than in their wild-type littermates when fed a CA diet. At this point, however, it is unclear whether or not these alterations are due to a direct role of the Gpbar1 gene, even though its expression in the liver is very low. Since the expression levels of the genes implicated in BA, cholesterol and phospholipid metabolism are not significantly altered in the gall-bladders of Gpbar1−/− mice, one can speculate that the changes in the hepatic gene expression levels are due to a signalling factor secreted from the gall-bladder.
Recently, Watanabe et al. [40] suggested that Gpbar1 regulates the energy expenditure through the induction of cAMP-dependent thyroid hormone activating enzyme D2 (type 2 iodothyroninedeiodinase). In their study, administration of BAs to mice caused increased energy expenditure in brown adipose tissue, preventing obesity. Diet supplementation with CA reversed 120 days of diet-induced weight gain in wild-type mice. In contrast, CA addition to a high-fat diet did not show an effect on mice deficient in D2 enzyme. In the same study, in vitro analysis of cultured cells suggested that these effects are mediated by increased cAMP production that stems from the binding of BAs with Gpbar1. Watanabe et al. [40] were unable to knock down the Gpbar1 signal with shRNAs (short hairpin RNAs) and these findings have not been confirmed in vivo. Our results do not reveal significant differences in weight gain between Gpbar1−/− mice and their wild-type littermates after 8 weeks on lithogenic diet, although Gpbar1−/− mice were slightly lighter than Gpbar1+/+ littermates at the end of the study (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/398/bj3980423add.htm). Further in vivo studies are required to define whether Gpbar1 function is required for the regulation of energy expenditure.
In summary, we have demonstrated that the targeted deletion of the Gpbar1 gene in mice is well tolerated and does not appear to cause any physiological abnormalities when the mice are not challenged. The marked reduction in gallstone development in Gpbar1−/− animals on a lithogenic diet suggests that Gpbar1 may represent a novel therapeutic target for prevention or treatment of cholelithiasis.
Online data
Acknowledgments
We thank Drs M. Bayne and H. Lan (Department of Discovery Technologies, Schering-Plough Research Institute, Kenilworth, NJ, U.S.A.) for helpful discussions and advice.
References
- 1.Chiang J. Y. Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G349–G356. doi: 10.1152/ajpgi.00417.2002. [DOI] [PubMed] [Google Scholar]
- 2.Chiang J. Y. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Russell D. W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 4.Davis R. A., Miyake J. H., Hui T. Y., Spann N. J. Regulation of cholesterol-7alpha-hydroxylase: barely missing a SHP. J. Lipid Res. 2002;43:533–543. [PubMed] [Google Scholar]
- 5.Fuchs M. Bile acid regulation of hepatic physiology: III. Regulation of bile acid synthesis: past progress and future challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G551–G557. doi: 10.1152/ajpgi.00468.2002. [DOI] [PubMed] [Google Scholar]
- 6.Redinger R. N. Nuclear receptors in cholesterol catabolism: molecular biology of the enterohepatic circulation of bile salts and its role in cholesterol homeostasis. J. Lab. Clin. Med. 2003;142:7–20. doi: 10.1016/S0022-2143(03)00088-X. [DOI] [PubMed] [Google Scholar]
- 7.Ory D. S. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home? Circ. Res. 2004;95:660–670. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- 8.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 9.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 11.Kliewer S. A., Moore J. T., Wade L., Staudinger J. L., Watson M. A., Jones S. A., McKee D. D., Oliver B. B., Willson T. M., Zetterstrom R. H., et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 12.Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin B., Kliewer S. A. Nuclear receptors. I. Nuclear receptors and bile acid homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;282:G926–G931. doi: 10.1152/ajpgi.00044.2002. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Itadani H., Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem. Biophys. Res. Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 15.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. A G-protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 16.Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 17.Wang D. Q., Paigen B., Carey M. C. Phenotypic characterization of Lith genes that determine susceptibility to cholesterol cholelithiasis in inbred mice: physical-chemistry of gallbladder bile. J. Lipid Res. 1997;38:1395–1411. [PubMed] [Google Scholar]
- 18.Allain C. C., Poon L. S., Chan C. S., Richmond W., Fu P. C. Enzymatic determination of total serum cholesterol. Clin. Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 19.Salisbury B. G., Davis H. R., Burrier R. E., Burnett D. A., Bowkow G., Caplen M. A., Clemmons A. L., Compton D. S., Hoos L. M., McGregor D. G., et al. Hypocholesterolemic activity of a novel inhibitor of cholesterol absorption, SCH 48461. Atherosclerosis. 1995;115:45–63. doi: 10.1016/0021-9150(94)05499-9. [DOI] [PubMed] [Google Scholar]
- 20.Folch J., Lees M., Sloane Stanley G. H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 21.Burrier R. E., Smith A. A., McGregor D. G., Hoos L. M., Zilli D. L., Davis H. R., Jr The effect of acyl CoA: cholesterol acyltransferase inhibition on the uptake, esterification and secretion of cholesterol by the hamster small intestine. J. Pharmacol. Exp. Ther. 1995;272:156–163. [PubMed] [Google Scholar]
- 22.Carey M. C. Critical tables for calculating the cholesterol saturation of native bile. J. Lipid Res. 1978;19:945–955. [PubMed] [Google Scholar]
- 23.Wang R., Lam P., Liu L., Forrest D., Yousef I. M., Mignault D., Phillips M. J., Ling V. Severe cholestasis induced by cholic acid feeding in knockout mice of sister of P-glycoprotein. Hepatology. 2003;38:1489–1499. doi: 10.1016/j.hep.2003.09.037. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Han Y., Kim C. S., Lee Y. K., Moore D. D. Resistance of SHP-null mice to bile acid-induced liver damage. J. Biol. Chem. 2003;278:44475–44481. doi: 10.1074/jbc.M305258200. [DOI] [PubMed] [Google Scholar]
- 25.Luo Y., Liang C. P., Tall A. R. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J. Biol. Chem. 2001;276:24767–24773. doi: 10.1074/jbc.M100912200. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyake J. H., Duong-Polk X. T., Taylor J. M., Du E. Z., Castellani L. W., Lusis A. J., Davis R. A. Transgenic expression of cholesterol-7-alpha-hydroxylase prevents atherosclerosis in C57BL/6J mice. Arterioscler. Thromb. Vasc. Biol. 2002;22:121–126. doi: 10.1161/hq0102.102588. [DOI] [PubMed] [Google Scholar]
- 28.Ito S., Fujimori T., Furuya A., Satoh J., Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D. Q., Afdhal N. H. Genetic analysis of cholesterol gallstone formation: searching for Lith (gallstone) genes. Curr. Gastroenterol. Rep. 2004;6:140–150. doi: 10.1007/s11894-004-0042-1. [DOI] [PubMed] [Google Scholar]
- 30.Admirand W. H., Small D. M. The physicochemical basis of cholesterol gallstone formation in man. J. Clin. Invest. 1968;47:1043–1052. doi: 10.1172/JCI105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D. Q., Carey M. C. Complete mapping of crystallization pathways during cholesterol precipitation from model bile: influence of physical-chemical variables of pathophysiologic relevance and identification of a stable liquid crystalline state in cold, dilute and hydrophilic bile salt-containing systems. J. Lipid Res. 1996;37:606–630. [PubMed] [Google Scholar]
- 32.van Erpecum K. J. Cholesterol-gallstone formation: more than a biliary lipid defect? J. Lab. Clin. Med. 2004;144:121–123. doi: 10.1016/j.lab.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Moschetta A., Kliewer S. A. Weaving betaKlotho into bile acid metabolism. J. Clin. Invest. 2005;115:2075–2077. doi: 10.1172/JCI26046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angelin B. Telling the liver (not) to make bile acids: a new voice from the gut? Cell Metab. 2005;2:209–210. doi: 10.1016/j.cmet.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 36.Kerr T. A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D. W., Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., Moore D. D. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 38.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Yu C., Wang F., Jin C., Huang X., McKeehan W. L. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J. Biol. Chem. 2005;280:17707–17714. doi: 10.1074/jbc.M411771200. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature (London) 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.