Abstract
The LASS (longevity assurance homologue) family members are highly conserved from yeasts to mammals. Five mouse and human LASS family members, namely LASS1, LASS2, LASS4, LASS5 and LASS6, have been identified and characterized. In the present study we cloned two transcriptional variants of hitherto-uncharacterized mouse LASS3 cDNA, which encode a 384-amino-acid protein (LASS3) and a 419-amino-acid protein (LASS3-long). In vivo, [3H]dihydrosphingosine labelling and electrospray-ionization MS revealed that overproduction of either LASS3 isoform results in increases in several ceramide species, with some preference toward those having middle- to long-chain-fatty acyl-CoAs. A similar substrate preference was observed in an in vitro (dihydro)ceramide synthase assay. These results indicate that LASS3 possesses (dihydro)ceramide synthesis activity with relatively broad substrate specificity. We also found that, except for a weak display in skin, LASS3 mRNA expression is limited almost solely to testis, implying that LASS3 plays an important role in this gland.
Abbreviations: ESI-MS, electrospray-ionization MS; FBS, fetal bovine serum; FB1, fumonisin B1; 3×FLAG, triple FLAG epitope; HA, haemagglutinin; HEK, human embryonic kidney; LASS, longevity assurance homologue; RT, reverse transcription; TM, transmembrane
INTRODUCTION
Ceramide, the structural backbone of sphingolipids, is also a bioactive lipid that functions in cellular processes such as apoptosis, differentiation and the cell cycle [1–3]. It is produced either through the conversion of sphingosine in the salvage pathway, or by de novo synthesis, in which dihydroceramide is desaturated [4]. Both pathways are dependent on (dihydro)ceramide synthase, which catalyses an amide linkage between a fatty acyl-CoA and a sphingoid base (sphingosine for ceramide and dihydrosphingosine for dihydroceramide).
Past studies of (dihydro)ceramide synthesis have identified roles for proteins belonging to a family named LASS (longevity assurance homologue), which includes yeast Lag1 and its homologue Lac1 [5,6], and six mammalian LASS family members (LASS1–LASS6) [4]. In particular, Lag1, Lac1 and the recently identified single-span membrane protein Lip1 were identified as forming a hetero-oligomer that acts as a yeast (dihydro)ceramide synthase [7]. Although Δlag1 cells and Δlac1 cells each produce dihydroceramide normally, Δlag1Δlac1 cells and Δlip1 cells completely lack acyl-CoA-dependent (dihydro)ceramide synthase activities [5,6]. No mammalian Lip1 homologue has been identified; however, purified LASS5 alone does exhibit (dihydro)-ceramide synthase activity [8]. This finding suggests that, in the yeast (dihydro)ceramide synthase complex, the LASS family members Lag1 and Lac1 are the catalytic subunits.
To date, five of the six mammalian LASS family members (LASS1, LASS2, LASS4, LASS5 and LASS6) have been characterized [9–12]. Overproduction of a single LASS protein results in an increase in one or more specific ceramide species, which differ in fatty-acid chain length (LASS1, C18:0; LASS5 and LASS6, C16:0; and LASS2 and LASS4, C22:0 and C24:0) [9]. Thus LASS proteins possess specific fatty acyl-CoA preferences. By contrast, all can use sphingosine or dihydrosphingosine as the long-chain base substrate. Furthermore, LASS family members exhibit different tissue-specific expression patterns [9,12]. The presence of specific LASS proteins in certain tissues, together with the availability of fatty acyl-CoAs, may contribute to the production of tissue-specific ceramides.
In the present study we have cloned two transcriptional variants of mouse LASS3 cDNA. These LASS3 isoforms differ in transcriptional initiation sites and thus produce two polypeptides of different length, LASS3 and LASS3-long. When expressed in HEK-293T (human embryonic kidney 293T) cells, both LASS3 isoforms increased the production of several ceramide species, including C18:0- and C24:0-ceramides. Thus LASS3 exhibits a weaker substrate preference than the other LASS proteins [9–12]. In addition, the expression pattern of LASS3 was found to be highly testis-specific, implying that LASS3 has important roles in testis function, such as sperm formation and androgen production.
MATERIALS AND METHODS
Cell culture and transfection
HEK-293T cells were grown in Dulbecco's modified Eagle's medium containing 10% (v/v) FBS (fetal bovine serum) in a humidified atmosphere of 5% CO2 at 37 °C. Cells were transfected with the indicated cDNAs using Lipofectamine® Plus (Invitrogen, Carlsbad, CA, U.S.A.), according to the manufacturer's directions. Lysates of transiently transfected cells were prepared 24 h after transfection.
Yeast strains and media
Saccharomyces cerevisiae strains used included the diploid strain KA31 (MATa/α ura3/ura3 leu2/leu2 his3/his3 trp1/trp1) and the haploid strain KA31-1A (MATa ura3 lue2 his3 trp1) [13]. KA31 was used to prepare the heterozygous diploid strain YMY08 (LIP1/Δlip1::LEU2), and KA31-1A was used for the haploid strain YMY16 (Δlip1::LEU2). The cells were grown either in YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone and 2% (w/v) glucose] or in YPGal medium [1% (w/v) yeast extract, 2% (w/v) peptone, and 2% (w/v) galactose].
The Δlip1::LEU2 construct used to prepare the above cell lines was constructed as follows. The pGEM-Lip1 plasmid was first constructed using a pGEM-T Easy vector (Promega) and a DNA fragment that had been amplified from the yeast genomic DNA using primers (sLip-U1 5′-GAAGTACGACGAAGAATTTG-3′ and sLip-L1 5′-CGAGAGGAAGCACAAAGGC-3′). An SmaI site was introduced into the pGEM-Lip1 plasmid within the LIP1 gene using the QuikChange™ kit (Stratagene) and primers (SmaIU 5′-GATAATGTTCCACCCGGGCACTACCCAATTCACG-3′ and SmaIL 5′-CGTGAATTGGGTAGTGCCCGGGTGGAACATTATC-3′), producing the pGEM-Lip1 (SmaI) plasmid. The 138 bp HindIII–SmaI region in the pGEM-Lip1 (SmaI) plasmid was then replaced with the LEU2 marker. The resulting Δlip1::LEU2 construct was used for homologous recombination with the LIP1 gene.
Cloning of the mouse LASS3 gene and reverse transcription (RT)-PCR
To identify the mouse LASS3 gene, the GenBank™ database was searched for sequences similar to the human LASS3 gene (GenBank™ accession number BC034970) and a cDNA sequence (GenBank™ accession number XM_907730) that shared high similarity was found. Although this sequence had only been a predicted one on the basis of the mouse genomic sequence, we confirmed the existence of its mRNA using RT-PCR. We also found another, shorter, transcript of the LASS3 gene. We named these the long and short forms of LASS3 transcripts LASS3-long and LASS3 respectively. RT-PCR was performed on mouse testis cDNA (TaKaRa Bio, Shiga, Japan) using LA-Taq™ DNA polymerase with a GC buffer (TaKaRa Bio) and primers mRT-1 (5′-GGTCTCTGCAAGAATTTCAGGGCTTACG-3′) and mLASS3 L1 (5′-CATCATGATCTTCCAGGTCTGAC-3′) for LASS3-long or mRT-2 (5′-CGAAGCTAACATATCCTTTTCTTCATTG-3′) and mLASS3 L1 for LASS3. To prepare mammalian expression plasmids encoding LASS3 or LASS3-long, these two mouse LASS3 isoforms were each amplified from mouse skin total RNA using the SuperScript® One-Step RT-PCR system with Platinum® Taq (Invitrogen) and primers mLASS3-BamHI-U1 (5′-CGGATCCATGTTTCAGACGTTTAGAAAATGG-3′) and mLASS3-L2 (5′-CTAACGGCCATGCTGACCATTG-3′) for LASS3 or mLASS3-BamHI-U2 (5′-CGGATCCATGAGGTGTGTTGCTCATGAG-3′) and mLASS3-L2 for LASS3-long. The amplified fragments were cloned into a pGEM-T Easy vector to generate pGEM-LASS3 (BamHI) and pGEM-LASS3-L (BamHI).
The pcDNA3-HA-LASS3 and pcDNA3-HA-LASS3-L plasmids, which encode N-terminal HA (haemagglutinin)-tagged LASS3 and LASS3-long respectively, were constructed by cloning the 1.2 kb BamHI–NotI fragment of pGEM-LASS3 (BamHI) or 1.3 kb BamHI–NotI fragment of pGEM-LASS3-L (BamHI) into the BamHI–NotI site of the pcDNA3-HA-1 vector [14]. The pCE-puro-3×FLAG-LASS3 plasmid, which encodes an N-terminal triple FLAG (3×FLAG)-epitope-tagged LASS3, was constructed by cloning the 1.2 kb BamHI–NotI fragment of pGEM-LASS3 (BamHI) into the BamHI–NotI site of the pCE-puro 3×FLAG-1 vector [15]. The pcDNA3-HA-LASS6 plasmid, which encodes N-terminal HA-tagged LASS6, was constructed as described previously [9].
pAKNF316 (URA3 marker) is a yeast vector designed for N-terminal, 3×FLAG-tagged protein expression under the control of the TDH3 promoter. pAKNF-3×FLAG-LASS3 was constructed by cloning a 1.2 kb BamHI–NotI fragment of the pGEM-LASS3 (BamHI) plasmid into the BamHI–NotI site of pAKNF316. The pYES2 plasmid (Invitrogen) is a 2 μ-based cloning vector (URA3 marker) under the control of GAL1 promoter. pYES2-3×FLAG-LASS3 was constructed by cloning the 1.3 kb EcoRI–EcoRI fragment of pCE-puro 3×FLAG-LASS3 into the EcoRI site of pYES2.
(Dihydro)ceramide synthase and [3H]dihydrosphingosine labelling assays
An in vitro (dihydro)ceramide synthase assay was performed as described previously [9]. HEK-293T cells transfected with the indicated plasmids were suspended in buffer A [50 mM Hepes/NaOH (pH 7.5), 1×protease inhibitor mixture (Complete™ EDTA free; Roche Diagnostics, Indianapolis, IN, U.S.A.) and 0.5 mM dithiothreitol] and lysed by sonication. The resulting total cell lysates (50 μg of protein) were mixed with 5 μM dihydrosphingosine (Biomol, Plymouth Meeting, PA, U.S.A.), 0.2 μCi of [4,5-3H]dihydrosphingosine (50 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO, U.S.A.), 25 μM fatty acylCoA, 1 mM MgCl2 and 0.1% digitonin in buffer B [50 mM Hepes/NaOH (pH 7.5) and 0.5 mM dithiothreitol] in a final volume of 100 μl and incubated at 37 °C for 30–60 min. The lipids were extracted by the method of Bligh and Dyer [16]. The dried lipids were resuspended in 30 μl of chloroform/methanol (2:1, v/v), and separated on Silica Gel 60 TLC plates (Merck, Whitestation, NJ, U.S.A.), using chloroform/methanol/2 M ammonium hydroxide (40:10:1, by vol.) as the solvent system.
In labelling assays, HEK-293T cells transfected with plasmids were incubated for 6 h with or without 20 μM FB1 (fumonisin B1, a mycotoxin produced by the fungus Fusarium moniliforme), then metabolically labelled by incubation with 1.0 μCi of [4,5-3H]-dihydrosphingosine at 37 °C for 3 h. The cells were washed with PBS, followed by lipid extraction as described previously [17]. The labelled lipids were separated on Silica Gel 60 high-performance TLC plates (Merck) using a solvent system of chloroform/methanol/acetic acid (190:9:1, by vol.). The plates were sprayed with the fluorographic reagent En3Hance™ (PerkinElmer Life Sciences, Woodbridge, Ont., Canada), dried, and exposed to X-ray films at −80 °C.
Northern-blot analysis
Analysis was performed on 20 μg of total RNA from adult mouse tissues (Seegene, Seoul, Korea). The LASS3 probe was prepared from a DNA fragment corresponding to the C-terminal 600 bp of the LASS3 open reading frame, which had been amplified from pGEM-LASS3 using the primers mLASS3-U3 5′-CGCTCATGTCATCCACCACCTG-3′ and mLASS3-L2. The amplified fragment was then labelled with [32P]dCTP using a Random Primer DNA labeling Kit (TaKaRa Bio). Hybridization was carried out in ExpressHyb buffer (TaKaRa Bio) for 4 h at 68 °C.
Immunoblotting
Total cell lysates used in the (dihydro)ceramide synthase assay were also analysed by immunoblotting, which was performed as described previously [9]. The anti-HA antibody HA7 (1:2000 dilution; Sigma, St. Louis, MO, U.S.A.) was used as the primary antibody. A 1:5000 dilution of horseradish-peroxidase-conjugated donkey anti-mouse IgG F(ab′)2 fragment (Amersham Biosciences, Piscataway, NJ, U.S.A.) was used as the secondary antibody. Labelling was detected using an enhanced chemiluminescence (ECL®) system kit; Amersham Biosciences).
ESI-MS (electrospray-ionization MS)
HEK-293T cells were grown in 100-mm-diameter plastic culture dishes and transfected with the indicated plasmids. At 24 h after transfection, the cells were incubated with or without 20 μM FB1 for a further 24 h, then washed twice with PBS. The lipids were extracted twice in 10 ml of chloroform/methanol (2:1, v/v), each time at 40 °C for 1 h, then evaporated under nitrogen. Lipids were resolved by TLC on Silica Gel 60 plates with chloroform/methanol/acetic acid (190:9:1, by vol.). Ceramide bands co-migrating with a ceramide standard were scraped from the TLC plates, extracted with chloroform/methanol (2:1, v/v) and evaporated. Each ceramide obtained was then dissolved in chloroform/methanol (3:2, v/v), which included C14:0-ceramide (C14:0 fatty acid with sphingosine) as an internal standard, and subjected to ESI-MS. ESI-MS measurements were performed in negative mode using an MStation (JMS-700TZ). Measurement conditions for the ESI-MS were: needle voltage, 2.0 kV; ring lens voltage, 80.0 V; resolution, 3000; and sample flow, 40 μl/min.
RESULTS
Identification of mouse LASS3
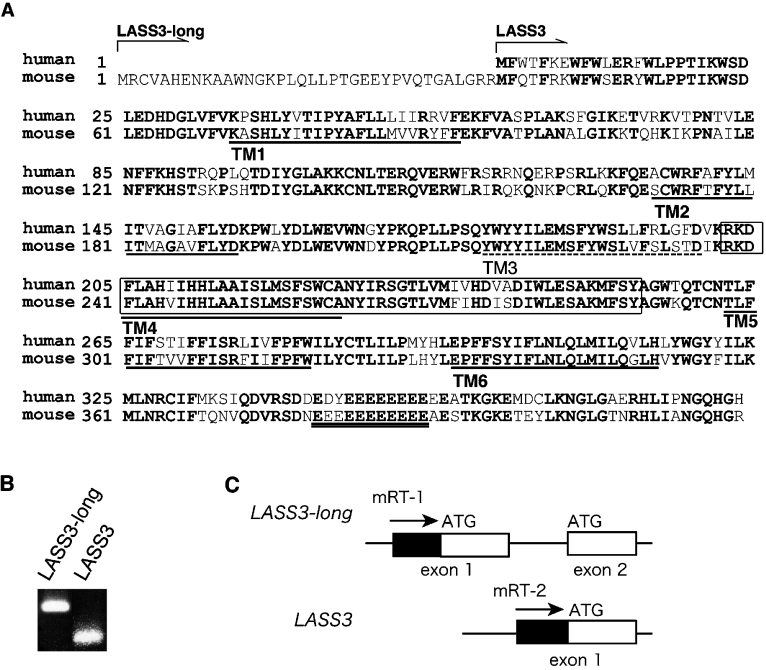
Although five of the six mammalian LASS members have previously been characterized [9–12], LASS3 has not. Therefore, we prepared mouse LASS3 cDNA for comparison with other mouse LASS cDNAs which we had prepared and analysed in a previous study [9]. To identify the mouse LASS3 gene, the GenBank™ database was searched for sequences similar to the human LASS3 gene (GenBank™ accession number BC034970). The cDNA predicted by the mouse genomic sequence XM_907730 shared high similarity with the human LASS3 gene. RT-PCR analysis revealed that the corresponding mRNA does exist in mouse testis (Figure 1B); we named this cDNA LASS3-long. LASS3-long cDNA encodes 419 amino acids, a sequence longer than the human LASS3 protein, with 36 additional amino acids at the N-terminus (Figure 1A). Further comparison between the genomic structures of the mouse LASS3-long gene and the human LASS3 gene revealed that the mouse LASS3-long gene contains an additional exon not present in the human LASS3 gene (Figure 1C). Transcription of this exon results in the production of the added amino acids at the N-terminus of the protein. However, LASS3-long contains a methionine residue (Met37) that corresponds to the first methionine residue of human LASS3. Therefore we examined the possibility that translation of another LASS3 protein is also initiated at the Met37 as a result of an alternative transcription initiation site. We prepared a primer corresponding to the 5′-upstream sequence of the exon 2 of LASS3-long and performed RT-PCR on testis cDNA. As shown in Figure 1(B), the short cDNA, called simply LASS3, was indeed amplified. This result suggests that the mouse has two isoforms of LASS3 mRNA that differ in their transcription initiation sites.
Figure 1. Amino acid sequence of mouse LASS3.
(A) Comparison of the mouse and human LASS3 amino acid sequences (GenBank™ accession numbers, DQ646881/DQ358087 and BC034970, respectively) is shown. LASS3-long contains 36 additional amino acid residues at the N-terminus. Conserved amino acids are shown in bold. The Lag1 motif is enclosed by a box. The clusters of Glu residues, characteristic of LASS3 proteins, are indicated by double underlining. The TopPredII program [18] predicted that LASS3 comprises up to six TM spanning domains, shown here underlined. TM1, TM2, TM4 and TM5 each exhibited high probability scores (TM1, 1.1948; TM2; 1.4104, TM4, 1.3427; TM5, 1.8896), whereas the scores for TM3 and TM6 were relatively low (1.0630 and 1.0260 respectively). Taking the topology model of LASS6 [9] into consideration, we propose that only five domains, TM1, TM2, TM4, TM5 and TM6 (continuous underlining) span the membrane and that TM3 (broken underlining) does not. (B) The mRNA expression of LASS3 and LASS3-long was determined by RT-PCR using mouse testis cDNA as detailed in the Materials and methods section. (C) The first and second exons of LASS3-long, the first exon of LASS3, and the primers used in (B), are shown.
The mouse LASS3 gene encodes 383 or 419 amino acids with a predicted molecular mass of 46.1 or 50.0 kDa. LASS3 shares 78.6% identity and 92.7% similarity with human LASS3. Of the other mouse LASS members, it exhibits the highest homology with LASS2 (53.2% identity, 68.9% similarity) and the lowest homology with LASS1 (26.1% identity, 36.0% similarity). Like other LASS members, LASS3 contains a highly conserved Lag1 motif (Figure 1A, box). In contrast with the N-glycosylated LASS2, LASS5 and LASS6, LASS3 has no putative N-glycosylation site [9]. The TopPredII program [18] predicted that LASS3 has up to six TM (transmembrane) spanning domains. In light of our recently published topology model for mouse LASS6 [9], we propose that LASS3 traverses the membrane five times via domains TM1, TM2, TM4, TM5 and TM6 (Figure 1A, continuous underline). In addition, clusters of Glu residues, characteristic of LASS3 proteins, were observed in the C-terminal hydrophilic region (Figure 1A, double underline).
(Dihydro)ceramide synthase activity of LASS3
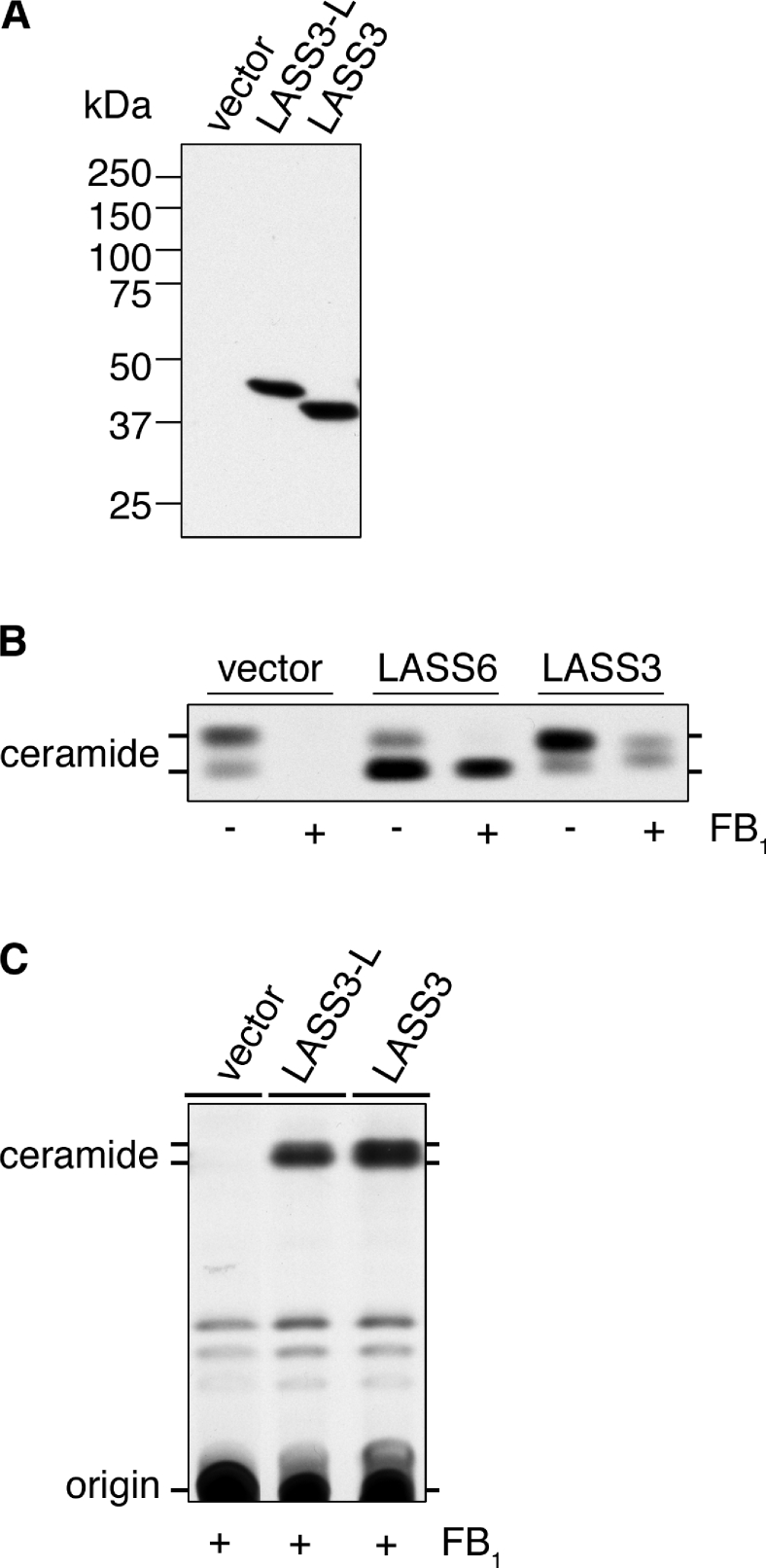
To investigate the enzyme activity of LASS3, HEK-293T cells were transiently transfected with pcDNA3-HA-LASS3 and pcDNA3-HA-LASS3-L, encoding N-terminally HA-tagged LASS3 (HA-LASS3) and LASS3-long (HA-LASS3-long) respectively. Immunoblotting with an anti-HA antibody confirmed the expression of these proteins (Figure 2A). Using these cells, we examined the involvement of LASS3 and LASS3-long in ceramide synthesis as determined by a [3H]dihydrosphingosine labelling assay (Figures 2B and 2C). Cells transfected with the control vector produced two ceramide bands corresponding to long-chain (upper) and short-chain (lower) ceramides (Figure 2B). These bands disappeared in the presence of FB1, a ceramide synthase inhibitor known to inhibit endogenous ceramide synthase activity but not the activity of overproduced LASS members (for an unknown reason) [9,10,12]. Consistent with a previous report [9], overproduction of the positive control LASS6 increased short-chain ceramide in the presence or absence of FB1 (Figure 2B), but with FB1 treatment the long-chain ceramide band produced by endogenous ceramide synthase disappeared. Overproducing LASS3, on the other hand, induced production of long-chain ceramide in the absence of FB1, and production of middle- and long-chain ceramides in the presence of FB1. These bands migrated faster than the ceramide band produced by LASS6 that was previously determined to be C16:0 [9]. These results suggest that LASS3 proteins are involved in the synthesis of middle- to long-chain ceramides. Furthermore, overproduction of LASS3-long (Figure 2C) produced ceramide bands similar to those observed in LASS3-overproducing cells in both level and chain length, suggesting that the two proteins are indistinguishable in enzyme activity and substrate specificity.
Figure 2. (Dihydro)ceramide synthase activity of LASS3.
HEK-293T cells were transfected with pcDNA3-HA (vector), pcDNA3-HA-LASS3 (LASS3), pcDNA3-HA-LASS3-L [LASS3-long (LASS3-L)] or pcDNA3-HA-LASS6 (LASS6) as indicated. (A) Total cell lysates (5 μg of protein) were separated on an SDS/10%-(w/v)-polyacrylamide gel then transferred to a membrane. Immunoblotting was then performed using an anti-HA antibody. (B) Transfected cells were incubated with 1 μCi of [3H]dihydrosphingosine for 3 h in the absence or presence of FB1 (20 μM). Lipids were extracted, separated by TLC, and visualized by autoradiography. (C) Transfected cells were incubated with 1 μCi of [3H]dihydrosphingosine for 3 h in the presence of FB1 (20 μM). Lipids were extracted, separated by TLC, and detected by X-ray film.
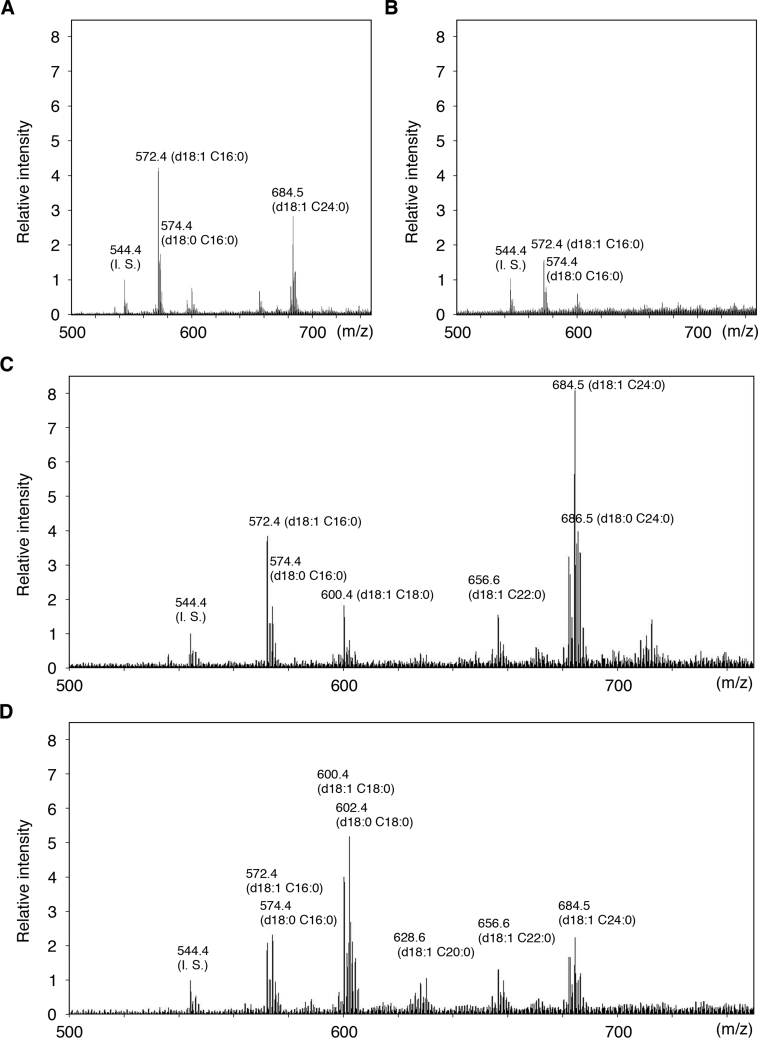
We examined the ceramide species produced by LASS3 in more precise detail. We prepared ceramides from LASS3-overproducing cells grown in the absence and presence of FB1 and analysed the lipids by ESI-MS. In samples from vector-transfected cells (Figure 3A), three major peaks were detected that corresponded to two C16:0-ceramide species [C16:0 fatty acid with a d18:1 (sphingosine) or a d18:0 (dihydrosphingosine) sphingoid base] and one C24:0-ceramide (C24:0 fatty acid with sphingosine). C18:0- and C22:0-ceramides were also detected as minor peaks. The addition of FB1 resulted in marked decreases in all ceramide species (Figure 3B). Overproduction of LASS3 in the absence of FB1 caused increased levels of C18:0-, C22:0- and, most notably, C24:0-ceramides (Figure 3C). In the presence of FB1, the LASS3-overproducing cells exhibited increases in C18:0-, C20:0-, C22:0- and C24:0-ceramides (Figure 3D). In this case, though, the increase in C18:0-ceramides was most prominent. These data indicate that LASS3 exhibits relatively low substrate specificity toward fatty acyl-CoAs. Similar results were obtained for LASS3-long (results not shown), therefore we used only LASS3 for further analysis.
Figure 3. Fatty acid composition of ceramides in mouse LASS3-transfected cells.
HEK-293T cells transfected with pcDNA3-HA (vector) (A and B) or pcDNA3-HA-LASS3 (C and D) were incubated for 24 h in the absence (A and C) or presence (B and D) of 20 μM FB1. Total lipids were prepared from the cells and separated by TLC. Ceramide bands were scraped from the TLC plate, extracted, and analysed by ESI-MS using the negative-ion mode. The measured m/z negative molecular ion [M+Cl]− signals as well as the assigned ceramides species are shown. C14:0-ceramide was added as an internal standard (I. S.). Values are relative to those associated with the internal standard.
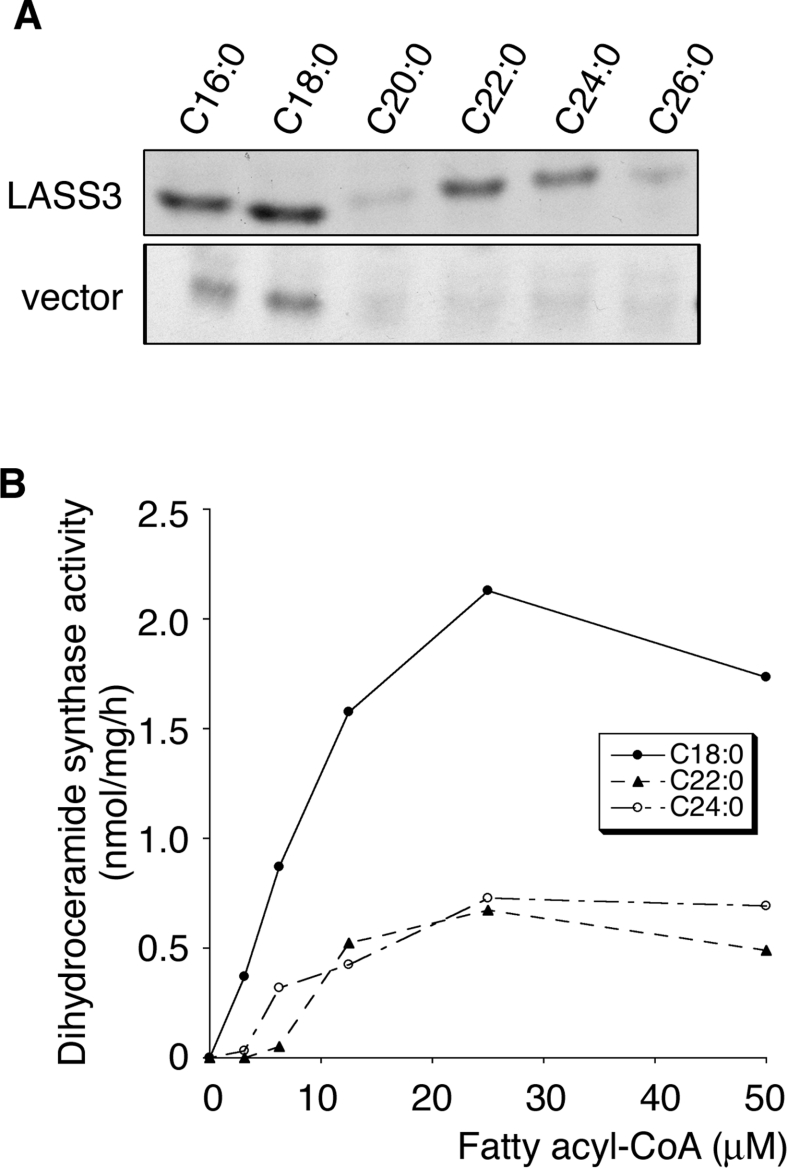
In vivo results might reflect not only the substrate preference of LASS3, but also the availability of cellular fatty acyl-CoAs. We therefore performed an in vitro (dihydro)ceramide synthase assay using various fatty acyl-CoAs and [3H]dihydrosphingosine (Figure 4A). Total cell lysates prepared from vector-transfected cells exhibited the highest activity toward C16:0-CoA and C18:0-CoA. On the other hand, lysates prepared from LASS3-overproducing cells showed significant activity toward several fatty acyl-CoAs, including C18:0-, C22:0-, and C24:0-CoA, with the highest activity for C18:0 (Figure 4A). C16:0-, C20:0-, and C26:0-CoA were also used as substrates by LASS3, albeit weak ones (Figure 4A). We further investigated the ceramide synthase activity of LASS3-overproducing cells using increasing concentrations of C18:0-, C22:0-, or C24:0-CoA (Figure 4B). LASS3-overproducing cells synthesized ceramide from C18:0-CoA with the highest activity at all concentrations tested. This in vitro result is quite consistent with the above in vivo results and, together, they indicate that LASS3 has distinct, but broad, substrate preference with regard to fatty acyl-CoAs. This is in contrast with other LASS family members with more specific substrate specificities [9].
Figure 4. In vitro (dihydro)ceramide synthase activity of LASS3 toward fatty acyl-CoAs of various lengths.
(A) Total cell lysates (50 μg of protein) prepared from HEK-293T cells transfected with pcDNA3-HA (vector) or pcDNA3-HA-LASS3 (LASS3) were incubated for 60 min at 37 °C with 0.2 μCi of [3H]dihydrosphingosine (50 Ci/mmol) in 5 μM unlabelled dihydrosphingosine and the indicated fatty acyl-CoA (25 μM) in assay buffer containing 0.1% digitonin. Lipids were extracted, separated by TLC, and visualized by autoradiography. (B) Total lysates (50 μg of protein) were prepared from HEK-293T cells transfected with pcDNA3-HA-LASS3. Lysates were incubated for 30 min at 37 °C with 0.2 μCi of [3H]dihydrosphingosine in 5 μM dihydrosphingosine and increasing concentrations of the indicated fatty acyl-CoA. Lipids were detected by X-ray film and the density measured using the Image J program (http://rsb.info.nih.gov/ij/index.html).
Testis-specific expression of LASS3
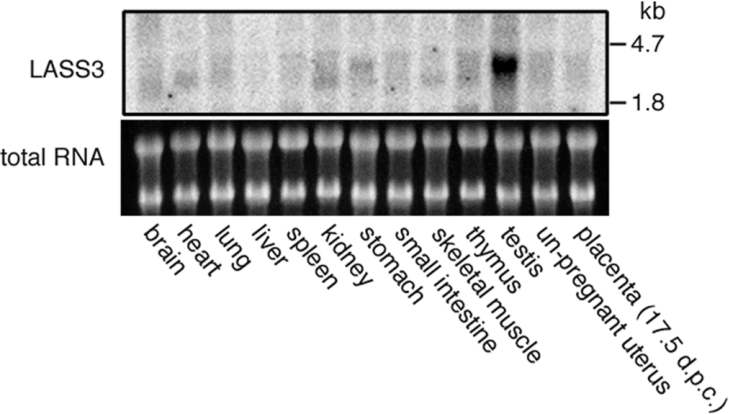
LASS family members have been shown to differ in tissue-specific expression patterns [9,12]. To see the tissue distribution of mouse LASS3, we performed a Northern-blot analysis using total RNAs from 13 different tissues (Figure 5). A strong 3.4 kb band of LASS3 was detected in testis. This band was barely detectable in skin (results not shown) and not observed in female-specific tissues such as ovary and placenta (Figure 5). Thus LASS3 is a highly testis-specific (dihydro)ceramide synthase.
Figure 5. Tissue distribution of mouse LASS3 mRNA.
A 32P-labelled LASS3 probe was hybridized to total RNA from 13 different mouse tissues (20 μg/lane) (upper panel). The lower panel shows a photograph of the ethidium bromide-stained gel, demonstrating similar levels of 18 S and 24 S ribosomal RNAs. d.p.c.; days post coitum.
LASS3 activity in the absence of Lip1 in yeast
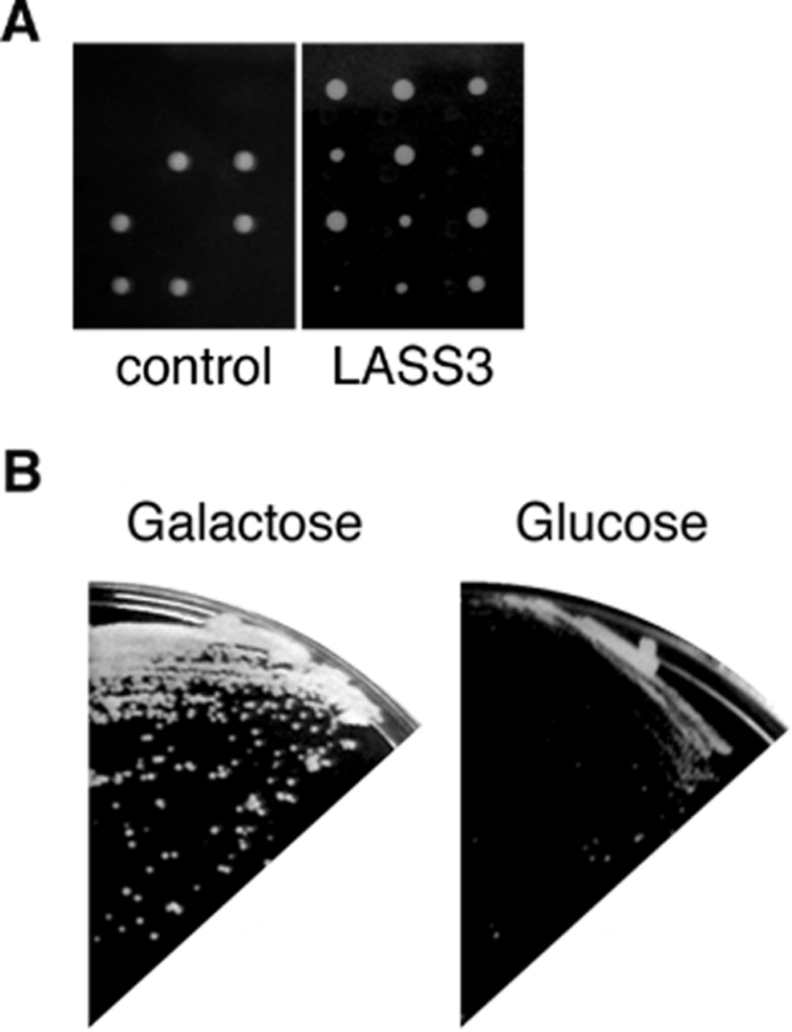
It has remained unclear whether LASS members are (dihydro)-ceramide synthases or regulators. In yeast, Lip1 forms a complex with Lag1 or Lac1 and is essential for (dihydro)ceramide synthesis activity [7]. However, in mammals there is no known Lip1 homologue, and it has been suggested that LASS family members alone are responsible for mammalian (dihydro)ceramide synthesis activity [8]. To test this hypothesis further, we examined the requirement of Lip1 for the (dihydro)ceramide synthesis activity of LASS3 in yeast. We introduced the pAKNF-3xFLAG-LASS3 plasmid (URA3 marker), which encodes 3×FLAG-LASS3 under the control of the TDH3 promoter, into the yeast diploid strain YMY08 (LIP1/Δlip1::LEU2). The resulting diploid strain was sporulated, and tetrads were dissected. After incubation on YPD plates for 2–3 days, we detected two large and two small colonies (Figure 6A). By contrast, a diploid strain bearing only the vector (Figure 6A) or the plasmid expressing yeast Lag1 (results not shown) produced only two colonies. Judging from their leucine and uracil auxotrophy, the two slow-growth colonies were Δlip1 cells bearing pAKNF-3xFLAG-LASS3 (results not shown).
Figure 6. LASS3 can function in the absence of Lip1.
(A) Four tetrads were dissected from the heterozygous diploid yeast strain YMY08 (Δlip1::LEU2/LIP1), harbouring a vector (left panel) or pAK-3xFLAG-LASS3 (3×FLAG-LASS3; right panel). The spores were grown on a YPD plate for 3 days at 30 °C. (B) YMY16 cells (Δlip1::LEU2) bearing pYES-3xFLAG-LASS3 were grown either on a YPGal plate (2% galactose, left panel) or a YPD plate (2% glucose, right panel) for 3 days at 30 °C.
We also confirmed the rescue of Δlip1 cells by LASS3 by another expression system. Using the yeast haploid strain KA31-1A, which harbours the plasmid pYES2-3×FLAG-LASS3 under the control of the inducible GAL1 promoter, we replaced the LIP1 gene with Δlip1::LEU2. The resulting strain (YMY16), bearing pYES2-3xFLAG-LASS3, was able to grow on medium that included galactose, but not on that which included glucose (Figure 6B). These results suggest that mouse LASS3 can function even in the absence of Lip1.
DISCUSSION
In the present study we have cloned and characterized the mouse LASS3 gene. This gene is the last known mammalian LASS gene to be characterized, and thus the analysis of the known LASS family members has been completed by this study. Each mammalian LASS protein exhibits a characteristic fatty acyl-CoA substrate preference, as well as specific tissue expression. LASS3 produces C18:0-ceramide preferentially, but also other middle- to long-chain ceramide species. Thus LASS3 is a (dihydro)ceramide synthase with relatively broad substrate specificity.
ESI-MS analysis demonstrated that LASS3 overproduction stimulated the generation of the same set of ceramide species in the absence or presence of FB1, although the ceramide compositions differed. C24:0-ceramide was increased the most in the absence of FB1, but C18:0-ceramide increased in its presence (Figure 3). However, in in vitro (dihydro)ceramide synthase assays with a variety of fatty acyl-CoAs, lysates prepared from LASS3-overproducing cells grown without FB1 exhibited the highest activity toward C18:0-CoA (Figure 4). Thus it is highly likely that the difference of the LASS3 effects on the ceramide species in vivo between FB1-treated and untreated cells is caused by altered availability of certain fatty acyl-CoAs.
In mammals, ceramides exhibit a tissue-dependent bias regarding their amide-linked fatty acid chain length. In addition to the availability of certain fatty acyl-CoA species, differences in the expression levels of LASS family members may also contribute to tissue-specific ceramides. For example, C16:0 is the most abundant fatty acid moiety among short-chain ceramides in most tissues [19], yet C18:0-ceramide is the major short-chain ceramide in brain [19,20], which exhibits high expression levels of the C18:0-specific (dihydro)ceramide synthase LASS1 [10]. In the present study we found that LASS3 exhibits a testis-specific distribution pattern (Figure 5). We previously found that LASS5 and LASS6 are also expressed in testis, but little LASS2, and no LASS1 or LASS4 [9]. Since LASS5 and LASS6 are specific for short-chain ceramides, LASS3 may play an important rôle in the production of long-chain ceramides in this gland. Gangliosides from bovine and boar testis are known to induce many ceramides with long-chain (20–24 carbon atoms) fatty acids and fewer with short-chain [21,22]. In addition, mouse testis glycosphingolipids that are essential for spermatogenesis and male mouse fertility reportedly carry polyunsaturated very-long-chain and hydroxylated fatty acids [23]. Unfortunately, these fatty acids were not available to test the ability of LASS3 to use them as substrates. However, it is quite possible that LASS3 is responsible for the production of these unique ceramide species, considering its broad substrate specificity.
In yeast, Lag1p/Lac1p co-immunoprecipitates with an additional, essential subunit, Lip1p [7]. In mammals, however, no homologue of Lip1p has been found. Purified LASS5 has been shown to exhibit (dihydro)ceramide synthesis activity in vitro in the absence of any additional protein [8]. In the present study, LASS3 was able to function in the Δlip1 cells (Figure 6), further suggesting that no Lip1 or similar protein is required for its (dihydro)ceramide synthase activity.
Although it is now known that each LASS member produces ceramides with specific chain-lengths, knowledge regarding the physiological function of each ceramide species is still limited. Ceramide is known to function in several cellular processes, such as apoptosis, the cell cycle and differentiation [1–3]. By overproducing certain LASS proteins, specific functions for each ceramide species might be determined. Future studies using knockout models for each LASS gene will also be needed.
Acknowledgments
We thank Seiko Oka (Center for Instrumental Analysis in our University) for assistance with the ESI-MS, and Dr Elizabeth A. Sweeney for editing and preparing the manuscript prior to submission.
References
- 1.Futerman A. H., Hannun Y. A. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolesnick R., Fuks Z. Radiation and ceramide-induced apoptosis. Oncogene. 2003;22:5897–5906. doi: 10.1038/sj.onc.1206702. [DOI] [PubMed] [Google Scholar]
- 3.Ogretmen B., Hannun Y. A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 4.Futerman A. H., Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15:312–318. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallee B., Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahiri S., Futerman A. H. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J. Biol. Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 9.Mizutani Y., Kihara A., Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr, Futerman A. H. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine [C18-(dihydro)ceramide] synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 11.Guillas I., Jiang J. C., Vionnet C., Roubaty C., Uldry D., Chuard R., Wang J., Jazwinski S. M., Conzelmann A. Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 12.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr, Futerman A. H. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 2003;278:43452–43459. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 13.Irie K., Takase M., Lee K. S., Levin D. E., Araki H., Matsumoto K., Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogawa C., Kihara A., Gokoh M., Igarashi Y. Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J. Biol. Chem. 2003;278:1268–1272. doi: 10.1074/jbc.M209514200. [DOI] [PubMed] [Google Scholar]
- 15.Kihara A., Anada Y., Igarashi Y. Mouse sphingosine kinase isoforms SPHK1a and SPHK1b differ in enzymatic traits including stability, localization, modification, and oligomerization. J. Biol. Chem. 2005;281:4532–4539. doi: 10.1074/jbc.M510308200. [DOI] [PubMed] [Google Scholar]
- 16.Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Med. Sci. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 17.Mizutani Y., Kihara A., Igarashi Y. Identification of the human sphingolipid C4-hydroxylase, hDES2, and its up-regulation during keratinocyte differentiation. FEBS Lett. 2004;563:93–97. doi: 10.1016/S0014-5793(04)00274-1. [DOI] [PubMed] [Google Scholar]
- 18.Claros M. G., von Heijne G. Prediction of transmembrane segments in integral membrane proteins, and the putative topologies, using several algorithms. CABIOS. 1994;10:685–686. [Google Scholar]
- 19.Pettus B. J., Baes M., Busman M., Hannun Y. A., Van Veldhoven P. P. Mass spectrometric analysis of ceramide perturbations in brain and fibroblasts of mice and human patients with peroxisomal disorders. Rapid. Commun. Mass Spectrom. 2004;18:1569–1574. doi: 10.1002/rcm.1520. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien J. S., Rouser G. The fatty acid composition of brain sphingolipids: sphingomyelin, ceramide, cerebroside, and cerebroside sulfate. J. Lipid Res. 1964;5:339–342. [PubMed] [Google Scholar]
- 21.Suzuki A., Ishizuka I., Yamakawa T. Isolation and characterization of a ganglioside containing fucose from boar testis. J. Biochem. (Tokyo) 1975;78:947–954. doi: 10.1093/oxfordjournals.jbchem.a131001. [DOI] [PubMed] [Google Scholar]
- 22.Bushway A. A., Clegg E. D., Keenan T. W. Composition and synthesis of gangliosides in bovine testis, sperm and seminal plasma. Biol. Reprod. 1977;17:432–442. doi: 10.1095/biolreprod17.3.432. [DOI] [PubMed] [Google Scholar]
- 23.Sandhoff R., Geyer R., Jennemann R., Paret C., Kiss E., Yamashita T., Gorgas K., Sijmonsma T. P., Iwamori M., Finaz C., et al. Novel class of glycosphingolipids involved in male fertility. J. Biol. Chem. 2005;280:27310–27318. doi: 10.1074/jbc.M502775200. [DOI] [PubMed] [Google Scholar]