Abstract
Matrix metallopeptidase-12 (MMP-12) binds three calcium ions and a zinc ion, in addition to the catalytic zinc ion. These ions are thought to have a structural role, stabilizing the active conformation of the enzyme. To characterize the importance of Ca2+ binding for MMP-12 activity and the properties of the different Ca2+ sites, the activity as a function of [Ca2+] and the effect of pH was investigated. The enzymatic activity was directly correlated to calcium binding and a Langmuir isotherm for three binding sites described the activity as a function of [Ca2+]. The affinities for two of the binding sites were quantified at several pH values. At pH 7.5, the KD was 0.1 mM for the high-affinity binding site, 5 mM for the intermediate-affinity binding site and >100 mM for the low-affinity binding site. For all three sites, the affinity for calcium decreased with reduced pH, in accordance with the loss of interactions upon protonation of the calcium-co-ordinating aspartate and glutamate carboxylates at acidic pH. The pKa values of the calcium binding sites with the highest and intermediate affinities were determined to be 4.3 and 6.5 respectively. Optimal pH for catalysis was above 7.5. The low-, intermediate- and high-affinity binding sites were assigned on the basis of analysis of three-dimensional-structures of MMP-12. The strong correlation between MMP-12 activity and calcium binding for the physiologically relevant [Ca2+] and pH ranges studied suggest that Ca2+ may be involved in controlling the activity of MMP-12.
Keywords: active conformation, affinity, calcium binding, human macrophage elastase, matrix metallopeptidase-12 (MMP-12), pH-dependency
Abbreviations: Dpa, N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl; Mca, (7-methoxycoumarin-4-yl)acetyl; MMP-12, matrix metallopeptidase-12; rfu, relative fluorescence units
INTRODUCTION
MMP-12 (matrix metallopeptidase-12, also known as human macrophage elastase) is a metalloendopeptidase (EC 3.4.24.65) belonging to the matrixin subfamily M10A, as classified in the MEROPS peptidase database [1]. MMP-12 is of interest as a drug target as it is believed to be involved in many diseases, such as chronic obstructive pulmonary disease, rheumatoid arthritis and multiple sclerosis [2–4]. The degradation of extra cellular matrix by MMP-12 is an essential part of these diseases and therefore researchers are designing potential MMP-12 inhibitors and testing their suitability as drugs.
In addition to the catalytic zinc ion, MMP-12 has a zinc ion and three calcium ions that are believed to stabilize the structure. Although there are some crystal structures and kinetic data available for this enzyme, its calcium dependence has been poorly characterized. Moreover, the physiological relevance of calcium binding to MMP-12 is not known, although calcium appears to structurally stabilize the active conformation of the enzyme. This also stabilizes the enzyme by preventing autohydrolysis.
More than 20 MMPs have been identified and calcium binding is a common property of this class of enzymes, illustrated by the presence of calcium binding sites in the three-dimensional structures of all eleven MMPs that have been solved and deposited in the Protein Data Bank [5]. Surprisingly, the calcium dependency of MMPs is generally overlooked by researchers, despite the use of unphysiologically high [Ca2+] in MMP activity assays. The question of whether calcium is regulatory or merely a structural requirement for activity of these enzymes has not been given much attention. This can be compared with the interest in the role of calcium binding to the calpains, a family of calcium-activated cytoplasmic cysteine proteases that require 6–3000 times higher [Ca2+] for half maximal activity than the highest cytoplasmic Ca2+ concentration. This has caused a discussion among calpainologists as to whether there are additional activation mechanisms or if calpains simply work at sub-maximal velocities in vivo [6]. It seems that such a discussion concerning the role of calcium for MMP-activity has not arisen. The aim of this study was therefore to investigate the dependence of MMP-12 activity on calcium binding and to characterize the differences in the binding of calcium to the three binding sites of MMP-12. By understanding the characteristics of the binding and how it is influenced by the concentration of calcium and other environmental factors, it would be possible to initiate a discussion of the biological relevance of the calcium requirement of MMPs.
EXPERIMENTAL
MMP-12 activity assay
MMP-12 was obtained from Medivir AB (Huddinge, Sweden) where it was produced essentially as described by Morales et al. [7]. The protein consisted of the catalytic domain, comprising amino acids 100–263. The catalytic activity was determined in 96-well plates in a final volume of 150 μl, where 120 μl of buffer (50 mM Mes, 50 mM Mops or 50 mM sodium acetate, pH 4.5–7.5), 10 μl of CaCl2 dissolved in water and 10 μl of the internally quenched fluorogenic peptide substrate Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 (where Dpa is N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl and Mca is (7-methoxycoumarin-4-yl)acetyl; RnD Systems Europe Ltd), dissolved in DMSO to 150 μM, was incubated at 30 °C for 5 min. The reaction was initiated by the addition of 10 μl MMP-12. The final concentration of CaCl2 varied from 21 μM to 200 mM while the final concentration of DMSO was a constant 6.7% (v/v), and that of MMP-12 and substrate was 70 nM and 10 μM respectively. The enzyme stock solution (Tris/HCl, pH 7.5 and 5 mM CaCl2) was diluted in assay buffer without CaCl2 just prior to initiating the reaction. The residual CaCl2 from the enzyme was accounted for in the data analysis. Plates were read on a Fluoroskan Ascent (Thermo Labsystems Oy, Helsinki, Finland) using λex=320 nm and λem=460 nm for 500 s.
Assessment of auto hydrolysis
MMP-12 (1.5 μM) was incubated in assay buffer with 180 mM CaCl2 or 0.5 mM CaCl2 at 30 °C for up to 6 min. Aliquots were transferred to loading buffer [10 mM Tris/HCl, pH 8.0, 1 mM EDTA, 5% 2-mercaptoethanol and 2.5% (w/v) SDS] and denatured by heating at 95 °C for 5 min. The samples were separated by SDS/PAGE (20% gels) using a PhastSystem apparatus (Pharmacia, Uppsala, Sweden) and the proteins were visualized by Coomassie Blue or silver staining according to the PhastSystem instructions.
Data evaluation
The initial slopes of the progress curves (reaction velocities) were calculated using the Ascent Software 2.6 (Thermo Labsystems Oy). All further data analysis was performed using GraFit 5.0.8 (Erithacus Software Limited, Surrey, U.K.) and Statistica 7.1 (Statsoft Inc., Tulsa, OK, U.S.A.).
Models for one to three binding sites (Langmuir isotherm for multiple non-equivalent binding sites) could be fitted to the experimental data. The equations used were:
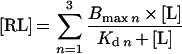 |
(1) |
where n is the number of binding sites for Ca2+, Bmax is the total binding capacity, [RL] is the amount of complex, [L] is the concentration of Ca2+ and Kd is the equilibrium dissociation constant. The velocity of the catalysed reaction was assumed to be proportional to [RL].
A model for a singly ionising system [8] was fitted to the Kd data at different pH using:
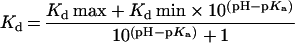 |
(2) |
The activity of the enzyme as a function of pH was modelled using:
 |
(3) |
where Lim1, Lim2 and Lim3 are the lower, middle and upper limits of activity respectively.
RESULTS
To determine the importance of Ca2+ for the activity of MMP-12, the dependence of activity as a function of [Ca2+] was investigated. Since the interaction of Ca2+ and the enzymatic activity are pH dependent, the pH dependence was also investigated. Thus an experimental design where both [Ca2+] and pH were varied was applied. A buffer containing three different buffer substances, selected to cover the pH range 4–7.5, which do not interact with calcium [9], was used. The activity was measured with up to 16 different [Ca2+] at each pH studied. This generated a data set where the pH-dependency of both enzyme activity and calcium binding could be extracted.
Since many proteases are unstable and subjected to inactivation as a result of autohydrolysis, two control experiments were performed. In the first, the activity of the enzyme as a function of time and [Ca2+] was explored. Figure 1 illustrates that there was a detectable inactivation, and resulting non-linearity, over a period of 500 s. However, the initial period (up to 120 s) of the progress curves, used to calculate initial velocities, was linear for all [Ca2+] tested. In a second experiment, the degree of autohydrolysis was explored as a function of time and [Ca2+]. SDS/PAGE analysis showed that MMP-12 was subject to autohydrolysis during the time of the experiments, however, it was only observed when the gels were silver stained (Figure 2) and not when they were Coomassie Blue stained (results not shown). In addition, 20-fold more enzyme was used in this experiment compared with the activity measurements. Consequently, the degree of autohydrolysis was very limited during the time of the measurements. In both experiments, high [Ca2+] stabilized the enzyme, indicating that MMP-12 loses catalytic activity with time in a [Ca2+]-dependent fashion. However, the degree of inactivation appeared to be significantly larger than the degree of autohydrolysis, suggesting that the decrease in catalytic activity is, at least initially, due to factors other than autohydrolysis.
Figure 1. Progress curves for MMP-12 catalysis at different [Ca2+].
Product formation was detected in rfu (relative fluorescence units) as a function of time for five different [Ca2+] shown to the right of the curves in mM. The data has been rescaled so that all curves begin at 0 rfu.
Figure 2. Auto hydrolysis of MMP-12 as a function time and [Ca2+].
An SDS/PAGE gel of MMP-12 incubated at 30 °C for 1, 2 or 6 min with 180 or 0.5 mM CaCl2. The gel was developed by silver staining.
pH-dependence of MMP-12 activity
An initial analysis of the data simply involved investigating the activity of MMP-12 as a function of pH-dependency. The activity was strongly pH-dependent, with the highest activity at pH 7.5, which was the highest pH used in the present study (Figure 3A). The relationship between activity and pH differed for high and low [Ca2+], indicating that calcium binding had a major impact on activity. At low [Ca2+] (0.16 mM), eqn (2) gave a satisfactory description of the data, demonstrating that a single pKa value determined the pH profile. At higher [Ca2+] (1.26–200 mM) an equation for two pKa values (eqn 3) was needed to describe the data. The pKa values could not be reliably quantified since the number of data points was not enough for the complexity of the equations used. However, the analysis showed that there was a higher complexity in the pH-dependence of MMP-12 activity at high [Ca2+] compared with low [Ca2+] and that the pH profile was therefore not simply determined by the pH-dependence of the catalytic mechanism.
Figure 3. MMP-12 activity dependence on [Ca2+] and pH.
(A) pH-profiles of MMP-12 activity. Initial velocities [v, in rfu (relative fluorescent units)/s] of substrate cleavage by MMP-12 at pH 4.5–7.5 and four different [Ca2+]. The solid line is the theoretical curve obtained by non-linear regression using a model representing a singly ionizing system (eqn 2) and the dashed lines using a model with two ionizing groups (eqn 3). (B) Initial velocities of substrate cleavage by MMP-12 and different [Ca2+] at pH 7.5. The dotted line represents a non-linear regression fit of the one binding site model (eqn 1; n=1), the dashed line represents a fit of the two binding site model (eqn 1; n=2) and the solid line represent a fit of the three binding site model (eqn 1; n=3). The same data is plotted in a direct plot (B), and with log[Ca2+] (C). ■, 0.16 mM Ca2+; □, 1.26 mM Ca2+; ●, 33.6 mM Ca2+; ○, 200 mM Ca2+.
Determination of affinities
Analysis of the activity of MMP-12 as a function of the concentration of calcium showed that the activity was directly correlated to the binding of calcium (Figure 3B and 3C). Since the enzyme is known to have three binding sites for calcium, a three-site model was chosen for the modelling. This was substantiated by a comparison of different models for the data at pH 7.5 (Figure 3B and 3C), which showed that a model for a single binding site did not describe the data satisfactorily and that a three-site model was slightly better than a two-site model. However, reliable KD values were only obtained for two sites (Table 1) since the affinity of calcium for the third binding site was found to be very low (>100 mM) at pH 7.5 and the quantification was not consistent. The three-site model also gave the best fit for data at pH 7.0, but at pH 5.0, 5.5, 6.0, and 6.5 it was no better than a two-site model, while at pH 4.5 a single-site model was sufficient. The simplest model that could be used to describe the data was therefore used to quantify the KD values (Table 1). The data revealed that the three calcium ions interacting with MMP-12 had significantly different affinities and that the affinities were highly pH-dependent. Consequently, the contribution of calcium binding to the intermediate- and low-affinity sites was pH-dependent. Thus the use of less complex binding models at lower pH values is supported by the fact that the low affinity site was not significantly occupied at pH values below 7 and the intermediate-affinity site at pH 4.5.
Table 1. Affinities for interactions between Ca2+ and MMP-12 at different pH-values.
Data of the type shown in Figure 3(b) was analysed by non-linear regression using a three-site model (eqn 1, n=3) for pH 7.0 and 7.5, a two-site model (eqn 1, n=2) for pH 5.0–6.5 and a one site model (eqn 1, n=1) for pH 4.5. Hence, no value for KD2 was determined at pH 4.5. Errors were calculated as standard errors from the standard deviations of regressions from two different measurements.
| pH | KD1 (mM) | KD2 (mM) |
|---|---|---|
| 7.5 | 0.10±0.035 | 4.6±9.3 |
| 7.0 | 0.16±0.038 | 6.5±12 |
| 6.5 | 0.19±0.039 | 31±10 |
| 6.0 | 0.23±0.068 | 57±13 |
| 5.5 | 0.75±0.14 | 63±18 |
| 5.0 | 2.9±0.74 | 69±59 |
| 4.5 | 6.3±1.0 | Not determined |
Determination of pKa values
Examination of the pH-dependence of the KD values for the two binding sites with highest affinities for calcium (Table 1) showed that the relationship could be described by a model representing a singly ionizing system (Figure 4). (Equations for more than one ionizable group did not fit the experimentally determined KD values.) It was thus possible to estimate the apparent pKa values of 4.3±0.3 for the high-affinity site and 6.5±0.2 for the intermediate-affinity site. The reduction of enzymatic activity at acidic pH did not allow reliable measurements below pH 4.5 and hence a possible titration of further carboxylic acids in the calcium binding sites was not practicable. Interestingly, the highest [Ca2+] used at pH 4.5 inhibited the enzymatic activity (results not shown).
Figure 4. Determination of pKa values for the calcium binding sites of MMP-12.
KD values for the high-affinity site (A) and the intermediate-affinity site (B) plotted against pH (data from Table 1). The line is the theoretical curve obtained by non-linear regression using a model representing a singly ionising system (eqn 2).
DISCUSSION
Validity of experimentally determined values
The affinities of the interactions between calcium and MMP-12 were obtained by simply measuring the enzyme activity as a function of [Ca2+]. The activity was used as a reporter of the amount of Ca2+ bound, under the assumption that calcium binding and activity are directly correlated. The data analysis by itself, however, supports this because of the very good fit of the models to the data. The question of whether the calcium is regulatory or merely a structural requirement for activity for this enzyme does not matter in that respect. In both cases the fraction of active enzyme is measured. The model for three binding sites gave the best fit at the highest pH-values, which is in accordance with the structural knowledge that there are three calcium binding sites, although the analysis yielded too high standard errors for the KD value of the third site to be presented.
pH
The pH-dependency of the MMP-12 activity was more complex at higher [Ca2+] compared with the lower concentrations. The pH was able to influence the activity directly by changing the ionization state of the catalytic glutamate and other residues involved in catalysis, and indirectly by affecting calcium binding and thereby the structure of the enzyme. Hence, the results suggest that at low [Ca2+] the pH-dependency of activity is governed by catalysis, while at high [Ca2+] the pH-dependency is the sum of the effect on catalysis and calcium binding.
The pKa values determined for the calcium binding sites are probably an overall pKa for all carboxylic acids contributing to calcium binding, which means that all carboxylic acids in one calcium-binding site have about the same pKa. However, the pKa values may also be valid for one unique carboxylic acid and in that case the remaining carboxylic acids would titrate at pH lower than 4 in the high-affinity site and lower than 5 in the intermediate-affinity site. It is probably not possible to see further titration and thus find the second pKa, because titration of a second glutamate or aspartate will totally disrupt the interaction between calcium and the binding site, resulting in immeasurable affinities, as is the case for the low-affinity site below pH 7.0 and for the intermediate site at pH 4.5. Hence, at pH lower than 5 only one calcium-binding site is functional.
The low-affinity site appears to have a pKa that is higher than 6.5, which is hard to explain by a simple titration of an aspartic acid residue. They have typical pKa values in proteins between 2 and 6, but this can of course be perturbed. Although in this case, the aspartic acid residue interacts with an ion, which should make it more acidic and not more basic [8]. The titration of the low-affinity site could therefore perhaps be attributed to some other amino acids(s) that affect the structure of the whole calcium-binding site so that calcium binding is lost. Calcium binding is not only dependent on the inner-sphere ligands, but also on long-range electrostatic interactions and charge balancing by distant residues [10]. Moreover, the binding sites are usually supported by a hydrogen-bonding network stabilizing their structure. All these factors are also pH-dependent and could of-course contribute to the high apparent pKa of this site.
Assignment of Ca2+ binding sites
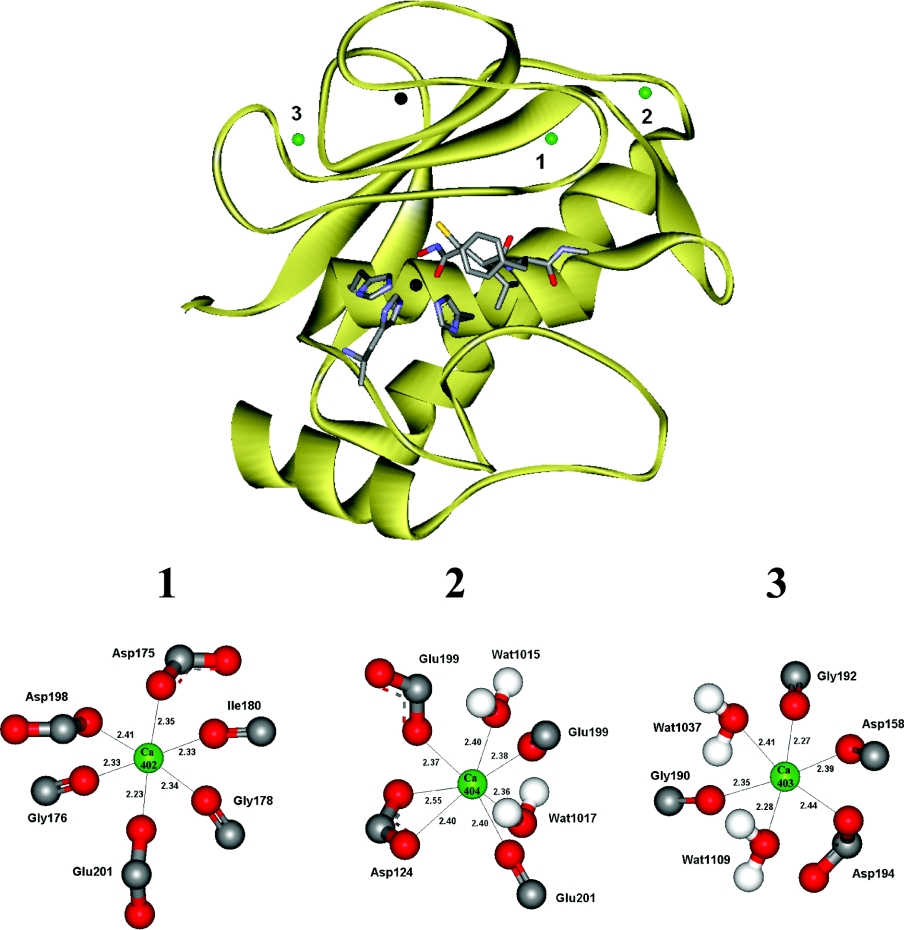
By studying the three-dimensional structure (PDB ID: 1JK3) [11] of MMP-12, it was evident that the composition and structure of the three calcium-binding sites are different (Figure 5). Site 1 binds the calcium ion designated Ca402 in the structure file. In this site, calcium is co-ordinated by six oxygen atoms, which are the contributions of three backbone carbonyls and three carboxylate oxygen atoms. This site has a nearly ideal octahedral geometry and the closest contacts with the calcium ion. Site 2 binds Ca404, which interacts with five oxygen atoms, contributed by two backbone carbonyls and three carboxylate oxygen atoms. In addition, the calcium interacts with two water molecules that complete the pentagonal bipyramid geometry of the binding site. Finally, site 3 binds Ca403, which is octahedrally co-ordinated by two water molecules and four oxygen atoms, contributed by three backbone carbonyls and one carboxylate oxygen atom. Consequently it only has four protein contacts. We propose that site 1 represents the high-affinity site and note that it is located closest to the catalytic site, holding strand IV and the S-loop in place (Figure 5). These are important for substrate binding and specificity. Furthermore, the low affinity site is proposed to be represented by site 3. Accordingly, site 2 is proposed to be the intermediate-affinity binding site, located adjacent to the high affinity site. The identification of inner sphere ligands may not be sufficient for ranking the sites with respect to their affinities to calcium, since charge balancing by distant residues and conformational changes induced by calcium binding can have a large effect on the affinity for calcium ions. Nevertheless, there is a correlation between the number of water molecules in the binding site and affinity [12]. Still, the proposed assignment should be tested by a more specific method, for example, where individual residues in the binding sites are mutated and the effect on the different affinities determined.
Figure 5. Calcium binding sites of MMP-12.
The three-dimensional structure for MMP-12 (produced using the coordinates from the PDB-file 1JK3 by the program WebLab ViewerLite 3.20) showing the three calcium ions (green) and their binding sites, numbered from 1–3. The active site is represented by one of the two zinc ions (black), three zinc-co-ordinating histidines, and the inhibitor batimastat. Close up pictures of the calcium binding sites are shown below. Ca402 has six protein contacts while Ca404 and Ca403 have five and four contacts respectively. Site1 is the proposed high-affinity binding site that binds Ca402; site 2 the intermediate-affinity binding site that binds Ca404; site 3 the low-affinity binding site that binds Ca403. Wat, water molecule.
The calcium binding properties of MMP-12 observed in this study seems to be a common property among the MMPs. For example, the structures of MMP-2 (PDB ID: 1QIB), MMP-3 (PDB ID: 1B8Y) and MMP-9 (PDB ID: 1GKC) reveal three calcium binding sites corresponding to those of MMP-12 in all three structures [5]. MMP-8 (PDB ID: 1I76) has two corresponding calcium binding sites but the intermediate-affinity site is missing from its structure.
Biological relevance
The common opinion is that for MMPs the Ca2+ stabilizes their structure and is not regulatory in the sense that calcium, for example, triggers proteolytic activity by inducing a conformational change and thus switches the enzyme from an inactive to an active conformation. There is support for the structural- requirement hypothesis in this study, that is that binding is not co-operative as it usually is for small ligands with activity regulating properties and also that keeping MMP-12 in a buffer with low [Ca2+] for extended periods of time results in an irreversible loss of activity. The latter is probably due to partial unfolding and/or autohydrolysis at low [Ca2+]. On the other hand, if Ca2+ was merely important for structure, then it would be expected that all three binding sites would have high affinity for calcium. The [Ca2+] of the extracellular space is around 1 mM [13], which from the results of this study, would result in a 50% MMP-12 activity at pH 7.5. The degree of bound calcium would be 92% at the high-affinity site, 6.6% at the intermediate-affinity site and <1% at the low-affinity site. For comparison, the conditions normally used for assays (pH 7.5 and 200 mM Ca2+) would result in 99.9% calcium bound at the high-affinity site, 93% at the intermediate-affinity site and 66% at the low-affinity site. The discrepancy is obvious, so how can the extracellular calcium level keep the enzyme folded and protected from autohydrolysis with such low fractional occupancy? Although the concentration of calcium in the extracellular space is tightly regulated, it can be significantly different in microenvironments and parts of tissues during certain circumstances. One such circumstance, related to this work, is inflammation, where the [Ca2+] may be several fold higher than normal [14]. Macrophages, the very cells expressing MMP-12, are attracted to sites of inflammation by sensing the high [Ca2+]. There is accumulating evidence that the activity of extracellular proteins that bind calcium can be regulated by local changes in [Ca2+]. An example is lipoprotein lipase, an enzyme that is only active as a dimer and whose dimerization is influenced by calcium [15]. Also collagen binding by basement membrane protein BM-40 is thought to be regulated by calcium [16] and the activity of bpDNAse is affected by calcium [17].
Our interpretation is consequently that the physiological explanation of the calcium requirement of MMP-12 is that the activity of MMP-12 is regulated by the [Ca2+] so that it is active at sites of inflammation or other calcium-rich microenvironments and not elsewhere where it would have a deleterious effect. Releasing stable proteases in tissues could lead to a lot of damage, as for example by the snake venom metallopeptidases (haemorrhagins), a group of peptidases that may have evolved from an ancestor common to both the haemorrhagins and the MMPs [18]. While the venom proteases are very stable and have disulphide bridges to stabilize the structure, the less-stable MMPs could have evolved by using calcium as structural stabilizers instead of disulphides, and thus programming them to ‘self-destruct’ at low [Ca2+] by a fine-tuned mechanism using the balance between extracellular [Ca2+] and the affinity between calcium and its binding sites in the protease. The calcium requirement thus provides a safety mechanism so that proteolysis is limited to a certain space and time. Furthermore, calcium-dependent degradation of TIMP-2, one of the four natural inhibitors of MMPs, has been demonstrated [19]. This also suggests that [Ca2+] is a factor in the regulation of extracellular matrix turnover by MMP-activity. So, the answer to the question of whether the calcium is regulatory or merely a structural requirement for activity for this enzyme could be that calcium is both a structural stabilizer and has a regulatory function in the sense that it limits MMP-12 activity to microenvironments of high [Ca2+].
Conclusions
The current study has revealed that the affinities for the interaction between calcium and MMP-12 and the pH-dependence of the interactions are in a range where small variations in the physiological conditions would have a major impact on MMP-12 activity. This suggests that MMP-12 activity may be regulated by extracellular [Ca2+]. The relevance for MMP-12 involvement in disease and how the regulation mechanism influences the potential of MMP-12 as a drug target is an obvious continuation of the current study.
Acknowledgments
MMP-12 was a gift from Medivir AB, Huddinge, Sweden. This work was supported by the Swedish Agency for Innovation Systems (VINNOVA).
References
- 1.Rawlings N. D., Tolle D. P., Barrett A. J. MEROPS: the peptidase database. Nucl. Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snider G. L. Understanding inflammation in chronic obstructive pulmonary disease: the process begins. Am. J. Respir. Crit. Care Med. 2003;167:1045–1046. doi: 10.1164/rccm.2302002. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg G. A. Matrix metalloproteinases and neuroinflammation in multiple sclerosis. Neuroscientist. 2002;8:586–595. doi: 10.1177/1073858402238517. [DOI] [PubMed] [Google Scholar]
- 4.Liu M., Sun H., Wang X., Koike T., Mishima H., Ikeda K., Watanabe T., Ochiai N., Fan J. Association of increased expression of macrophage elastase (matrix metalloproteinase 12) with rheumatoid arthritis. Arthritis Rheum. 2004;50:3112–3117. doi: 10.1002/art.20567. [DOI] [PubMed] [Google Scholar]
- 5.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich P. The intriguing Ca2+ requirement of calpain activation. Biochem. Biophys. Res. Commun. 2004;323:1131–1133. doi: 10.1016/j.bbrc.2004.08.194. [DOI] [PubMed] [Google Scholar]
- 7.Morales R., Perrier S., Florent J. M., Beltra J., Dufour S., De Mendez I., Manceau P., Tertre A., Moreau F., Compere D., et al. Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12. J. Mol. Biol. 2004;341:1063–1076. doi: 10.1016/j.jmb.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 8.Fersht A. New York: W. H. Freeman and Company; 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. [Google Scholar]
- 9.Yu Q., Kandegedara A., Xu Y., Rorabacher D. B. Avoiding interferences from Good's buffers: A contiguous series of noncomplexing tertiary amine buffers covering the entire range of pH 3–11. Anal. Biochem. 1997;253:50–56. doi: 10.1006/abio.1997.2349. [DOI] [PubMed] [Google Scholar]
- 10.Pidcock E., Moore G. R. Structural characteristics of protein binding sites for calcium and lanthanide ions. J. Biol. Inorg. Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- 11.Lang R., Kocourek A., Braun M., Tschesche H., Huber R., Bode W., Maskos K. Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 Å crystal structure. J. Mol. Biol. 2001;312:731–742. doi: 10.1006/jmbi.2001.4954. [DOI] [PubMed] [Google Scholar]
- 12.McPhalen C. A., Strynadka N. C., James M. N. Calcium-binding sites in proteins: a structural perspective. Adv. Protein Chem. 1991;42:77–144. doi: 10.1016/s0065-3233(08)60535-5. [DOI] [PubMed] [Google Scholar]
- 13.Fraústo da Silva J. J. R., Williams R. J. P. Oxford: Oxford University Press; 1993. The biological chemistry of the elements: the inorganic chemistry of life. [Google Scholar]
- 14.Brown E. M., MacLeod R. J. Extracellular calcium sensing and extracellular calcium signaling. Physiol. Rev. 2001;81:239–297. doi: 10.1152/physrev.2001.81.1.239. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Lookene A., Wu G., Olivecrona G. Calcium triggers folding of lipoprotein lipase into active dimers. J. Biol. Chem. 2005;280:42580–42591. doi: 10.1074/jbc.M507252200. [DOI] [PubMed] [Google Scholar]
- 16.Maurer P., Mayer U., Bruch M., Jeno P., Mann K., Landwehr R., Engel J., Timpl R. High-affinity and low-affinity calcium binding and stability of the multidomain extracellular 40-kDa basement membrane glycoprotein (BM-40/SPARC/osteonectin) Eur. J. Biochem. 1992;205:233–240. doi: 10.1111/j.1432-1033.1992.tb16773.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen C. Y., Lu S. C., Liao T. H. The distinctive functions of the two structural calcium atoms in bovine pancreatic deoxyribonuclease. Protein Sci. 2002;11:659–668. doi: 10.1110/ps.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moura-da-Silva A. M., Theakston R. D., Crampton J. M. Evolution of disintegrin cysteine-rich and mammalian matrix-degrading metalloproteinases: gene duplication and divergence of a common ancestor rather than convergent evolution. J. Mol. Evol. 1996;43:263–269. doi: 10.1007/BF02338834. [DOI] [PubMed] [Google Scholar]
- 19.Munshi H. G., Wu Y. I., Ariztia E. V., Stack M. S. Calcium regulation of matrix metalloproteinase-mediated migration in oral squamous cell carcinoma cells. J. Biol. Chem. 2002;277:41480–41488. doi: 10.1074/jbc.M207695200. [DOI] [PubMed] [Google Scholar]