Abstract
HNF-4 (hepatocyte nuclear factor 4) is a key regulator of liver-specific gene expression in mammals. We have shown previously that the activity of the human APOC3 (apolipoprotein C-III) promoter is positively regulated by the anti-inflammatory cytokine TGFβ (transforming growth factor β) and its effectors Smad3 (similar to mothers against decapentaplegic 3) and Smad4 proteins via physical and functional interactions between Smads and HNF-4. We now show that the pro-inflammatory cytokine TNFα (tumour necrosis factor α) antagonizes TGFβ for the regulation of APOC3 gene expression in hepatocytes. TNFα was a strong inhibitor of the activity of apolipoprotein promoters that harbour HNF-4 binding sites and this inhibition required HNF-4. Using specific inhibitors of TNFα-induced signalling pathways, it was shown that inhibition of the APOC3 promoter by TNFα involved NF-κB (nuclear factor κB). Latent membrane protein 1 of the Epstein–Barr virus, which is an established potent activator of NF-κB as well as wild-type forms of various NF-κB signalling mediators, also inhibited strongly the APOC3 promoter and the transactivation function of HNF-4. TNFα had no effect on the stability or the nuclear localization of HNF-4 in HepG2 cells, but inhibited the binding of HNF-4 to the proximal APOC3 HRE (hormone response element). Using the yeast-transactivator-GAL4 system, we showed that both AF-1 and AF-2 (activation functions 1 and 2) of HNF-4 are inhibited by TNFα and that this inhibition was abolished by overexpression of different HNF-4 co-activators, including PGC-1 (peroxisome-proliferator-activated-receptor-γ co-activator 1), CBP [CREB (cAMP-response-element-binding protein) binding protein] and SRC3 (steroid receptor co-activator 3). In summary, our findings indicate that TNFα, or other factors that trigger an NF-κB response in hepatic cells, inhibit the transcriptional activity of the APOC3 and other HNF-4-dependent promoters and that this inhibition could be accounted for by a decrease in DNA binding and the down-regulation of the transactivation potential of the AF-1 and AF-2 domains of HNF-4.
Keywords: apolipoprotein C-III (APOC3), co-activators hepatocyte nuclear factor 4 (HNF-4), nuclear factor κB (NF-κB), transcriptional inhibition, tumour necrosis factor α (TNFα)
Abbreviations: AF, activation function; APOA1, apolipoprotein A-I; APOA4, apolipoprotein A-IV; APOC3, apolipoprotein C-III; CBP, CREB (cAMP-response-element-binding protein) binding protein; DBD, DNA-binding domain; DMEM, Dulbecco's modified Eagle's medium; EAR, erbA-related protein; ERK, extracellular-signal-regulated kinase; FBS, fetal bovine serum; GRIP1, glucocorticoid receptor interacting protein-1; HEK 293T, human embryonic kidney 293T; hyb, hybrid; HNF-4, hepatocyte nuclear factor 4; HRE, hormone response element; IκB-ND, inhibitor of NF-κB non-degradable; IKKβ, inhibitor of NF-κB kinase β; IMBB-FORTH, Institute of Molecular Biology and Biotechnology–Foundation of Research and Technology Hellas; LBD, ligand binding domain; LMP1, latent membrane protein 1; MEK-1, MAPK (mitogen-activated protein kinase)/ERK kinase; NF-κB, nuclear factor κB; NIK, NF-κB-inducing kinase; ONPG, o-nitrophenyl galactopyranoside; pERK, phospho-ERK; PGC-1, peroxisome-proliferator-activated-receptor-γ co-activator 1; RXRα, retinoid X receptor α; Smad, similar to mothers against decapentaplegic; TGFβ, transforming growth factor β; TNFα, tumour necrosis factor α; SRC3, steroid receptor co-activator 3; TIF2, transcriptional intermediary factor 2
INTRODUCTION
HNF-4 (hepatocyte nuclear factor 4) belongs to the hormone nuclear receptor gene superfamily of transcription factors on the basis of its domain architecture, which is characterized by the presence of an N-terminal AF-1 (activation function 1), a DBD (DNA-binding domain) of the zinc-finger type and a multifunctional LBD (ligand-binding domain) which harbours AF-2 and is connected with the DBD by a hinge region [1–4]. Although HNF-4 belongs to a subclass of nuclear receptors termed ‘orphan receptors’, it was recently shown that fatty acids may constitute endogenous ligands of HNF-4 [5–7]. HNF-4 also harbours an inhibitory C-terminal domain which modulates the binding of co-activators to the adjacent AF-2 [4,8]. The AF-1 and AF-2 domains of HNF-4 have been shown to interact with different members of the p160 family of co-activators, including SRC1 (steroid receptor co-activator 1) and GRIP1 (glucocorticoid receptor interacting protein-1)/TIF2 (transcriptional intermediary factor 2) [9–11], as well as with the histone acetyltransferase CBP [CREB (cAMP-response-element-binding protein) binding protein] [12]. CBP acetylates HNF-4, and this modification has been shown to regulate several functions of HNF-4, including DNA binding, nuclear accumulation and interaction with co-activators [13]. In addition, HNF-4 interacts with PGC-1 (peroxisome-proliferator activated-receptor-γ co-activator 1), a protein that plays important roles in hepatic gluconeogenesis [11,14].
HNF-4 is predominantly expressed in the liver, intestine, pancreas and kidney and regulates the expression of genes and gene networks with important physiological roles in the above organs [2]. Conditional inactivation of HNF-4 in the liver of mice caused weight loss, increased rate of mortality and lipid abnormalities due to impaired expression of HNF-4-regulated genes involved in lipid and bile acid metabolism and transport [15]. Among the genes that were transcriptionally silenced by the inactivation of HNF-4 in the liver was the APOC3 (apolipoprotein C-III) gene [15].
APOC3 has been implicated in the modulation of binding of apolipoprotein E-containing lipoproteins to cell receptors and their subsequent catabolism [16–20]. The importance of APOC3 in triacylglycerol homoeostasis is supported by studies in transgenic mice, in which overexpression of the APOC3 gene was found to be associated with severe hypertriacylglycerolaemia (‘hypertriglyceridaemia’) due to the defective clearance of triacylglycerol-rich lipoprotein remnants [21,22].
The human APOC3 gene is closely linked to the human APOA1 and APOA4 (apolipoproteins A-I and A-IV) genes on the long arm of chromosome 11 [23]. In vitro mutagenesis established that three HREs (hormone-response elements), located in the proximal promoter and enhancer, as well as three Sp1 (stimulating protein-1)-binding sites located in the APOC3 enhancer, are important for the APOC3 gene expression in hepatic cells [24–28]. Two of the above HREs (elements B and I) bind HNF-4 and other orphan and ligand-dependent nuclear receptors [25–28].
Previous studies have demonstrated that the APOC3 gene is down-regulated during the acute-phase response, owing to the action of pro-inflammatory cytokines such as TNFα (tumour-necrosis factor-α) and interleukin-1 [29,30]. Transcription factors found previously to mediate this process include the AP-1 (activation protein-1) proteins c-Jun and ATF-2 (activating transcription factor 2), as well as C/EBPδ (CAAT/enhancer binding protein δ) [30,31]. Natural extinguishing of the acute-phase response occurs in part because of the production of anti-inflammatory cytokines such as interleukin-10, interleukin-13 and TGFβ (transforming growth factor β) [32]. TGFβ and its signalling mediators, the Smad (similar to mothers against decapentaplegic) proteins, are potent anti-inflammatory molecules in mammals [33–36].
We have shown recently that TGFβ and its signal transducers, the Smad proteins, transactivate the APOC3 gene promoter by interacting physically and functionally with HNF-4, which binds to the proximal APOC3 HRE (element B) [37,38]. We now show that the pro-inflammatory cytokine TNFα antagonizes TGFβ for the regulation of APOC3 gene expression in hepatocytes. Inhibition of the APOC3 promoter by TNFα requires the participation of the NF-κB (nuclear factor κB) pathway, which affects the DNA binding and transactivation potential of HNF-4.
MATERIALS AND METHODS
Materials
All reagents for cell culture, including DMEM (Dulbecco's modified Eagle's medium), FBS (fetal bovine serum), trypsin/EDTA and PBS were purchased from Life Technologies. ONPG (o-nitrophenyl galactopyranoside), PMSF, the anti-FLAG (M2) mouse monoclonal antibody and the anti-β-tubulin mouse monoclonal antibody were purchased from Sigma. The anti-HNF-4 (C-19) goat polyclonal antibody and anti-NF-κB p65(A) rabbit polyclonal antibody were purchased from Santa Cruz Biotechnology. The anti-ERK [anti-(extracellular-signal-regulated kinase)] and anti-pERK [anti-phospho-ERK] antibodies were gifts from Dr G. Mavrothalassitis [IMBB-FORTH (Institute of Molecular Biology and Biotechnology, Foundation of Research and Technology Hellas), Heraklion, Crete, Greece]. The rabbit anti-HNF-4 antibody was kindly provided by Dr Iannis Talianidis (IMBB-FORTH). The dNTPs, poly(dI/dC) and the G-Sepharose beads were purchased from Amersham Pharmacia. The luciferase assay kit and random hexamers were purchased from Promega. The MEK [MAPK (mitogen-activated protein kinase)/ERK kinase]-1 inhibitor U0126 was purchased from Upstate Biotechnology. Human recombinant TNFα was purchased from Roche and Minotech. Human recombinant TGFβ was purchased from R&D Systems. The Super Signal West Pico Chemiluminescent Substrate was purchased from Pierce. Streptavidin Dynabeads were purchased from Dynal Biotech. Superscript reverse transcriptase was purchased from Invitrogen. Oligonucleotides were synthesized at the Microchemistry Laboratory of FORTH.
Cell culture, transient transfections and reporter assays
Human hepatoma HepG2 cells, HEK (human embryonic kidney) 293T cells and COS-7 fibroblasts were cultured in DMEM, supplemented with 10% (v/v) FBS and penicillin/streptomycin, in a 37 °C, 5%-CO2 incubator. Human recombinant TNFα (0, 250, 500 or 1000 units), TGFβ (0, 40, 80 pM) or the MEK1 inhibitor U0126 (10 μM) were added to the cell-culture medium for different time periods. Transient transfections were performed by the Ca3(PO4)2 co-precipitation method using 6 μg of DNA/well when the transfections were performed in six-well plates or 35 μg/plate when transfections were performed in P-100 plates. Luciferase assays were performed using the Luciferase assay kit from Promega Corp. according to the manufacturer's instructions. Normalization for transfection efficiency was performed using β-galactosidase assays.
Plasmids
The apolipoprotein promoter plasmids APOC3 (−890/+24)-Luc, APOC3 (−686/+24)-Luc, APOC3 (−99/+24)-Luc, APOC2 (−550/+18)-Luc, APOA4 (−700/+24)-Luc, the NF-κB responsive vector (NF-κB)3-Luc and expression vectors for LMP1 (latent membrane protein 1), NIK (NF-κB-inducing kinase), IKKβ (inhibitor of NF-κB kinase β), IκB-ND (inhibitor of NF-κB non-degradable), p65, p50, p50/p65 hyb (hybrid) Smad proteins (Smad3 and Smad4) have been described previously [24,38–41]. Expression vectors for wild-type or truncated forms of rat HNF-4a1 in expression vector pCDNA1-amp or fused with the DBD of GAL4 were kindly provided by Dr Margarita Hadzopoulou-Cladaras (Department of Genetics, Development and Molecular Biology, Laboratory of Developmental Biology, School of Biology, Aristotle University of Thessaloniki, Thessaloniki, Greece). The expression vector for the human PGC-1 co-activator was kindly given by Dr Anastasia Kralli (Department of Cell Biology, The Scripps Research Institute, La Jolla, CA, U.S.A.). The expression vector for the human CBP co-activator was kindly given by Dr Ioannis Talianidis (IMBB–FORTH). The expression vector for the human SRC3 was kindly given by Dr Hinrich Gronemeyer [IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Illkirch, Strasbourg, France].
Reverse-transcription PCR
HepG2 cells were treated with 1,000 units of TNFα in the absence or in the presence of TGFβ (40 and 80 pM) for 24 h or left untreated. Total RNAs were prepared and equal concentrations of RNAs were used for cDNA synthesis. The synthesis of cDNAs was performed using Superscript reverse transcriptase and random haxamers as primers. The cDNAs were used for PCR amplifications with primers corresponding to the human APOC3 cDNA. The sequences of the primers were as follows: APOC3 forward primer, 5′ AGGAGTCCCAGGTGGCCCAGCAG 3′; APOC3 reverse primer, 5′ CACGGCTGAAGTTGGTCTGACCTCA 3′.
Indirect immunofluorescence
HepG2 cells or transfected COS-7 cells were seeded on glass coverslips coated with 0.1% gelatin. Cells were washed three times on a slow rotating platform with PBS+/+ (PBS plus 0.9 mM CaCl2 and 0.5 mM MgCl2) and fixed with 3% p-formaldehyde in PBS+/+ for 5 min at room temperature. Cells were washed three times with PBS+/+ and permeabilized with 0.5% Triton X-100 in buffer 1 (10×buffer 1 is 137 mM NaCl, 5 mM KCl, 1 mM Na2HPO4, 0.4 mM KH2PO4, 5.5 mM glucose, 4 mM NaHCO3, 2 mM MgCl2, 2 mM EDTA, 2 mM EGTA and 20 mM Mes, pH 6.0–6.5) for 5 min at room temperature. Cells were washed three times with PBS+/+, blocked with PBS +/+/1.5% FBS and incubated with the primary antibody (1:200 dilution, in PBS+/+/1.5% FBS) for 30 min at 4 °C. Cells were washed three times with PBS+/+/1.5% FBS and incubated with the secondary antibody (1:50 dilution in PBS +/+/1.5% FBS) for 30 min at 4 °C in the dark. Cells were washed three times with PBS+/+ in the dark and mounted on glass slides using mounting solution (glycerol/PBS, 1:1, v/v). Cells were observed using a Leica SP confocal fluorescent microscope.
Western blotting
For Western-blotting analysis of endogenous (HepG2) or transfected (HEK-293T) proteins, cells were trypsinized, pelleted by centrifugation at 2000 rev./min (350 g) for 5 min, washed once with ice-cold PBS, re-centrifuged as described above and resuspended in lysis buffer [50 mM Tris/HCl (pH 7.4), 150 mM Nacl, 1% Nonidet P40, 0.25% sodium deoxycholate and 1 mM EGTA] supplemented with protease inhibitors (1 mM PMSF, 1 μg/ml pepstatin and 1 μg/ml leupeptin). Extracts were allowed to rotate at 4 °C for 30 min and centrifuged at 13000 rev./min (15800 g) for 5 min at 4 °C. Protein concentration was measured using the Bio-Rad DC Protein Assay kit, and equal amounts were loaded on SDS/10.5%-(w/v)-polyacrylamide gels, followed by electrotransfer to Protran 0.45-μm-pore-size nitrocellulose transfer membrane (Schleicher & Schuell BioScience). Immunoblotting was performed using appropriate monoclonal or polyclonal antibodies, followed by incubation with horseradish-peroxidase-conjugated secondary antibodies. Proteins were visualized by enhanced chemiluminescence.
Chromatin immunoprecipitations
The chromatin immunoprecipitation assay was performed as described previously [42], using chromatin from HepG2 cells and a rabbit polyclonal antibody towards human HNF-4. Immunoprecipitated chromatin was analysed by PCR using primers corresponding to the proximal (−233/−21) and distal (−882/−518) regions of the human APOC3 promoter. The proximal APOC3 promoter primers were: P1: 5′ CAG GCC CAC CCC CAG TTC CTG AGC TCA 3′; P2: 5′ CCT GTT TTA TAT CAT CTC CAG GGC AGC AGG C 3′. The distal APOC3 promoter primers were: D1: 5′ AGT TGC TCC CAC AGC CAG GGG GCA GT 3′; D2: 5′ TCT CAC AGC CCC TCC CAG CAC CTC CAT 3′. The products of the PCR amplifications (35 cycles) were analysed by agarose-gel electrophoresis and ethidium bromide staining.
DNA affinity precipitation
For DNA affinity precipitation, nuclear extracts from HepG2 cells that had been treated with TNFα (1000 units) for 24 h or from untreated HepG2 cells were used. Dynabeads were washed once with 1×B&W buffer [5 mM Tris/HCl (pH 7.5), 0.5 mM EDTA and 1 mM NaCl], mixed with 0.58 μM of biotinylated oligonucleotide and incubated at room temperature (25 °C) for 15 min. The oligonucleotide-coupled beads were washed twice with 1×B&W buffer and once with 1×BBRC buffer (10% glycerol, 10 mM Tris/HCl, pH 7.5, 50 mM KCl, 4 mM MgCl2 and 0.2 mM EDTA).
The protein–DNA binding reactions were allowed to proceed for 30 min on ice in a buffer containing 10% (v/v) glycerol, 20 mM Hepes (pH 7.9), 40 mM KCl, 20 mM MgCl2, 4 mM spermidine, 100 μg/ml BSA, 0.02 mM zinc acetate, 0.05% Nonidet P40 and 0.5 mM dithiothreitol. Each reaction mixture included 30 μg of nuclear extracts, 3 μg of competitor poly(dI/dC) and the biotinylated oligonucleotide-coupled Dynabeads, which were prepared as described above in a total reaction volume of 50 μl. Biotinylated oligonucleotides corresponding to the −92/−67 region of the human APOC3 promoter [28] and the −86/−70 region of the human p21Cip1 promoter [43] were utilized. HNF-4 bound to the oligonucleotides was detected by SDS/PAGE and immunoblotting using a polyclonal anti-HNF-4 antibody.
RESULTS
Antagonistic effects between TGFβ and TNFα on the activity of the human APOC3 promoter in HepG2 cells
Negative cross-talk between the TGFβ and TNFα signalling pathways on the activity of the human APOC3 promoter was investigated by transactivation assays in human hepatoma-derived HepG2 cells. For this purpose, HepG2 cells were transfected with a plasmid bearing the firefly luciferase reporter gene under the control of the promoter of the human APOC3 gene between nucleotides −890 and +24 (APOC3 −890/+24-Luc) and were treated with increasing amounts of recombinant human TNFα (250, 500 and 1000 units) for 24 h. As shown in Figure 1(A), TNFα treatment inhibited the activity of the APOC3 promoter in a dose-dependent manner. This finding was in agreement with previous observations showing that TNFα inhibited the transcription of the human APOC3 gene in HepG2 cells [29].
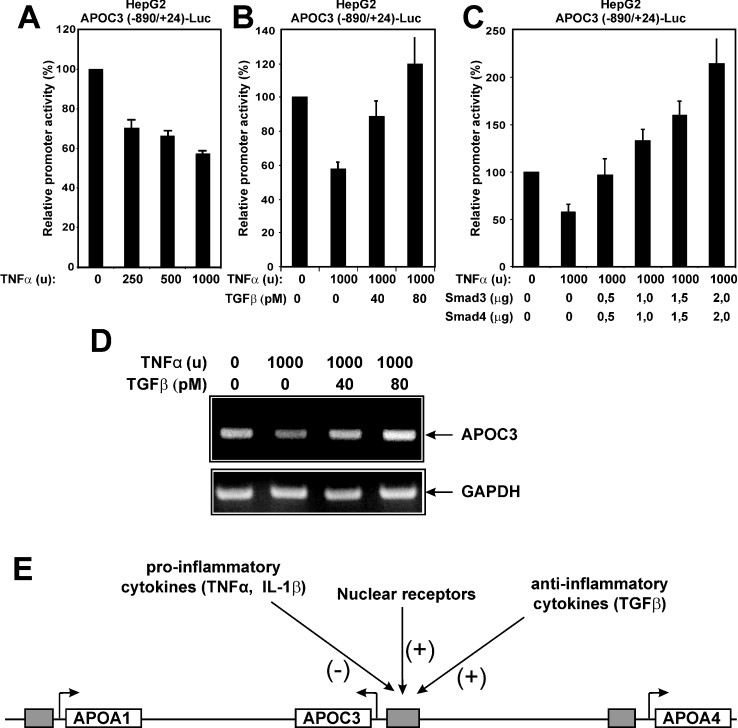
Figure 1. Antagonistic effects of pro- and anti-inflammatory cytokines on the activity of the human APOC3 promoter in HepG2 cells.
(A) Human hepatoma HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg) and treated with different doses of human recombinant TNFα (0, 250, 500 and 1000 units) for 24 h. Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (B) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg) and treated with TNFα (1000 units) in the absence or in the presence of increasing doses of human recombinant TGFβ1 (0, 40 and 80 pM) for 24 h. Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (C) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg) along with increasing concentrations of expression vectors for human Smad3 and Smad4 (0, 0.5, 1.0, 1.5 and 2.0 μg) and treated with a constant dose of TNFα (1000 units) for 24 h. Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (D) HepG2 cells were treated with TNFα (1000 units) in the absence or in the presence of increasing doses of human recombinant TGFβ1 (0, 40 and 80 pM) for 24 h. The expression of the APOC3 gene was analysed by reverse transcription PCR. The expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for normalization of RNA concentration in the various samples. (E) Schematic representation of the human APOA1–APOC3–APOA4 gene cluster and factors that regulate the expression of the APOC3 gene in a positive (+) or a negative (−) manner. The arrows show the direction of transcription of each gene and the grey boxes represent the promoter regions of the three genes of the cluster. Abbreviation: IL-1β, interleukin 1β.
It was next shown that the inhibitory effect of TNFα on the APOC3 promoter could be overcome by the simultaneous treatment of HepG2 cells with increasing doses of human recombinant TGFβ (Figure 1B), suggesting a negative cross-talk between the two pathways operating on the APOC3 promoter. Inhibition of APOC3 promoter activity by TNFα could also be abrogated by overexpression of Smad3 and Smad4 proteins, which are the key effectors of the TGFβ pathway (Figure 1C). In agreement with the data of Figure 1(B), treatment of HepG2 cells with TNFα (1000 units) for 24 h caused a reduction in the steady-state mRNA levels of the human APOC3 gene and this inhibition could be overcome by the simultaneous treatment of HepG2 cells with increasing doses (40 and 80 pM) of TGFβ (Figure 1D).
In conclusion, the results shown in Figure 1 combined with our previous findings [27,28,37,38] indicate that the activity of the human APOC3 promoter in HepG2 cells can be modulated by positive and negative signals which are produced by anti-inflammatory and pro-inflammatory cytokines respectively. This is shown schematically in Figure 1(E).
Inhibition of APOC3 promoter activity by TNFα is mediated by NF-κB
To identify and characterize the signalling pathway(s) that is (are) responsible for the TNFα-induced inhibition of APOC3 promoter activity in HepG2 cells, two inhibitors that specifically block well-characterized and distinct TNFα-induced signalling cascades were utilized: a mutant that blocks the cascade leading to NF-κB activation (a dominant negative, non-degradable form of the IκB inhibitor that keeps NF-κB in the cytoplasm, IκB-ND) and a chemical inhibitor (the MEK1 inhibitor U0126) that blocks the cascade leading from TNFα to ERK. First, the activation of the above signalling cascades by TNFα in HepG2 cells was shown by well-established assays such as TNFα-induced p65 nuclear translocation (Figure 2A) and the TNFα-induced phosphorylation of ERK (Figure 2B). Then the effect of the two inhibitors on the constitutive as well as on the TNFα-inducible activity of the APOC3 promoter was evaluated. As shown in Figure 2(C), inhibition of the APOC3 promoter by TNFα was totally abolished in the presence of the IκB-ND inhibitor. In fact, a strong activation of the APOC3 promoter was observed by the NF-κB inhibitor, even in the absence of TNFα, a finding indicative of the existence of strong endogenous negative signals in HepG2 cells that constitutively inhibit APOC3 promoter activity via the NF-κB pathway. By contrast, the U0126 inhibitor of the MEK1/ERK pathway had only a minor positive effect on both the basal and the TNFα-inhibited APOC3 promoter activity (Figure 2C).
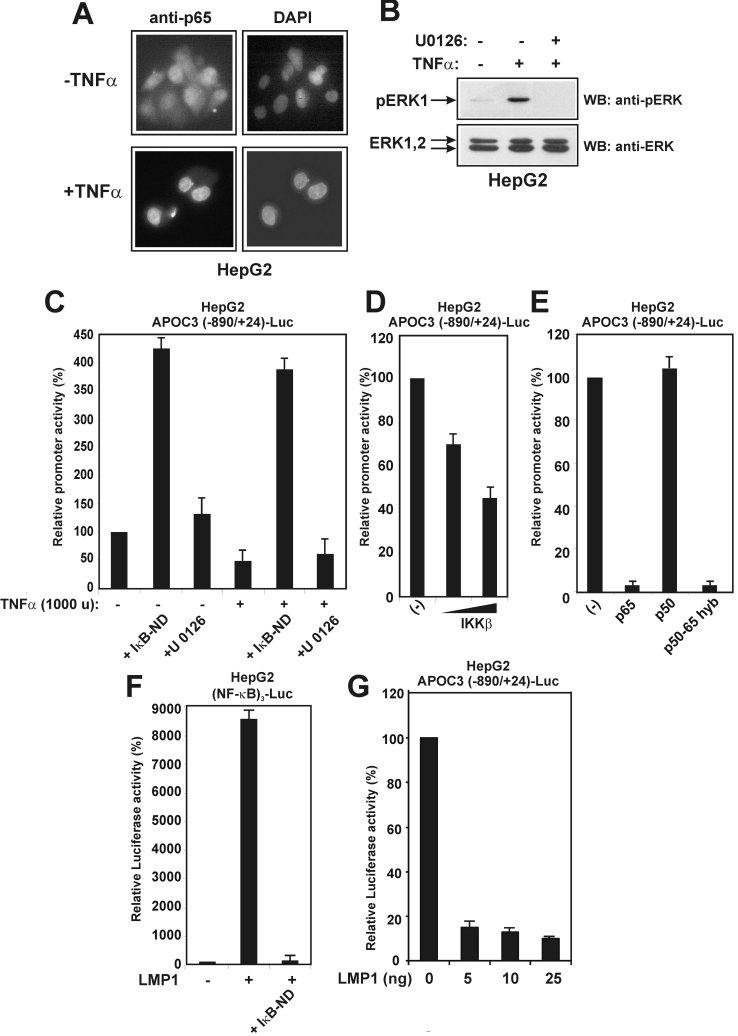
Figure 2. Inhibition of APOC3 promoter activity by TNFα is mediated by NF-κB.
(A) TNFα activates NF-κB in HepG2 cells. HepG2 cells were serum-starved for 16 h and treated with TNFα (1000 units) for 4 h or left untreated. The intracellular distribution of the p65 subunit of NF-κB was examined by indirect immunofluorescence using an anti-p65 antibody followed by a secondary FITC-conjugated antibody. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). (B) TNFα activates the MEK1/ERK pathway in HepG2 cells. HepG2 cells were pretreated with the MEK1 inhibitor U0126 (10 μM) for 24 h, followed by a short treatment with TNFα (1000 units) for 15 min. Cell extracts were analysed by Western blotting for pERK or total ERK using the corresponding anti-ERK antibodies followed by secondary horseradish-peroxidase-conjugated antibodies. Arrows show the position of pERK and ERK1/2 proteins. (C) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg) and treated with TNFα (1000 units) in the presence or in the absence of an expression vector for the non-degradable form of IκBα, IκB-ND (2.0 μg), or the MEK1 inhibitor U0126 (10 μM) for 24 h. Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (D) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg), along with increasing concentrations of an expression vector for IKKβ (0, 1.0 and 2.0 μg). Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (E) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg), along with expression vectors for p65, p50 or a p50/p65 hyb protein (2.0 μg each). Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (F) HepG2 cells were transiently transfected with the (NF-κB)3-Luc reporter plasmid (2 μg), along with an expression vector for LMP1 (25 ng) in the presence or in the absence of an expression vector for IκB-ND (2.0 μg). Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram. (G) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg), along with increasing concentrations of an expression vector for LMP1 (0, 5, 10 and 25 ng). Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown as a histogram.
Next, the effect of selected NF-κB signalling effectors on APOC3 promoter activity in HepG2 cells was investigated by transactivation assays. Figure 2(D) shows that overexpression of IKKβ kinase inhibited APOC3 promoter activity in a dose-dependent manner, and Figure 2(E) shows that overexpression of the p65/RelA subunit of NF-κB totally abolished APOC3 promoter activity, whereas the p50 subunit had no effect. Finally, a p65 hyb protein in which the RHD (Rel homology domain) of p65 was exchanged for the corresponding domain of p50 (p50/p65 hyb) was as effective as full-length p65 in inhibiting the APOC3 promoter, suggesting that the inhibition is mediated by the C-terminal transactivation domain of p65.
As an additional confirmation of the inhibitory role played by NF-κB in APOC3 promoter regulation, the LMP1 of Epstein–Barr virus was utilized in transactivation assays. LMP1 is a protein homologous with the TNFα receptor that has been shown previously to be a potent activator of the NF-κB pathway in various cell types [44]. In a control experiment, it was established that LMP1 strongly activated a NF-κB-responsive promoter [(NF-κB)3] in HepG2 cells and this activation was inhibited by the dominant-negative IκB-ND mutant (Figure 2F). More importantly, it was shown that a small amount of LMP1 was sufficient to fully block APOC3 promoter activity in HepG2 cells (Figure 2G).
In summary, the findings of Figure 2 indicate that activation of the NF-κB pathway in HepG2 cells by TNFα causes a strong inhibition of the APOC3 promoter activity. The molecular mechanism of this inhibition was the subject of the subsequent experiments presented below.
The proximal HRE of the APOC3 promoter is sufficient to mediate inhibition by TNFα
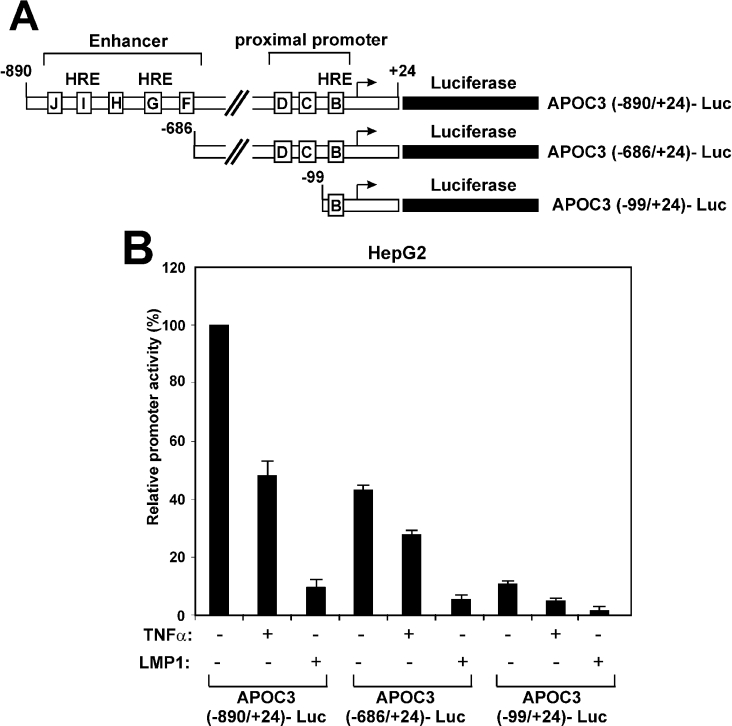
To identify the region of the APOC3 promoter that responds to TNFα, two truncated APOC3 promoter fragments were utilized in transactivation experiments in HepG2 cells. One mutant (APOC3 −686/+24) lacks the region between nucleotides −890 and −687, which includes the distal enhancer elements F–J (Figure 3A). The second mutant (APOC3 −99/+24) contains the region between nucleotides −99 and +24, which includes only the proximal element B (Figure 3A). Element B was shown previously to harbour an HRE that binds a selection of orphan and ligand-dependent nuclear receptors, including HNF-4 [27,28]. Mutagenesis of this element abolished APOC3 promoter activity both in vitro and in vivo [27,28].
Figure 3. The proximal HRE of the APOC3 promoter is sufficient to mediate inhibition by TNFα.
(A) Schematic representation of the APOC3 promoter–luciferase constructs used in the transactivation experiments of (B). Regulatory elements in the APOC3 proximal promoter and the enhancer are shown by boxes labelled B–J. (B) HepG2 cells were transiently transfected with the various APOC3-Luc reporter plasmid (2 μg) shown at the bottom of the graph and were treated with TNFα (1000 units) for 24 h or co-transfected with an expression vector for LMP1 (25 ng). Cell extracts were analysed for luciferase activity. Normalized relative APOC3 promoter activity is shown by a histogram.
As shown in Figure 3(B), both APOC3 promoter mutants were inhibited by TNFα and LMP1 to approximately the same extent as was the full-length APOC3 promoter. The minimal region of the APOC3 promoter required for TNFα or LMP1-induced inhibition is the −99/+24, since further deletion of the APOC3 promoter to nucleotide −55 abolished both the constitutive and the TNFα-inhibited APOC3 promoter activity (results not shown).
The results shown in Figure 3 indicate that inhibition of the APOC3 promoter activity by TNFα could be mediated, at least in part, by interfering with the activity of nuclear receptors that bind to the proximal APOC3 HRE.
TNFα inhibits the HNF-4-mediated transactivation of the APOC3 and APOC2 promoters
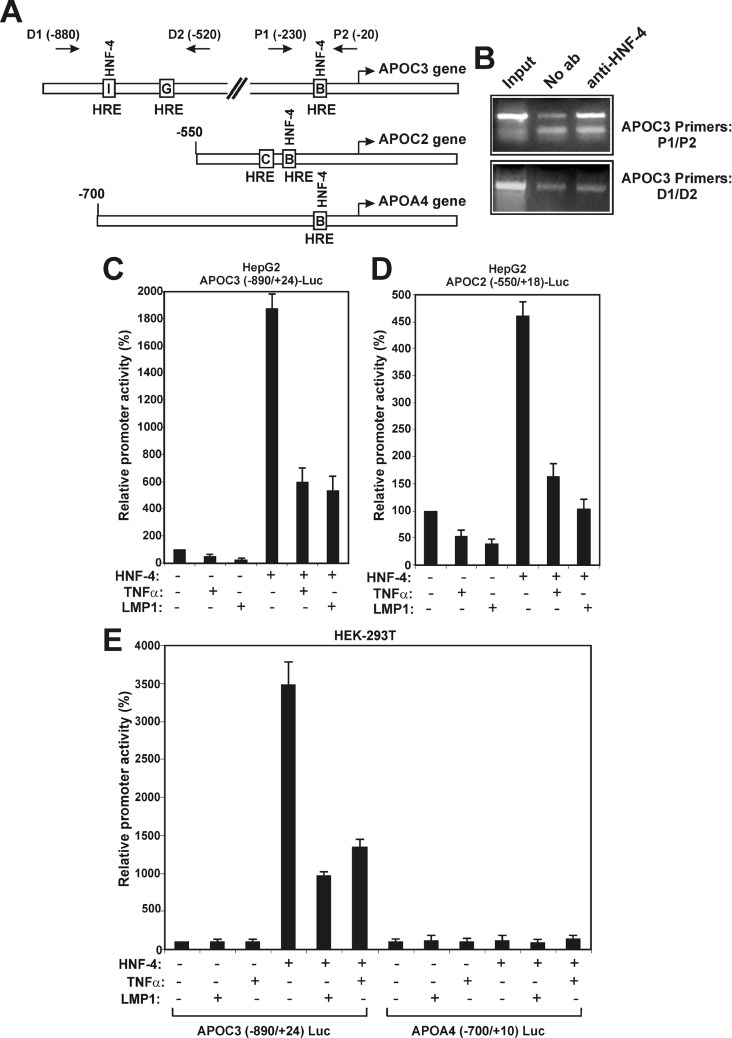
Previous in vivo studies have established that HNF-4 is the single most important regulator of the expression of the APOC3 gene, as well as of other apolipoprotein genes in hepatocytes [15]. It was shown previously that HNF-4 binds with high affinity to the proximal HRE B of the APOC3 promoter in vitro [28]. The recruitment of HNF-4 to the APOC3 promoter in vivo was established here by a chromatin-immunoprecipitation experiment. As shown in Figure 4(B), HNF-4 was recruited to the proximal APOC3 promoter region −230/−20 that includes HRE B, but, surprisingly, it failed to bind to the distal region −880/−520 that harbours the distal HREs G and I.
Figure 4. TNFα inhibits the HNF-4-mediated transactivation of the APOC3 and APOC2 promoters.
(A) Schematic representation of the human APOC3, APOC2 and APOA4 promoter regions, showing the locations of the HREs that bind HNF-4 in each promoter. The location of the oligonucleotide primer sets on the APOC3 promoter that were utilized in the chromatinimmunoprecipitation assays of (B) are shown by arrows labelled D1 and D2 (distal primers) and P1 and P2 (proximal primers). (B) Recruitment of HNF-4 to the proximal APOC3 promoter in vivo. The chromatin-immunoprecipitation assay was performed using chromatin from HepG2 cells as described in the Materials and methods section, using primers corresponding to the proximal (P1 and P2) or the distal (D1 and D2) regions of the APOC3 promoter in the absence (second lane) or in the presence (third lane) of an anti-HNF-4 antibody. Non-immunoprecipitated chromatin was included as a positive control (first lane labelled ‘Input’). (C and D) HepG2 cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid (2 μg) (C) or with the APOC2 (−550/+18)-Luc plasmid (2 μg) (D) and were treated with TNFα (1000 units) for 24 h or were co-transfected with an expression vector for LMP1 (25 ng). The same experiment was repeated in the presence of an expression vector for HNF-4 (2 μg). Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram. (E) HEK-293T cells were transiently transfected with the APOC3 (−890/+24)-Luc reporter plasmid or with the APOA4 (−700/+24)-Luc plasmid (2 μg) and were treated with TNFα (1000 units) for 24 h or co-transfected with an expression vector for LMP1 (25 ng). The same experiment was repeated in the presence of an expression vector for HNF-4 (2 μg). Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram.
It was then shown that HNF-4 strongly transactivated the APOC3 (−890/+24) promoter in HepG2 cells and that this transactivation was inhibited by TNFα or LMP1 expression (Figure 4C). A similarly negative effect of TNFα and LMP1 on HNF-4-mediated transactivation was observed using the APOC2 promoter (Figure 4D), which also contains a binding site for HNF-4 (HRE B; Figure 4A). To confirm the requirement of HNF-4 for the TNFα and LMP1-induced inhibition of the APOC3 promoter, transactivation experiments were performed in HEK-293T cells, which lack endogenous HNF-4. As shown in Figure 4(E), TNFα or LMP1 had no effect on the activity of the APOC3 promoter in HEK-293T cells, whereas they strongly inhibited the HNF-4-mediated transactivation of the same promoter. As a negative control, the human APOA4 promoter (−700/+10) was utilized. This promoter cannot be transactivated by HNF-4, despite the presence of a HNF-4 binding site in the proximal region (Figures 4A and 4E) [40].
In summary, the results shown in Figure 4 indicate that TNFα and other NF-κB activating factors can inhibit the activity of a certain set of promoters harbouring HNF-4 binding sites and that HNF-4 is required for this inhibition.
TNFα does not affect the nuclear localization or the stability of HNF-4
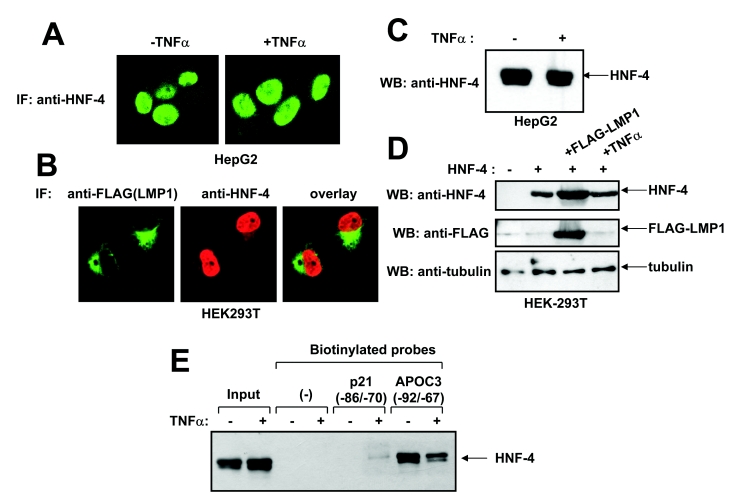
To investigate further the mechanism of inhibition of HNF-4-mediated transactivation by TNFα, the effect of TNFα and LMP1 on HNF-4 nuclear localization and stability was examined. Using immunofluorescence analysis, it was shown that the constitutive nuclear accumulation of HNF-4 in HepG2 cells was not affected following treatment with TNFα for 24 h (Figure 5A). In a double-immunofluorescence experiment, it was shown that, similar to TNFα, LMP1 had no effect on HNF-4 nuclear localization in HEK293T cells which had been co-transfected with both factors (Figure 5B). Immunoblotting analysis clearly showed that neither TNFα nor LMP1 affected the stability of endogenous HNF-4 in HepG2 cells (Figure 5C) or exogenous HNF-4 in HEK-293T cells (Figure 5D).
Figure 5. TNFα does not affect the nuclear localization or the stability, but inhibits the DNA-binding activity of HNF-4.
(A) HepG2 cells were plated in coverslips and treated with TNFα (1000 units) for 24 h or left untreated. The intracellular localization of HNF-4 was observed by indirect immunofluorescence (‘IF’) using an anti-HNF-4 polyclonal antibody followed by a secondary FITC-conjugated antibody. (B) HEK-293T cells were transiently co-transfected with expression vectors for HNF-4 and FLAG–LMP1 and placed on coverslips. The intracellular localization of HNF-4 and FLAG–LMP1 proteins was observed by indirect immunofluorescence using an anti-HNF-4 polyclonal antibody and an anti-FLAG mouse monoclonal antibody followed by the appropriate secondary FITC- or Rhodamine-conjugated antibodies. In (A) and (B), cells were observed using a Leica SP confocal fluorescence microscope. (C) HepG2 cells were treated with TNFα (1000 units) for 24 h or left untreated. The levels of expression of endogenous HNF-4 protein was determined by Western blotting (WB) using an anti-HNF-4 goat polyclonal antibody followed by a secondary horseradish-peroxidase-conjugated antibody. (D) HEK-293T cells were transiently transfected with an expression vector for HNF-4 and were treated with TNFα (1000 units) for 24 h or co-transfected with an expression vector for FLAG–LMP1. The levels of expression of exogenous HNF-4, FLAG–LMP1 and tubulin (control) in the transfected cells was determined by Western blotting using an anti-HNF-4 goat polyclonal antibody, an anti-FLAG mouse monoclonal antibody and a mouse monoclonal anti-β-tubulin antibody respectively, followed by the appropriate secondary horseradish-peroxidase-conjugated antibodies. (E) DNA-affinity precipitation experiment using nuclear extracts from HepG2 cells that had been treated with TNFα (1000 units) for 24 h or from untreated HepG2 cells and biotinylated oligonucleotides corresponding to the −92/−67 region of the human APOC3 promoter or the −86/−70 region of the human p21Cip1 promoter. HNF-4 bound to the oligonucleotides was detected by SDS/PAGE and immunoblotting using a polyclonal anti-HNF-4 antibody.
TNFα modifies the DNA-binding properties of HNF-4
It was next examined whether TNFα affected the DNA-binding properties of HNF-4. For this purpose a DNA-affinity precipitation experiment was performed using endogenous HNF-4 in HepG2 cells and a biotinylated double-stranded oligonucleotide corresponding to the −92/−67 region of the APOC3 promoter that includes the proximal HRE. As shown in Figure 5(E), HNF-4 bound efficiently to the proximal APOC3 (−92/−67) oligonucleotide, and this binding was inhibited by TNFα. In control experiments it was shown that HNF-4 did not bind to the streptavidin Dynabeads or to a double-stranded synthetic oligonucleotide corresponding to the −86/−70 region of the human p21Cip1 promoter [43].
TNFα and NF-κB inhibit the transcriptional activation function of HNF-4 by modulating the activity of AF-1 and AF-2 domains
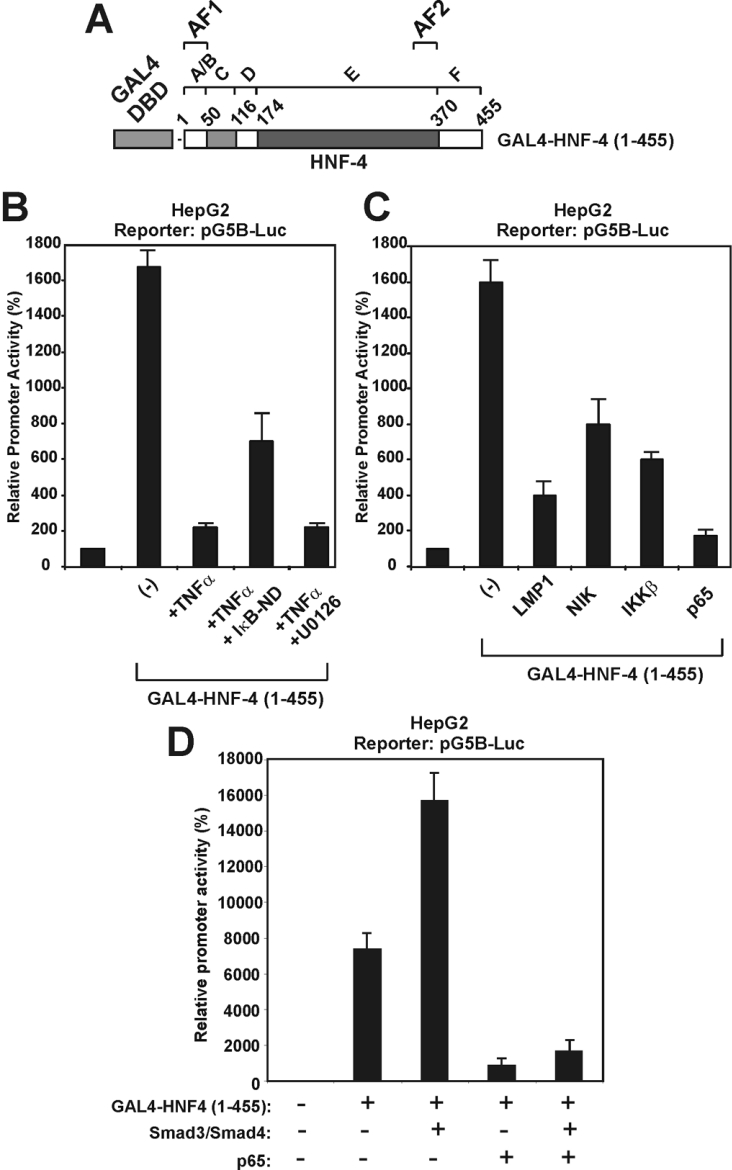
In the next series of experiments, the effects of the NF-κB pathway on nuclear HNF-4 functions was investigated using the well-characterized GAL4 system. For this purpose, a fusion protein consisting of full-length rat HNF-4a1 (amino acids 1–455) fused with the DBD of the yeast transactivator GAL4 (amino acids 1–147) (Figure 6A) was transiently expressed in HepG2 cells and its ability to transactivate the GAL4-responsive artificial promoter G5B (consisting of five tandem GAL4-binding sites) in the presence or in the absence of different NF-κB pathway activators and effectors was evaluated by luciferase assays. As shown in Figure 6(B), TNFα strongly inhibited the transcriptional activity of GAL4-HNF-4 in HepG2 cells. This repression could be partially reversed by the dominant-negative IκB-ND mutant, but not by the MEK1 inhibitor U0126, in agreement with the results shown in Figure 2(C). The transcriptional activity of GAL4–HNF-4 was also inhibited by LMP1, as well as by different effectors of the TNFα and LMP1 pathways, including the NIK, the IKKβ kinase and the p65/RelA subunit of NF-κB (Figure 6C). Finally, it was shown that p65 inhibited not only the constitutive, but also the Smad-inducible, transcriptional activity of HNF-4 (Figure 6D). The latter findings suggested that the antagonistic effects between TGFβ and TNFα on the APOC3 promoter shown in Figure 1(B) could be accounted for by antagonistic interactions between their signalling effectors (NF-κB and Smad proteins) with HNF-4, which binds to the APOC3 promoter.
Figure 6. Different effectors of the NF-κB pathway inhibit the transcriptional activation function of HNF-4 in a GAL4-based transactivation system.
(A) Schematic representation of GAL4-HNF-4 (1–455) protein that was utilized in the transactivation experiments of (B)–(D). Functional domains in HNF-4 are shown by the letters A–F. (B) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with an expression vector for GAL4-HNF-4 (1–455) (100 ng) and were treated with TNFα (1000 units) or the MEK1 inhibitor U0126 for 24 h as shown at the bottom of the graph. The dominant-negative form of IκB (IκB-ND) (2 μg) was also utilized in one of the assays, along with TNFα. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram. (C) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with an expression vector for GAL4-HNF-4 (1–455) (100 ng) in the absence or in the presence of expression vectors for LMP1 (25 ng), NIK (2 μg), IKKβ (2 μg) and p65 (2 μg), as shown at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram. (D) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with an expression vector for GAL4-HNF-4 (1–455) (100 ng) in the absence or in the presence of expression vectors for Smad3, Smad4 and p65 (2 μg) as indicated at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram.
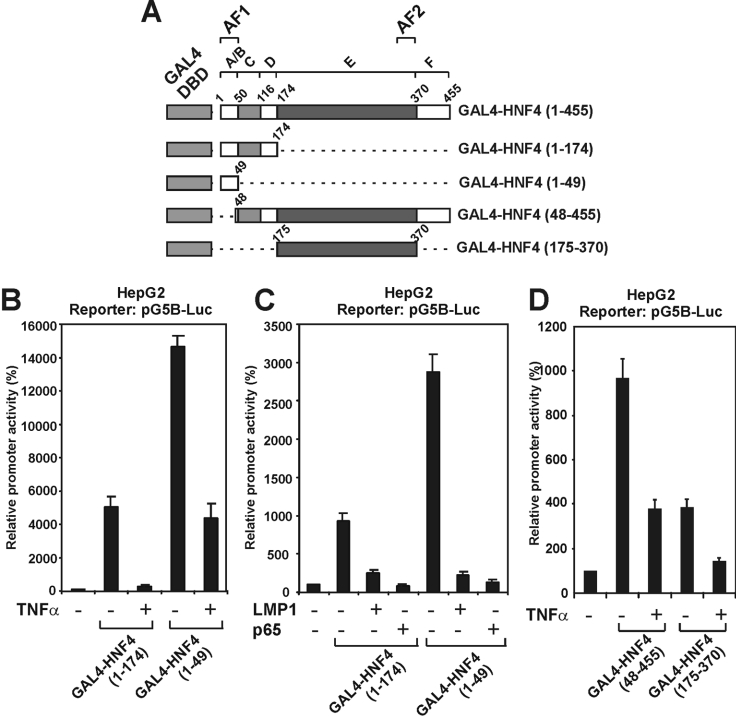
Using various truncated forms of HNF-4 fused with the DBD of GAL4 (Figure 7A), it was shown that both transactivation domains of HNF-4 (AF-1 and AF-2) respond to the inhibitory effects of TNFα. As shown in Figures 7(B) and 7(C), TNFα, LMP1 and p65 strongly inhibited the transcriptional activity of HNF-4 mutants 1–174 and 1–49 which contain only the transactivation domain AF-1. TNFα had a similar inhibitory effect on two HNF-4 mutants lacking the AF-1 domain, but retaining the AF-2 domain (mutants 48–455 and 175–370).
Figure 7. Both transactivation domains of HNF-4 (AF1 and AF2) are inhibited by TNFα and the NF-κB pathway.
(A) Schematic representation of the various truncated GAL4-HNF-4 proteins that were utilized in the transactivation experiments of (B)–(D). (B) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with expression vectors for GAL4-HNF-4 (1–174) or GAL4-HNF-4 (1–49) (100 ng) and were treated with TNFα (1000 units) for 24 h or left untreated as shown at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram. (C) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with expression vectors for GAL4-HNF-4 (1–174) or GAL4-HNF-4 (1–49) (100 ng) in the absence or in the presence of expression vectors for LMP1 (25 ng) or p65 (2 μg) as shown at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram. (D) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg) along with expression vectors for GAL4-HNF-4 (48-455) or GAL4-HNF-4 (175–370) (100 ng) and were treated with TNFα (1000 units) for 24 h or left untreated as shown at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram.
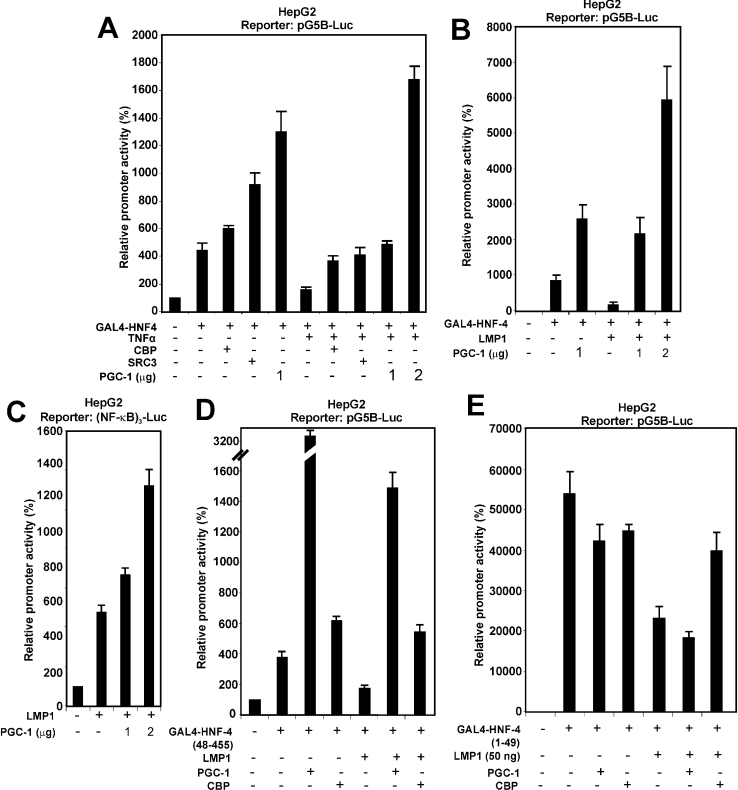
Different co-activators have been shown previously to activate the transactivation function of the AF-1 and AF-2 domains of HNF-4. Among them are co-activators which show preference for the AF-2, domain such as PGC-1 and co-activators that act via both domains such as CBP/p300 and SRC3 [9–14]. The findings of the present study, shown in Figures 6 and 7, suggested that TNFα, via the NF-κB pathway, interferes with the nuclear functions of HNF-4 and specifically with the transactivation functions of AF-1 and AF-2. The involvement of co-activators in the mechanism of repression of HNF-4 by TNFα was investigated by transactivation assays in HepG2 cells. Figure 8(A) shows that the inhibitory effect of TNFα on GAL4-HNF-4 could be totally abolished by overexpression of the HNF-4 co-activators CBP, SRC3 and PGC-1. PGC-1, in particular, not only abolished inhibition by TNFα, but induced a strong co-activation of GAL4–HNF-4 (4-fold above basal uninduced levels) (Figure 8A, last bar). PGC-1 had a similar positive effect under conditions of GAL4–HNF-4 inhibition by LMP1 (Figure 8B, third and fourth bars).
Figure 8. Inhibition of HNF-4 activity by TNFα may be accounted for by interference with the recruitment or the functions of AF-1 and AF-2 HNF-4 co-activators.
(A) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with an expression vector for GAL4-HNF-4 (100 ng) and were treated with TNFα (1000 units) for 24 h in the absence or in the presence of expression vectors for the co-activators CBP, SRC3 (2 μg each) and two different concentrations of the PGC-1 co-activator (1 and 2 μg) as indicated at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram. (B) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with expression vectors for GAL4-HNF-4 (100 ng), LMP1 (25 ng) and two different concentrations of an expression vector for PGC-1 (1 and 2 μg). Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram. (C) HepG2 cells were transiently transfected with the (NF-κB)3-Luc reporter plasmid (2 μg), along with an expression vector for LMP1 (50 ng) and two different concentrations of an expression vector for PGC-1 (1 and 2 μg). Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown by a histogram. (D) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with expression vectors for GAL4-HNF-4 (48–455) (100 ng), LMP1 (50 ng), PGC-1 (2 μg) and CBP (2 μg), as indicated at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram. (E) HepG2 cells were transiently transfected with the pG5B-Luc reporter plasmid (2 μg), along with expression vectors for GAL4-HNF-4 (1–49) (100 ng), LMP1 (50 ng), PGC-1 (2 μg) and CBP (2 μg) as indicated at the bottom of the graph. Cell extracts were analysed for luciferase activity. Normalized relative promoter activity is shown as a histogram.
To investigate potential competition between HNF-4 and NF-κB for common co-activators, the ability of PGC-1 to function as a co-activator of LMP1 signalling to NF-κB was examined. As shown in Figure 8(C), PGC-1 strongly co-activated LMP1 in a transactivation assay in which the artificial promoter (NF-κB)3 was used, suggesting that PGC-1 may act as a co-activator of LMP1 signalling to NF-κB. Competition between HNF-4 and LMP1 effectors for PGC-1 was specific for the AF-2 domain of HNF-4 [compare GAL4–HNF-4 (48–455) lacking AF-1 in Figure 8D with GAL4–HNF-4 (1–49), which contains only AF-1, in Figure 8E]. By contrast, CBP was equally active on both AF-1 and AF-2 domains (Figures 8D and 8E).
The findings shown in Figure 8 suggested that TNFα and LMP1-induced inhibition of HNF-4 transcriptional activity could be accounted for, at least in part, by interference with the recruitment of co-activators that interact with both the AF-1 and AF-2 domains of HNF-4.
DISCUSSION
The present findings indicate that pro-inflammatory cytokines, such as TNFα, that activate multiple signalling pathways, including NF-κB, can inhibit the expression of liver-specific genes by a novel mechanism that involves the modification of the DNA-binding and transactivation properties of a hormone nuclear receptor such as HNF-4. This mechanism seems to be important for the negative cross-talk between the TNFα and the anti-inflammatory cytokine TGFβ on the expression of the APOC3 gene during the acute-phase response, a process by which organisms respond to various conditions such as infections, ischaemic necrosis and trauma [32]. This is not the first example of a negative cross-talk between the two cytokines. TGFβ/TNFα antagonism regulates the expression of the collagen genes and is very important for the control of tissue homoeostasis and repair [45]. However, in the case of the collagen genes, the JNK–MAPK pathway, rather than nuclear receptors, is involved in this antagonism [46].
The participation of nuclear receptors in the mechanism of transcriptional inhibition of the APOC3 gene by TNFα was suggested by the results of transactivation experiments in which deletion mutants of the APOC3 promoter were utilized. These experiments showed that the proximal HRE (element B) was sufficient to mediate repression by TNFα (Figure 3). Previous in vitro studies had established that this proximal HRE of the APOC3 promoter is of the DR1 type (direct repeat with one nucleotide spacing) and strongly binds HNF-4 as well as ARP-1 (apolipoprotein A-I regulatory protein 1), EAR-2 (erbA-related protein 2), EAR-3 and heterodimers of RXRα (retinoid X receptor α) with RARα (retinoic acid receptor α) and less efficiently binds homodimers of RARα and heterodimers of RXRα with T3Rβ (thyroid hormone receptor β) or PPARα (peroxisome-proliferator-activated receptor α) [27,28]. Among the nuclear receptors that bind to this HRE, HNF-4 seems to play the prominent role, as conditional inactivation of HNF-4 in the liver abolished the expression of the APOC3 gene [15]. However, in vivo evidence for the recruitment of HNF-4 to this HRE as well as the HRE of the APOC3 enhancer in hepatic cells was missing. By performing a chromatin-immunoprecipitation assay we were able to demonstrate for the first time here the constitutive recruitment of HNF-4 to the proximal HRE of the APOC3 promoter in HepG2 cells (Figure 4B). Surprisingly, HNF-4 was not found on the distal HRE of the APOC3 enhancer, which was shown previously to bind HNF-4 in vitro and to be crucial for the expression of the APOC3 and APOA1 genes in vitro and in vivo (Figure 4B) [27,28,47]. This finding could suggest that nuclear receptors other than HNF-4 are the predominant regulators of the APOC3 gene via the distal HRE, but this has to be confirmed by additional in vivo DNA binding experiments. The recruitment of nuclear receptors other than HNF-4 to the proximal HRE and their involvement in transcriptional responses to extracellular signal also need to be thoroughly investigated.
Using GAL4-HNF-4 fusion proteins and different NF-kB-inducing agents, such as TNFα and the LMP1 transforming protein of the Epstein–Barr virus, we provide strong evidence for the involvement of the NF-κB pathway in the nuclear functions of HNF-4. (Figure 6). This could be explained by different, not necessarily mutually exclusive, mechanisms, as follows. (i) An indirect mechanism, such as the transcriptional activation of an HNF-4 inhibitor by NF-κB. Such an inhibitor could be the SHP (small heterodimer partner), which was shown to physically interact with HNF-4 and inhibit its transcriptional activity [48]. This hypothesis is supported by the findings of Figure 2(E), which showed that inhibition of HNF-4 requires the transactivation domain of the p65/Rel subunit of NF-κB. (ii) Inhibition could be accomplished by interference of NF-κB with the recruitment or functions of different HNF-4 co-activators. HNF-4 has been shown to interact with members of the p160 family of co-activators, including SRC-1 and GRIP1/TIF2, as well as with the histone acetyltransferase CBP and with PGC-1, a co-activator that plays important roles in hepatic gluconeogenesis [8–14]. In agreement with this hypothesis are the results presented in Figure 8, which showed that inhibition of HNF-4 by TNFα or LMP1 was overcome by overexpression of different HNF-4 co-activators, including CBP, SRC3 and PGC1, which interact with distinct transactivation domains of HNF-4 (AF-1 and AF-2) and that at least one of these proteins (PGC-1) may function as co-activator of NF-κB signalling (Figure 8C).
In addition to the negative effect of the TNFα/NF-kB pathway on the transactivation properties of HNF-4, TNFα appears to modify the DNA binding properties of this transcription factor as well. As shown in Figure 5(E), HNF-4 present in nuclear extracts from HepG2 cells that had been pre-treated with TNFα has a lower affinity for the proximal APOC3 HRE than HNF-4 present in untreated cells. This may be due to a modification, such as phosphorylation of HNF-4 in the DBD, which could affect its affinity for DNA. In support of this hypothesis are the results of previous studies showing that the DNA-binding properties of HNF-4 can be modulated in a positive or a negative manner by modifications such as acetylation [13] and phosphorylation which is induced by different signal transduction pathways involving MAPK and JAKs (Janus kinases) [49,50].
In summary, the present as well as previous studies from our group showed that the orphan nuclear receptor HNF-4 may have an important role in hepatic gene regulation, not only as a constitutive transcriptional activator but also as a ‘molecular switch’ utilized by pro-inflammatory (TNFα, interleukin 1) or anti-inflammatory (TGFβ) cytokines in order to induce or inhibit the expression of a selection of HNF-4 target genes during conditions of inflammation. The mechanism of inhibition of HNF-4 transcriptional activity by NF-κB described here may also apply to other hormone nuclear receptors that are homologous with HNF-4 and which bind to the same regulatory elements as HNF-4, but this hypothesis requires additional experimentation. If true, such a mechanism may have wide implications for nuclear receptor physiology.
Acknowledgments
We thank Dr C. Stournaras, Dr G. Mavrothalassitis, Dr A. Eliopoulos and Dr C. Tsatsanis (The Medical School, University of Crete, Heraklion, Crete, Greece), Dr I. Talianidis, Dr A. Gafencu (Institute of Cellular Biology and Pathology Nicolae Simionescu, Bucharest, Romania) and Dr M. Hadzopoulou-Cladaras for reagents, protocols and discussions. We thank Dr G. Koutsodontis (The Medical School, University of Crete, Heraklion, Crete, Greece) for assistance in the chromatin immunoprecipitation experiment. This research was supported by a grant from the Greek Ministry of Development (PENED-2001) and internal funds from the IMBB of Crete.
References
- 1.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sladek F. M., Seidel S. D. Nuclear Receptors and Genetic Disease. In: Burris T. P., McCabe E., editors. San Francisco: Academic Press; 2001. pp. 309–361. [Google Scholar]
- 3.Giguere V. Orphan nuclear receptors: from gene to function. Endocrinol. Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 4.Hadzopoulou-Cladaras M., Kistanova E., Evagelopoulou C., Zeng S., Cladaras C., Ladias J. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J. Biol. Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- 5.Duda K., Chi Y. I., Shoelson S. E. Structural basis for HNF-4α activation by ligand and co-activator binding. J. Biol. Chem. 2004;279:23311–23316. doi: 10.1074/jbc.M400864200. [DOI] [PubMed] [Google Scholar]
- 6.Dhe-Paganon S., Duda K., Iwamoto M., Chi Y.-I., Shoelson S. E. Crystal structure of the HNF-4α ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 7.Wisely B. G., Miller A. B., Davis R. G., Thornquest A. D., Johnson R., Spitzer T., Sefler A., Shearer B., Moore J. T., Miller A. B., et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 8.Ruse M. D., Jr, Privalsky M. L., Sladek F. M. Competitive cofactor recruitment by orphan receptor hepatocyte nuclear factor 4α1: modulation by the F domain. Mol. Cell. Biol. 2002;22:1626–1638. doi: 10.1128/MCB.22.6.1626-1638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sladek F. M., Ruse M. D., Nepomuceno L., Huang S. M., Stallcup M. R. Modulation of transcriptional activation and co-activator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4α1. Mol. Cell. Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J. C., Stafford M. J., Granner D. K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J. Biol. Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iordanidou P., Aggelidou E., Demetriades C., Hadzopoulou-Cladaras M. Distinct amino acid residues may be involved in co-activator and ligand interactions in hepatocyte nuclear factor-4α. J. Biol. Chem. 2005;280:21810–21819. doi: 10.1074/jbc.M501221200. [DOI] [PubMed] [Google Scholar]
- 12.Dell H., Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional co-activator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J. Biol. Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- 13.Soutoglou E., Katrakili N., Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 14.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. Control of hepatic gluconeogenesis through the transcriptional co-activator PGC-1. Nature (London) 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 15.Hayhurst G. P., Lee Y. H., Lambert G., Ward J. M., Gonzalez F. J. Hepatocyte nuclear factor 4α (nuclear receptor 2α1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert P. N., Assmann G., Gotto A. M., Jr, Fredrickson D. S. The Metabolic Basis of Inherited Diseases. In: Stanbury J. B., Wyngaarden J. B., Fredrickson D. S., Goldstein J. L., Brown M. S., editors. New York: McGraw-Hill; 1982. pp. 589–651. [Google Scholar]
- 17.Brown W. V., Baginsky M. L. Inhibition of lipoprotein lipase by an apoprotein of human very low density lipoprotein. Biochem. Biophys. Res. Commun. 1972;46:375–382. doi: 10.1016/s0006-291x(72)80149-9. [DOI] [PubMed] [Google Scholar]
- 18.Shelburne F., Hanks J., Meyers W., Quarfordt S. Effect of apoproteins on hepatic uptake of triglyceride emulsions in the rat. J. Clin. Invest. 1980;65:652–658. doi: 10.1172/JCI109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Windler E., Chao X., Havel R. J. Regulation of the hepatic uptake of triglyceride-rich lipoproteins in the rat. Opposing effects of homologous apolipoprotein E and individual C apoproteins. J. Biol. Chem. 1980;255:8303–8307. [PubMed] [Google Scholar]
- 20.Krauss R. M., Herbert P. M., Levy R. I., Fredrickson D. S. Further observations on the activation and inhibition of lipoprotein lipase by apolipoproteins. Circ. Res. 1973;33:403–411. doi: 10.1161/01.res.33.4.403. [DOI] [PubMed] [Google Scholar]
- 21.Ito Y., Azrolan N., O'Connell A., Walsh A., Breslow J. L. Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science. 1990;249:790–793. doi: 10.1126/science.2167514. [DOI] [PubMed] [Google Scholar]
- 22.de Silva H. V., Lauer S. J., Wang J., Simonet W. S., Weisgraber K. H., Mahley R. W., Taylor J. M. Overexpression of human apolipoprotein C-III in transgenic mice results in an accumulation of apolipoprotein B48 remnants that is corrected by excess apolipoprotein E. J. Biol. Chem. 1994;269:2324–2335. [PubMed] [Google Scholar]
- 23.Karathanasis S. K. Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII and AIV genes. Proc. Natl. Acad. Sci. U.S.A. 1985;82:6374–6378. doi: 10.1073/pnas.82.19.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogami K., Hadzopoulou-Cladaras M., Cladaras C., Zannis V. I. Promoter elements and factors required for hepatic and intestinal transcription of the human ApoCIII gene. J. Biol. Chem. 1990;265:9808–9815. [PubMed] [Google Scholar]
- 25.Talianidis I., Tambakaki A., Toursounova J., Zannis V. I. Complex interactions between SP1 bound to multiple distal regulatory sites and HNF-4 bound to the proximal promoter lead to transcriptional activation of liver-specific human APOCIII gene. Biochemistry. 1995;34:10298–10309. doi: 10.1021/bi00032a025. [DOI] [PubMed] [Google Scholar]
- 26.Kardassis D., Tzameli I., Hadzopoulou-Cladaras M., Talianidis I., Zannis V. I. Distal apolipoprotein C-III regulatory elements F to J act as a general modular enhancer for proximal promoters that contain hormone response elements. Synergism between hepatic nuclear factor-4 molecules bound to the proximal promoter and distal enhancer sites. Arterioscler. Thromb. Vasc. Biol. 1997;17:222–232. doi: 10.1161/01.atv.17.1.222. [DOI] [PubMed] [Google Scholar]
- 27.Ladias J. A. A., Hadzopoulou-Cladaras M., Kardassis D., Cardot P., Cheng J., Zannis V. I., Cladaras C. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2 and EAR-3. J. Biol. Chem. 1992;267:15849–15860. [PubMed] [Google Scholar]
- 28.Lavrentiadou S. N., Hadzopoulou-Cladaras M., Kardassis D., Zannis V. I. Binding specificity and modulation of the human ApoCIII promoter activity by heterodimers of ligand-dependent nuclear receptors. Biochemistry. 1999;38:964–975. doi: 10.1021/bi981068i. [DOI] [PubMed] [Google Scholar]
- 29.Lacorte J. M., Ktistaki E, Beigneux A., Zannis V. I., Chambaz J., Talianidis I. Activation of CAAT enhancer-binding protein delta (C/EBPdelta) by interleukin-1 negatively influences apolipoprotein C-III expression. J. Biol. Chem. 1997;272:23578–23584. doi: 10.1074/jbc.272.38.23578. [DOI] [PubMed] [Google Scholar]
- 30.Lacorte J. M., Beigneux A., Parant M., Chambaz J. Repression of apoC-III gene expression by TNFα involves C/EBPδ/NF-IL6β via an IL-1 independent pathway. FEBS Lett. 1997;415:217–220. doi: 10.1016/s0014-5793(97)01127-7. [DOI] [PubMed] [Google Scholar]
- 31.Hadzopoulou-Cladaras M., Lavrentiadou S. N., Zannis V. I., Kardassis D. Transactivation of the ApoCIII promoter by ATF-2 and repression by members of the Jun family. Biochemistry. 1998;37:14078–14087. doi: 10.1021/bi9804176. [DOI] [PubMed] [Google Scholar]
- 32.Koj A. Termination of acute-phase response: role of some cytokines and anti-inflammatory drugs. Gen. Pharmacol. 1998;31:9–18. doi: 10.1016/s0306-3623(97)00435-7. [DOI] [PubMed] [Google Scholar]
- 33.Shull M. M., Ormsby I., Kier A. B., Pawlowski S., Diebold R. J., Yin M., Allen R., Sidman C., Proetzel G., Calvin D., et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature (London) 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moustakas A., Souchelnytskyi S., Heldin C. H. Smad regulation in TGF-β signal transduction. J. Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 35.Massague J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 36.ten Dijke P., Hill C. S. New insights into TGF-β–Smad signalling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Kardassis D., Pardali K., Zannis V. I. SMAD proteins transactivate the human ApoCIII promoter by interacting physically and functionally with hepatocyte nuclear factor 4. J. Biol. Chem. 2000;275:41405–41414. doi: 10.1074/jbc.M007896200. [DOI] [PubMed] [Google Scholar]
- 38.Chou W. C., Pocola V., Shiraishi K., Valcourt U., Moustakas A., Hadzopoulou-Cladaras M., Zannis V. I., Kardassis D. Mechanism of a transcriptional cross talk between transforming growth factor-β-regulated Smad3 and Smad4 proteins and orphan nuclear receptor hepatocyte nuclear factor-4. Mol. Biol. Cell. 2003;14:1279–1294. doi: 10.1091/mbc.E02-07-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prokova V., Mosialos G., Kardassis D. Inhibition of transforming growth factor β signalling and Smad-dependent activation of transcription by the latent membrane protein 1 of Epstein–Barr virus. J. Biol. Chem. 2002;277:9342–9350. doi: 10.1074/jbc.M109099200. [DOI] [PubMed] [Google Scholar]
- 40.Ktistaki E., Lacorte J. M., Katrakili N., Zannis V. I., Talianidis I. Transcriptional regulation of the apolipoprotein A-IV gene involves synergism between a proximal orphan receptor response element and a distant enhancer located in the upstream promoter region of the apolipoprotein C-III gene. Nucleic Acids Res. 1994;22:4689–4696. doi: 10.1093/nar/22.22.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorgia P., Zannis V. I., Kardassis D. A short proximal promoter and the distal hepatic control region-1 (HCR-1) contribute to the liver specificity of the human apolipoprotein C-II gene. Hepatic enhancement by HCR-1 requires two proximal hormone response elements which have different binding specificities for orphan receptors HNF-4, ARP-1 and EAR-2. J. Biol. Chem. 1998;273:4188–4196. doi: 10.1074/jbc.273.7.4188. [DOI] [PubMed] [Google Scholar]
- 42.Koutsodontis G., Kardassis D. Inhibition of p53-mediated transcriptional responses by mithramycin A. Oncogene. 2004;23:9190–9200. doi: 10.1038/sj.onc.1208141. [DOI] [PubMed] [Google Scholar]
- 43.Koutsodontis G., Moustakas A., Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–12784. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 44.Hatzivassiliou E., Mosialos G. Cellular signalling pathways engaged by the Epstein–Barr virus transforming protein LMP1. Front. Biosci. 2002;7:d319–d329. doi: 10.2741/hatziva. [DOI] [PubMed] [Google Scholar]
- 45.Schiller M., Javelaud D., Mauviel A. TGF-β-induced SMAD signalling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J. Dermatol. Sci. 2004;35:83–92. doi: 10.1016/j.jdermsci.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Verrecchia F., Wagner E. F., Mauviel A. Distinct involvement of the Jun-N-terminal kinase and NF-κB pathways in the repression of the human COL1A2 gene by TNF-α. EMBO Rep. 2002;3:1069–1074. doi: 10.1093/embo-reports/kvf219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kan H-Y., Georgopoulos S., Zannis V. I. A hormone response element in the human apolipoprotein CIII (ApoCIII) enhancer is essential for intestinal expression of the ApoA-I and ApoCIII genes and contributes to the hepatic expression of the two linked genes in transgenic mice. J. Biol. Chem. 2000;275:30423–30431. doi: 10.1074/jbc.M005641200. [DOI] [PubMed] [Google Scholar]
- 48.Lee Y. K., Dell H., Dowhan D. H., Hadzopoulou-Cladaras M., Moore D. D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Fabiani E., Mitro N., Anzulovich A. C., Pinelli A., Galli G., Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7α-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J. Biol. Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 50.Li X., Salisbury-Rowswell J., Murdock A. D., Forse R. A., Burke P. A. Hepatocyte nuclear factor 4 response to injury involves a rapid decrease in DNA binding and transactivation via a JAK2 signal transduction pathway. Biochem. J. 2002;368:203–211. doi: 10.1042/BJ20020233. [DOI] [PMC free article] [PubMed] [Google Scholar]