Abstract
In eukaryotic cells, the Ccr4–Not complex can regulate mRNA metabolism at various levels. Previously, we showed that promoter targeting of the CNOT2 subunit resulted in strong repression of RNA polymerase II transcription, which was sensitive to the HDAC (histone deacetylase) inhibitor, trichostatin A [Zwartjes, Jayne, van den Berg and Timmers (2004) J. Biol. Chem. 279, 10848–10854]. In the present study, the cofactor requirement for CNOT2-mediated repression was investigated. We found that coexpression of SMRT (silencing mediator for retinoic acid receptor and thyroid-hormone receptor) or NCoR (nuclear hormone receptor co-repressor) in combination with HDAC3 (or HDAC5 and HDAC6) augmented the repression by CNOT2. This repressive effect is mediated by the conserved Not-Box, which resides at the C-terminus of CNOT2 proteins. We observed physical interactions of CNOT2 with several subunits of the SMRT/NCoR–HDAC3 complex. Our results show that the SMRT/NCoR–HDAC3 complex is a cofactor of CNOT2-mediated repression and suggest that transcriptional regulation by the Ccr4–Not complex involves regulation of chromatin modification.
Keywords: Ccr4–Not complex, chromatin, histone deacetylase, silencing mediator for retinoic acid receptor and thyroid-hormone receptor (SMRT)/nuclear hormone receptor co-repressor (NCoR), protein–protein interaction, transcription regulation
Abbreviations: Bcl-6, B-cell lymphocytic-leukaemia proto-oncogene 6; Caf1, Ccr4-associated factor 1; CMV, cytomegalovirus; CRD1, cell cycle regulatory domain 1; DBD, DNA-binding domain; ERα, oestrogen receptor α; Gcn5, general control of amino acid synthesis protein 5; GPS2, G-protein pathway suppressor 2; HDAC, histone deacetylase; HEK-293T cells, HEK-293 cells (human embryonic kidney cells) expressing the large T-antigen of SV40 (simian virus 40); LexA, Lex A repressor; Sin3A, Swi-independent 3A; mSin3A, murine Sin3A; MyoD, myogenic differentiation antigen D; NCoR, nuclear hormone receptor co-repressor; pol II, RNA polymerase II; RD1, repression domain 1; RID, receptor interaction domain; RXRβ, retinoic X receptor β; SAGA, Spt-Ada-Gcn5-acetyltransferase; TFIID, transcription factor II D; SANT, SWI3, ADA2, N-CoR and TFIIIB; SMRT, silencing mediator for retinoic acid receptor and thyroid-hormone receptor; TBL1, transducin β-like protein 1; TBLR1, TBL-related 1; TSA, trichostatin A
INTRODUCTION
Transcription of eukaryotic genes by pol II (RNA polymerase II) involves the action of an extensive set of transcription factors and chromatin regulatory proteins. Basal transcription only requires general transcription factors including the TFIID (transcription factor II D) and Mediator complexes. In contrast, regulated transcription involves DNA-binding transcription factors, which can recruit co-repressors or co-activators to the promoter. These cofactors can affect transcription by interacting with the basal transcription machinery or with chromatin. Chromatin regulatory proteins can remodel nucleosomal structures in an ATP-dependent manner or they can alter post-translational modifications of the histone tails, which includes methylation, phosphorylation and acetylation/deacetylation [1,2].
The evolutionarily conserved Ccr4–Not complex controls gene expression both at the level of transcription initiation and of mRNA degradation [3,4]. The Not subunits of the complex were first genetically isolated in the yeast Saccharomyces cerevisiae as negative regulators inhibiting transcription from the TATA-less promoter of the HIS3 gene. Several genetic and biochemical interactions have been observed between yeast Not proteins and transcription initiation complexes like TFIID and Mediator [3,4]. The mammalian Ccr4–Not complex contains the orthologues of yeast Not1-4p proteins (CNOT1–CNOT4) and also of the CNOT6(hCcr4), CNOT7(hCaf1) (where Caf1 is Ccr4-associated factor 1), CNOT8(hPop2/Calif), CNOT9/RQCD1(hRcd1/hCaf40) and CNOT10(hCaf130) proteins. Besides the role in transcription regulation, the Ccr4–Not complex is involved in mRNA decay. Human CNOT6(hCcr4) and yeast Ccr4 possess a 3′–5′-exonuclease activity that shortens the poly(A) tails of mRNAs [3–5]. Together, this indicates that the Ccr4–Not complex exerts its functions both in the cytoplasm and in the nucleus. Interestingly, a substantial portion of Ccr4–Not proteins is localized in the cytoplasm [3–6] and cell-cycle progression regulates the distribution of human CNOT7(hCaf1) between the nucleus and the cytoplasm [7]. In addition, human CNOT4 displays ubiquitin-protein ligase activity in vitro, but the importance for gene transcription and/or mRNA decay is not exactly known [8].
Genetic and biochemical studies provided evidence that the Ccr4–Not complex can regulate pol II transcription [7,9–12]. Various subunits of the yeast Ccr4–Not complex are required for the expression of RNR genes after hydroxyurea treatment or DNA damage [9]. In mammalian cells, Ccr4–Not core subunits are involved in transcriptional activation by nuclear receptors [7,10,11]. In contrast, it was found that CNOT9/RQCD1(hRCD1/hCaf40) can interact with the c-Myb transcription factor and repress its transactivation function in human cells [12].
Previously, we reported that promoter targeting of CNOT2 or CNOT9/RQCD1(hRcd1/hCaf40) results in strong repression of pol II transcription [13]. The domain of approx. 100 residues at the C-terminus of CNOT2, called the Not-Box, is responsible for repression of promoter activity. This Not-Box is present in all orthologues of CNOT2 and CNOT3. CNOT2-mediated repression is sensitive to the HDAC (histone deacetylase) inhibitor, TSA (trichostatin A). In general, recruitment of HDACs to the promoter is associated with repression of transcription. HDACs can catalyse removal of acetyl groups from lysine residues at the N-terminal tails of histone proteins. This decreases chromatin accessibility and recruitment of bromodomain-containing co-activators. To date, 18 different HDACs have been described in human cells [14]. They are divided into four classes: class I regroups the yeast Rpd3 orthologues and consists of HDAC1, HDAC2, HDAC3 and HDAC8, class II consists of HDAC4, HDAC5, HDAC6, HDAC9 and HDAC10 and are the orthologues of yeast HDA1, class III is the SIRT2 (Sirtuin 2) NAD+-dependent HDAC family and class IV is represented by HDAC11. Biochemical studies demonstrated the existence of different HDAC-containing complexes in mammalian cells [16,17,20,21,32]. HDAC1 and HDAC2 exist in similar complexes including mSin3A [murine Sin3A (Swi-independent 3A)], Mi2/NuRD (nucleosome remodelling and histone deacetylation) or CoREST (repressor element 1-silencing transcription factor co-repressor). The Sin3A–HDAC complex was shown to mediate repression by a variety of DNA-binding transcription factors [15], like Mad1 (mitotic arrest deficient 1), MeCP2 (methylated CpG binding protein 2) and p53. HDAC3 is part of a complex containing the co-repressor proteins SMRT (silencing mediator of retinoid and thyroid hormone receptors)/NCoR (nuclear hormone receptor co-repressor), TBL1 (transducin β-like protein 1), TBLR1 (TBL-related 1) and GPS2 (G-protein pathway suppressor 2) [16]. Class II HDACs, like HDAC4, HDAC5 and HDAC7, can be associated with this complex by interacting with the SMRT/NCoR proteins [17]. Unliganded nuclear hormone receptors recruit SMRT/NCoR complexes to repress transcription, but transcription repressors such as Bcl-6 (B-cell lymphocytic-leukaemia proto-oncogene 6), MyoD (myogenic differentiation antigen D) or ETO (eight twenty-one) also use this complex as co-repressors.
In the present study, we decided to investigate further the involvement of HDACs in CNOT2-mediated repression. In transient transfections, we found that co-expression of SMRT (or NCoR) in combination with specific HDACs increased transcriptional repression by CNOT2. In addition, we provide evidence for physical interactions between CNOT2 and several proteins of the SMRT/NCoR–HDAC3 complex. Our results show that the SMRT/NCoR–HDAC3 complex is a cofactor for CNOT2-mediated repression.
EXPERIMENTAL
Cell lines
The U2OS (human osteosarcoma) and HEK-293T [HEK-293 cells (human embryonic kidney cells) expressing the large T-antigen of SV40 (simian virus 40)] cell lines were cultured in DMEM (Dulbecco's minimal Eagle's medium; BioWhittaker) supplemented with 10% (v/v) fetal calf serum, 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin.
Plasmids
The mammalian expression vectors pCMVDBD, pCGDCNOT2, pCGDCNOT2 1–255, pCGDCNOT2 256–540, pCGDCNOT2 1–334, pCGDCNOT2 334–540, pCGDCNOT2 437–540 and pCGDyNot2, which expressed Gal4DBD (where DBD is DNA-binding domain) fusion proteins under control of the CMV (cytomegalovirus) promoter have been described previously [13].
The expression vectors pCMX-NCoR and pCMX-SMRT were kindly provided by Dr J. Wong (Department of Molecular and Cellular Biology, Baylor College of Medecine, Houston, TX, U.S.A.). They encode C-terminal FLAG-tagged mouse NCoR and SMRT respectively, under the control of CMV promoter. The expression vectors pCMV-FLAG-HDAC1-7 were obtained from Dr E. Verdin (Gladstone Institute of Virology and Immunology, University of California, San Francisco, CA, U.S.A.) and have been described previously [18,19]. The expression vector pEBB-FLAG-mSin3A (obtained from Dr H. G. Stunnenberg (Department of Molecular Biology, University of Nijmegen, Nijmegen, The Netherlands) encodes the mSin3A and is under the control of the human EF-1α (elongation factor 1α) promoter.
The LexA (Lex A repressor) CNOT2-(437–540) plasmid was constructed by insertion of a BglII/XhoI PCR fragment encoding residues 437–540 from human CNOT2 into the BamHI/XhoI-digested pEG202NLS. B42-NCoR RD1 (repression domain 1), B42-NCoR RD2, B42-NCoR RD3, B42-NCoR RD4 and B42-NCoR RID (receptor interaction domain) were constructed by insertion of restriction digested fragments obtained from pGAD10-NCoR 1/2 (5–373), pGAD10-NCoR 9/10 (743–1064), pGAD10-NCoR 11/12 (1056–1487), pGAD10-NCoR 13/14 (1495–1965) and pGAD10-NCoR 15/16 (2002–2440) [20] respectively into pJG4-5. B42-NCoR SANT-(374–757) (where SANT is SWI3, ADA2, N-CoR and TFIIIB), B42-SMRT RD1-(1–365), B42-SMRT SANT-(366–750), B42-SMRT RD2-(751–1089), B42-SMRT RD3-(1090–1523), B42-SMRT RD4-(1528–2005) and B42-SMRT RID-(2038–2473) were constructed by insertion into pJG4-5 of the respective PCR fragments obtained using the pCMX-NCoR or pCMX-SMRT as templates. B42-HDAC3 was obtained by insertion of the PCR fragment corresponding to human HDAC3 into pJG4-5. A GPS2 clone was obtained from the IMAGE Consortium (Integrated Molecular Analysis of Genomes and their Expression Consortium; clone DKFZp434A0312Q2) and was used as template for PCR amplification using appropriate primers. The PCR fragment obtained was then inserted into pJG4-5. All plasmid constructs were verified by DNA sequencing across the cloning junctions. Further details on primer sequences and cloning strategy are available upon request.
Luciferase assay
U2OS or HEK-293T cells were plated on 3 cm dishes and transfected in duplicate by the calcium phosphate method. To correct for transfection efficiency, 50 ng (U2OS) or 5 ng (HEK-293T) of pCMV-Renilla-luciferase was co-transfected. Cell lysates were prepared 40 h after transfection, and luciferase activity was determined using the Dual-Luciferase Reporter Assay system (Promega) and a Lumat LB9507 luminometer (Berthold) according to the manufacturer's instructions. Luciferase values were corrected for transfection efficiency by determining the firefly luciferase/Renilla luciferase ratio. The fold repression by Gal4DBD fusion proteins was calculated by normalization of the firefly luciferase/Renilla luciferase ratios to the ratio obtained by transfection containing only reporter plasmids and the empty pcDNA3.1 vector.
Immunoprecipitation assays
HEK-293T cells were plated on 9 cm dishes and transfected with 10 μg of DNA using the poly(ethyleneimine) method [22]. Cells were lysed, 40 h after transfection, in 1 ml of lysis buffer [50 mM Hepes/KOH, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.1% Nonidet P40, 5% (v/v) glycerol, 1 mM PMSF and proteases inhibitors]. Lysates were cleared by centrifugation at 20000 g for 15 min at 4 °C. FLAG-tagged proteins and the associated proteins were immunoprecipitated overnight at 4 °C using 10 μl of anti-FLAG M2 agarose (Sigma). Beads were washed three times with lysis buffer. Proteins associated with the beads were released by adding SDS sample buffer [2% (w/v) SDS, 80 mM Tris/HCl, pH 6.8, and 10% glycerol] without 2-mercaptoethanol and by elution at 65 °C for 10 min. Before loading on SDS/PAGE, 5 mM 2-mercaptoethanol was added and the samples were boiled. Proteins were detected by immunoblotting using anti-Gal4DBD-RK5C1 (sc-510; Santa Cruz Biotechnology) and by using ECL® (enhanced chemiluminescence; Amersham Biosciences).
Yeast two-hybrid interaction assay
Growth and manipulation of yeast strain EGY48 was carried out as described in [23]. For quantitative determination of β-galactosidase activities, yeast transformants were disrupted with zirconia/silica beads using a mini beadbeater [Biospec Products; 1 min maximum speed at room temperature (20 °C)] in a buffer containing 100 mM Tris/HCl (pH 8.0), 20% glycerol, 1 mM 2-mercaptoethanol, 0.5% SDS and protease inhibitors. After removal of insoluble material by centrifugation, β-galactosidase activities were determined using the Galacto-Light Plus chemiluminescent reporter assay (Tropix) essentially according to the manufacturer's instructions and normalized to total protein content as determined by a Bradford protein assay (Bio-Rad).
RESULTS
Effects of SMRT, NCoR and HDAC overexpression on CNOT2-mediated repression
In a previous study, we investigated the transcriptional effects of Ccr4–Not subunits when targeted to a promoter in transient transfection/reporter assays [13]. We found that CNOT2 is able to repress promoter activity and that this repression is sensitive to TSA, suggesting a mechanism involving HDACs [13]. To study this in more detail, we tested the effect of co-expression of different HDACs, alone or in combination with their respective partners on CNOT2-mediated repression. For this, we employed plasmids expressing CNOT2 fused to the DBD of yeast transcription factor Gal4 and expression plasmids for FLAG-tagged versions of different HDACs (HDAC1–HDAC6), SMRT, NCoR or Sin3A. The plasmids were transiently transfected into HEK-293T cells together with a firefly luciferase reporter plasmid (7×TKLuc), carrying seven binding sites for Gal4 upstream of the minimal HSV (herpes simplex virus) thymidine kinase promoter, and a CMV promoter-controlled Renilla luciferase reporter for normalization of transfection efficiencies. A plasmid expressing only the DBD of Gal4 was included as a negative control in the experiments.
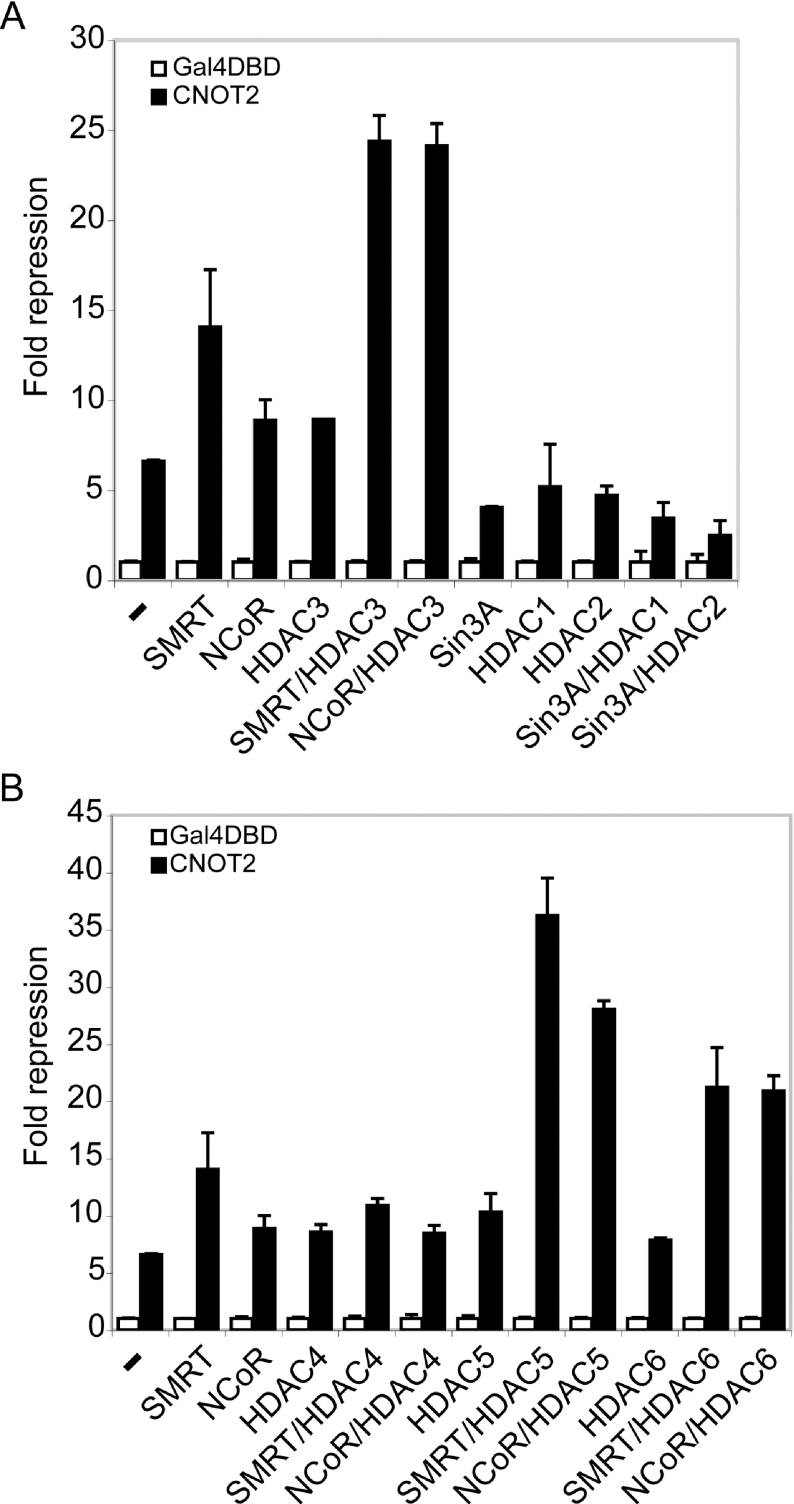
As expected, the Gal4-CNOT2 fusion plasmid was able to repress promoter activity (∼6-fold; Figure 1A). Co-transfection of Sin3A, HDAC1 or HDAC2 did not increase repression but rather resulted in relief of repression (Figure 1A). Whereas co-expression of NCoR or HDAC3 had little effect on CNOT2 repression, we observed an increase in repression upon SMRT expression (from 6- to 14-fold). Repression of promoter activity was further augmented when CNOT2 was co-expressed with SMRT in combination with HDAC3. This resulted in a 25-fold repression, suggesting a synergetic effect of the two proteins. Co-expression of NCoR and HDAC3 gave a similar effect. In contrast, co-expression of Sin3A and its cognate catalytic subunits (HDAC1 and HDAC2) did not augment CNOT2-mediated repression.
Figure 1. Overexpression of SMRT, NCoR and HDAC3 (or HDAC5 and HDAC6) augments transcriptional repression activities of CNOT2.
(A) Effect of overexpression of class I HDACs, SMRT, NCoR and Sin3A on the repression activities of CNOT2. HEK-293T cells were transiently co-transfected with 2 μg of p7xGalTKLuc reporter plasmid, 1 μg of pCMV expression plasmid for Gal4DBD (white bars) or Gal4–CNOT2 fusion protein (black bars) and 3 μg of pCMV expression plasmid for co-repressor proteins. For the purpose of clarity, for each cofactor combination, the value obtained for Gal4–CNOT2 repression was divided by the value obtained with the Gal4DBD protein. As a consequence, on the graph, the fold repression of Gal4DBD is 1 for each cofactor. The effect of the repressors on the activities of Gal4DBD alone is less than 2-fold, except for SMRT/HDAC3, HDAC4 combinations, Sin3A, HDAC1 and Sin3A/HDAC1. The experiments shown are representative of at least three independent assays, performed in duplicate. (B) Effect of overexpression of class II HDACs, SMRT and NCoR on CNOT2-mediated repression. The assays were performed and the results are displayed as in (A).
Our next step was to study the effect of class II HDACs. Overexpression of HDAC4, HDAC5 or HDAC6 did not augment CNOT2-mediated repression (Figure 1B). Overexpression of HDAC7 resulted in a strong non-specific repression and was excluded from further analysis (results not shown). HDAC4 and HDAC5 have been shown to interact with the SMRT/NCoR–HDAC3 complex [17]. Co-expression of HDAC5 but not of HDAC4 with SMRT or NCoR increased CNOT2-mediated repression (Figure 1B). Surprisingly, co-expression of HDAC6 and SMRT or NCoR also resulted in a strong specific effect. HDAC6 was shown to localize in the cytoplasm and to deacetylate tubulin [24–26]. However, HDAC6 has been shown to interact with p300 and to repress transcription via its CRD1 (cell cycle regulatory domain 1) motif [27,28], suggesting that HDAC6 could have a nuclear function. Immunoblot of transfected cell lysates confirmed that the HDAC, SMRT and NCoR proteins were expressed and were of the expected sizes (results not shown).
In conclusion, overexpression of the SMRT and NCoR co-repressor proteins with HDAC3, HDAC5 or HDAC6 augments CNOT2-mediated repression in HEK-293T cells. Similar observations were made when the human osteosarcoma U2OS cell line was used for transient transfection (results not shown).
Repression enhancement by SMRT or NCoR overexpression requires the Not-Box of CNOT2
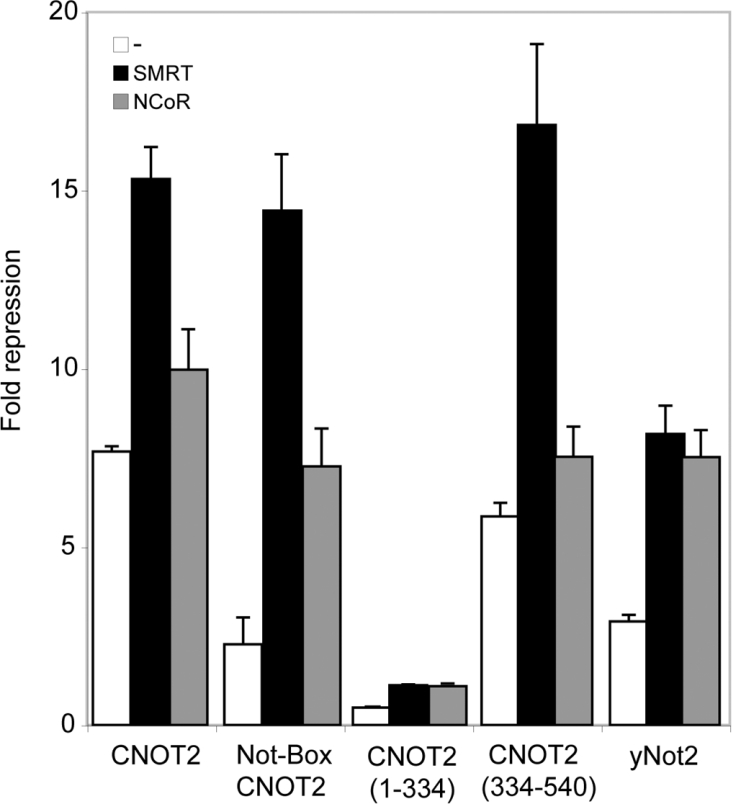
Previous analyses indicated that the Not-Box of CNOT2 constitutes the minimal repression domain [13]. To test whether the observed effects of SMRT or NCoR on CNOT2 repression require this Not-Box, we co-transfected U2OS cells with plasmids for different Gal4–CNOT2 fusion proteins and the firefly luciferase reporter plasmid 7×TKLuc. We used in this experiment three different constructs of Gal4–CNOT2: an N-terminal construct, CNOT2-(1–334), which was not able to repress promoter activity; the complementary construct containing the Not-Box, CNOT2-(334–540); and the isolated Not-Box, CNOT2-(437–540) [13]. These fusion proteins are expressed to similar levels [13]. We also included in this experiment Gal4 fusions with yeast Not2. As expected, transfection of all constructs except the CNOT2-(1–334) construct resulted in repression of promoter activity (Figure 2). Co-expression of SMRT clearly increased repression for all constructs, except for CNOT2-(1–334). The effect of SMRT overexpression is particularly strong for the C-terminal constructs of CNOT2. Again, co-expression of NCoR had little effect on the repression activity of full-length CNOT2, but repression by isolated Not-Box of CNOT2 was significantly enhanced by NCoR. Interestingly, the co-expression of SMRT or NCoR also augmented the repression mediated by yeast Not2.
Figure 2. Enhancement of repression by SMRT and NCoR requires the Not-Box.
U2OS cells were transiently transfected as described in the legend of Figure 1. The effects of SMRT and NCoR on Gal4DBD alone were 0.97- and 1.2-fold respectively. The assays were performed in duplicate. The experiment shown is representative of at least three independent assays.
In conclusion, the enhanced repression observed upon SMRT or NCoR overexpression is dependent on the presence of the Not-Box. It is important to note that this correlates well with previous results, which showed a stronger relief of repression by TSA with the Not-Box of CNOT2 than with the full-length CNOT2 [13].
CNOT2 physically interacts with HDAC3 in mammalian cells
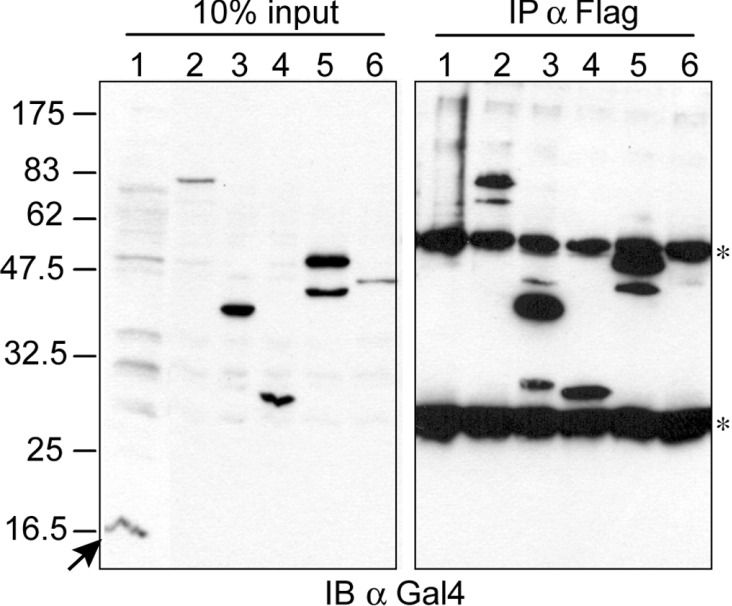
As a final step, potential physical interactions between CNOT2 and the SMRT/NCoR–HDAC3 complex subunits were tested in two different experimental set-ups. To test the interaction in human cells, we transfected HEK-293T cells with plasmids overexpressing FLAG-tagged HDAC3 and different Gal4–CNOT2 fusion proteins. FLAG–HDAC3 was immunoprecipitated from transfected lysates and the precipitates were analysed by immunoblotting using Gal4 antibodies. We observed strong co-precipitation of Gal4 fused to full-length CNOT2 (Figure 3, lane 2) and not of the Gal4 only control. In addition, CNOT2 constructs containing the Not-Box also interacted with HDAC3. Analysis of the CNOT2-(1–334) mutant was precluded by its co-migration of with the IgH chains of the FLAG antibody. Taken together, the co-immunoprecipitation analysis indicated that CNOT2 can interact, directly or indirectly, with HDAC3.
Figure 3. The Not-Box of CNOT2 interacts with HDAC3 in vivo.
HEK-293T cells were transiently transfected with plasmids expressing FLAG-tagged HDAC3 and different Gal4 fusion proteins: Gal4DBD (lane 1), Gal4–CNOT2 (lane 2), Gal4–CNOT2-(334–540) (lane 3), Gal4–CNOT2-(437–540) (lane 4), Gal4–CNOT2-(256–540) (lane 5) or Gal4–CNOT2-(1–255) (lane 6). FLAG-tagged HDAC3 was precipitated using anti-FLAG beads in the presence of 100 mM KCl and the interacting proteins were analysed by SDS/PAGE and immunoblotting using the RK5C1 antibody (right panel). Proteins present in 10% of the input were analysed in the same way (left panel). Asterisks indicate positions of IgG proteins (heavy and light chains) and the arrow indicates the position of Gal4DBD protein. Positions of co-migrating marker proteins are indicated by their molecular mass in kDa to the left of the Figure.
CNOT2 interacts with multiple subunits of the SMRT/NCoR–HDAC3 complex in the yeast two-hybrid assay
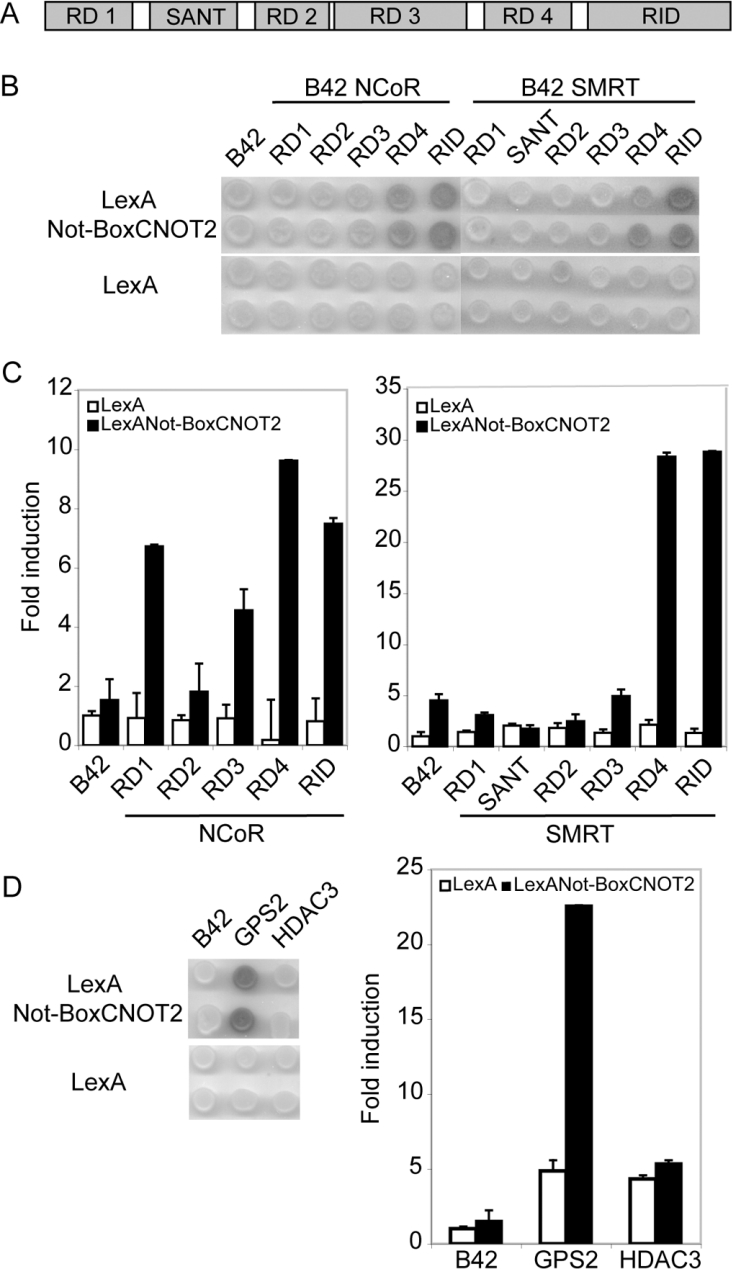
Next, we employed the yeast two-hybrid assay to investigate interaction of CNOT2 with subunits of the SMRT/NCoR–HDAC3 complex. In yeast, full-length CNOT2 acts as an activator when fused to the DBD of LexA [23]. Therefore we used a LexA fusion of the isolated Not-Box of CNOT2 as bait. Fragments of NCoR [20], and the corresponding fragments of SMRT (Figure 4A), were fused to the B42 activation domain. All B42 fusion proteins were expressed to similar levels with the exception of the NCoR SANT fragment, which was excluded from further analysis (results not shown). We observed interactions of the Not-Box with the two C-terminal fragments of NCoR and SMRT corresponding to the RID and RD4 (Figure 4B). Comparison of β-galactosidase activities indicated a stronger interaction with the SMRT fragments (Figure 4C). The interaction with other subunits of the SMRT/NCoR–HDAC3 complex was also tested. This showed that the Not-Box of CNOT2 interacts strongly with GPS2 but, surprisingly, not with HDAC3 (Figure 4D). B42 fusions with TBL1 or TBLR1 were expressed to very low levels precluding the analysis of these subunits (results not shown).
Figure 4. The Not-Box of CNOT2 interacts with different subunits of the SMRT/NCoR–HDAC3 complex in a yeast two-hybrid assay.
(A) Schematic drawing showing the known functional domains of SMRT and NCoR proteins. (B) The Not-Box of CNOT2 interacts with RD4 and with the RID of SMRT and NCoR. EGY48 cells were transformed with LexA and B42 expression plasmids together with a reporter plasmid. Galactosidase activity was detected by X-Gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside) staining of yeast cells. The LexA fusion proteins are indicated to the left and the B42 fusion proteins are indicated above the panel. Full-length CNOT2 could not be tested as the LexA fusion protein behaves as a strong transcriptional activator in this assay. (C) Quantification of β-galactosidase activity. Lysates of yeast cells were prepared as described in the Experimental section. The indicated fold induction is relative to the activity of yeast cells expressing LexA and B42 control. (D) The Not-Box of CNOT2 interacts with GPS2 but not HDAC3 in the yeast two-hybrid assay. The assay was performed and the interactions were quantified as in (B, C).
Taken together, the co-precipitation and the yeast two-hybrid analysis indicate that the Not-Box of CNOT2 can interact with several subunits of the SMRT/NCoR–HDAC3 complex.
DISCUSSION
CNOT2-mediated repression has been shown to be sensitive to the HDAC inhibitor TSA, suggesting involvement of HDACs [13]. Here, we present results indicating that the SMRT/NCoR–HDAC3 complex acts as a cofactor for repression by CNOT2. We observed that overexpression of SMRT or NCoR in combination with HDAC3 co-operatively enhanced CNOT2 repression (Figure 1). CNOT2 interacts with HDAC3 in mammalian cells (Figure 3), and by using the yeast two-hybrid assay we found interactions of the Not-Box of CNOT2 with GPS2 and with two regions of the NCoR and SMRT co-repressors including the RID (Figure 4).
Together, our results indicate that the SMRT/NCoR–HDAC3 complex can act as a cofactor for transcriptional repression by the CNOT2 subunit of the human Ccr4–Not complex and that direct protein–protein interactions are responsible for this.
The Ccr4–Not complex regulates gene expression at multiple levels
Genetic and biochemical studies demonstrated links between the Ccr4–Not complex and transcriptional repression in yeast. To circumvent the lack of cognate Ccr4–Not responsive promoters in mammalian cells, we used an artificial targeting approach to study the mechanism of CNOT2-mediated repression of transcription to identify the SMRT/NCoR–HDAC3 complex as a co-repressor. Several observations support the validity of this approach. Mutants of the Drosophila orthologue of CNOT2, Regena, display a position-variegation effect phenotype, which is linked to chromatin regulation [29]. Secondly, the repression function of CNOT2 resides in the Not-Box representing a highly conserved part of the protein. While full-length yeast Not2 inhibits transcription in mammalian cells, promoter targeting in yeast cells results in SAGA (Spt-Ada-Gcn5-acetyltransferase)-dependent transcriptional activation [30]. The SAGA complex harbours the Gcn5 (general control of amino acid synthesis protein 5) subunit, which can acetylate histone H3 tails [31]. Strikingly, yeast cells lack orthologous SMRT/NCoR–HDAC3 complexes, which can remove the acetyl groups from H3 tails [32].
Besides transcription initiation, the Ccr4–Not complex is also involved in mRNA degradation via the Ccr4p and Caf1 subunits, which were identified as the major cytoplasmic deadenylases in yeast [3,4]. Not2p, Not4p and Not5p were recently found to be involved in the stimulation of mRNA decapping, resulting in enhanced decay of specific transcripts [33]. It is tempting to speculate that the transcription and mRNA degradation functions of the Ccr4–Not complex could be linked. Interestingly, CNOT4 harbours a highly conserved RNA-recognition motif [23], which could act to bridge these separate functions.
The SMRT/NCoR–HDAC3 complex acts as a cofactor for CNOT2-mediated transcription repression
The NCoR and SMRT co-repressor proteins were first identified as cofactors for transcriptional repression by the unliganded receptors for thyroid hormone and retinoid acid hormones [16,34]. Subsequent analysis indicated that the homologous NCoR and SMRT co-repressors reside in similar protein complexes harbouring HDACs, in particular HDAC3. Within these large co-repressor proteins different domains carrying distinct functions have been recognized [16,34]. Several reports showed that the co-repressor function of SMRT/NCoR proteins is not restricted to the nuclear receptors. The RID represents a domain responsible for interaction with unliganded nuclear receptors, but it can also interact with a variety of transcription factors like MyoD and Bcl-6 [35,36]. In addition, CNOT2 interacts with the RD4 region (residues 1495–1965 or 1528–2005 of NCoR or SMRT respectively), which carries an autonomous repression function [21]. Interestingly, the RD4 region of NCoR also harbours one of the interaction sites for the TBL1 and TBLR1 subunits, which can interact directly with core nucleosomes [21]. The two-hybrid interaction of CNOT2 with SMRT was stronger than that with NCoR fragments (Figure 4). This agrees with observations that SMRT overexpression enhances CNOT2 repression, whereas NCoR overexpression had a more limited effect (Figures 1 and 2). We also found an interaction with GPS2 in yeast two-hybrid assays. GPS2, first known as a protein involved in the intracellular signalling pathway, was shown to be part of the SMRT/NCoR–HDAC3 complex [37]. Remarkably, HDAC3 did not interact in the two-hybrid assay. This could indicate that the CNOT2–HDAC3 interaction observed in mammalian cells is indirect.
In the present study, we focused on the HDAC3 and GPS2 proteins as subunits of SMRT–NCoR complexes. However, it has been reported that NCoR can also be part of different complexes including Sin3A, BRG1 (Brahma-related gene 1) and KAP1 (Kruppel-associated box-associated protein 1) complexes [34,38–40]. We observed that CNOT2-mediated repression was slightly relieved by overexpression of Sin3A, HDAC1 or HDAC2 (Figure 1). Possibly, overexpression of Sin3A complex subunits sequesters the SMRT/NCoR proteins. This would limit the amount of endogenous SMRT/NCoR–HDAC3 complex available for CNOT2, thus relieving transcriptional repression.
It is important to note that none among NCoR, SMRT and HDAC3 proteins have been found in biochemical purifications of the Ccr4–Not complex as stable interaction partners [41] (T. K. Albert and H. Th. M. Timmers, unpublished work). Conversely, the Ccr4–Not proteins have not been detected in preparations of NCoR–HDAC3 or SMRT–HDAC3 complexes [20,21,34,37,40]. This suggests that the interaction between Ccr4–Not and SMRT/NCoR-containing complexes is transient. Together, this leads us to a model in which promoter association of the Ccr4–Not complex recruits SMRT/NCoR–HDAC3 complexes via its CNOT2 subunit to selected promoter regions, resulting in local deacetylation and restriction of access of chromatin remodelling activities and transcription factors including pol II.
Involvement of other HDACs in CNOT2-mediated repression
Overexpression of HDAC5 and HDAC6 also enhanced CNOT2-mediated repression (Figure 1). These proteins have been classified as class IIa and IIb HDACs respectively [14]. In contrast with class I HDAC3, the HDAC4 and HDAC5 proteins reside in both nuclear and cytoplasmic compartments and are expressed in a tissue-specific manner. Both HDAC4 and HDAC5 are catalytically inactive by themselves, but they require association into SMRT/NCoR–HDAC3 complexes for activity [17]. Possibly, the HDAC5 protein behaves similarly in this assay, explaining the enhancement of CNOT2-mediated repression by HDAC5, but it remains surprising that HDAC4 does not display this effect (Figure 1).
The HDAC6 protein was believed to be a dedicated cytoplasmic HDAC, involved in regulating microtubule-dependent cell motility by deacetylating tubulin [24–26], and in linking protein acetylation and ubiquitination signalling pathways [42,43]. However, many studies show that HDAC6 can function also in the nucleus [27,44]. For example, HDAC6 interacts with the transcription factor Runx2 (Runt-related transcription factor 2) to repress the p21CIP1/WAF1 promoter [44]. Also, HDAC6 can be recruited by the repression domain of p300, named CRD1, when this domain is SUMO (small ubiquitin-related modifier-1)-modified [27]. HDAC6 could shuttle from the cytoplasm to the nucleus as was shown for other class II HDACs. An alternative explanation is that HDAC6 interacts with the cytoplasmic form of the Ccr4–Not complex and shuttles to the nucleus associated with this complex. However, further studies are needed to decipher the relationships of the HDAC5 and HDAC6 proteins with the SMRT/NCoR–HDAC3 and the Ccr4–Not complexes.
Targets for regulation of gene expression by the Ccr4–Not complex
Our observations support the view that the evolutionarily conserved Ccr4–Not complex can regulate gene expression at the promoter level [3,4], but the identity of natural promoters targeted by the Ccr4–Not complex in vivo has remained unknown. Several subunits of the human complex were shown to interact with DNA-sequence-specific transcription factors. Murine CNOT9/RQCD1(Rcd-1/Caf40) interacts with c-Myb transcription factor and can repress c-Myb and AP1 (activator protein 1)-dependent transcription activation [12]. CNOT9/RQCD1(Rcd-1/Caf40) can also interact with several retinoic acid receptors and the ATF-2 (activating transcription factor 2) transcription factor, and is involved in activating transcription from c-Jun promoter [10]. The CNOT7(mCaf1) subunit can interact with RXRβ (retinoic X receptor β) and ligand-dependent RXRβ-mediated transcription is decreased in CNOT7−/− cells [11]. CNOT7(hCaf1) can also interact with ERα (oestrogen receptor α). Depending on the promoter, this results in activation or repression by ERα activity [45]. Taken together, this indicates that the Ccr4–Not complex could be recruited by several transcription factors. Detailed studies are required to determine whether CNOT2 and the Ccr4–Not complex are important cofactors for these transcription factors. Genomic approaches, like mRNA expression profiling combined with genome-wide localization analysis, will help to identify target genes for the Ccr4–Not regulatory complex. Identification of bona fide promoter targets for the Ccr4–Not complex will allow rigorous testing of functional links between the CNOT2 protein and the SMRT/NCoR–HDAC3 complex, which have been uncovered by the present study.
Acknowledgments
We are grateful to Dr J. Wong, Dr H. G. Stunnenberg, Dr T. K. Albert (Department of Gene Expression, Institute of Molecular Immunology, GSF Research Centre for Environment and Health, Munich, Germany) and Dr E. Verdin for providing expression constructs and antibodies. We are especially grateful to Dr J. Wong for sharing of unpublished results and discussions during the initial phase of this work. We also thank members of our laboratory for help in the experiments and for critical discussions. H. Th. M. T. was supported financially by the Netherlands Organization for Scientific Research (NWO-MW Pionier no. 900-98-142 and NWO-CW no. 700-50-034) and European Union (Improving Human Potential RTN2-2001-00026). S. J. and C. G. M. Z. were supported by European Union and NWO-MW Pionier grants respectively.
References
- 1.Martens J. A., Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 2.Berger S. L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 3.Collart M. A., Timmers H. T. The eukaryotic Ccr4–not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 4.Denis C. L., Chen J. The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 6.Rouault J. P., Prevot D., Berthet C., Birot A. M., Billaud M., Magaud J. P., Corbo L. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J. Biol. Chem. 1998;273:22563–22569. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- 7.Morel A. P., Sentis S., Bianchin C., Le Romancer M., Jonard L., Rostan M. C., Rimokh R., Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 8.Albert T. K., Hanzawa H., Legtenberg Y. I., de Ruwe M. J., van den Heuvel F. A., Collart M. A., Boelens R., Timmers H. T. Identification of a ubiquitin-protein ligase subunit within the CCR4–NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder K. W., Winkler G. S., Timmers H. T. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4–Not complex. Nucleic Acids Res. 2005;33:6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiroi N., Ito T., Yamamoto H., Ochiya T., Jinno S., Okayama H. Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J. 2002;21:5235–5244. doi: 10.1093/emboj/cdf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T., Yao R., Ogawa T., Suzuki T., Ito C., Tsunekawa N., Inoue K., Ajima R., Miyasaka T., Yoshida Y., et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 2004;36:528–533. doi: 10.1038/ng1344. [DOI] [PubMed] [Google Scholar]
- 12.Haas M., Siegert M., Schurmann A., Sodeik B., Wolfes H. c-Myb protein interacts with Rcd-1, a component of the CCR4 transcription mediator complex. Biochemistry. 2004;43:8152–8159. doi: 10.1021/bi035857y. [DOI] [PubMed] [Google Scholar]
- 13.Zwartjes C. G., Jayne S., van den Berg D. L., Timmers H. T. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4–not complex. J. Biol. Chem. 2004;279:10848–10854. doi: 10.1074/jbc.M311747200. [DOI] [PubMed] [Google Scholar]
- 14.Yang X. J., Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- 16.Privalsky M. L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 17.Verdin E., Dequiedt F., Kasler H. G. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 18.Fischle W., Emiliani S., Hendzel M. J., Nagase T., Nomura N., Voelter W., Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 19.Fischle W., Dequiedt F., Fillion M., Hendzel M., Voelter W., Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon H. G., Chan D. W., Huang Z. Q., Li J., Fondell J. D., Qin J., Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., Behr J. P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albert T. K., Lemaire M., van Berkum N. L., Gentz R., Collart M. A., Timmers H. T. Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. HDAC6 is a microtubule-associated deacetylase. Nature (London) 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 26.Matsuyama A., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., Hay R. T. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 28.Ling L., Lobie P. E. RhoA/ROCK activation by growth hormone abrogates p300/histone deacetylase 6 repression of Stat5-mediated transcription. J. Biol. Chem. 2004;279:32737–32750. doi: 10.1074/jbc.M400601200. [DOI] [PubMed] [Google Scholar]
- 29.Frolov M. V., Benevolenskaya E. V., Birchler J. A. Regena (Rga), a Drosophila homolog of the global negative transcriptional regulator CDC36 (NOT2) from yeast, modifies gene expression and suppresses position effect variegation. Genetics. 1998;148:317–329. doi: 10.1093/genetics/148.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson J. D., Benson M., Howley P. M., Struhl K. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J. 1998;17:6714–6722. doi: 10.1093/emboj/17.22.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen M., Carrozza M. J., Lasonder E., Workman J. L., Logie C., Stunnenberg H. G. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Mol. Cell. Biol. 2004;24:2364–2372. doi: 10.1128/MCB.24.6.2364-2372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhlrad D., Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P. L., Shi Y. B. N-CoR–HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr. Top. Microbiol. Immunol. 2003;274:237–268. doi: 10.1007/978-3-642-55747-7_9. [DOI] [PubMed] [Google Scholar]
- 35.Dhordain P., Lin R. J., Quief S., Lantoine D., Kerckaert J. P., Evans R. M., Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey P., Downes M., Lau P., Harris J., Chen S. L., Hamamori Y., Sartorelli V., Muscat G. E. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol. Endocrinol. 1999;13:1155–1168. doi: 10.1210/mend.13.7.0305. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Kalkum M., Chait B. T., Roeder R. G. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 38.Nagy L., Kao H. Y., Chakravarti D., Lin R. J., Hassig C. A., Ayer D. E., Schreiber S. L., Evans R. M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 39.Heinzel T., Lavinsky R. M., Mullen T. M., Soderstrom M., Laherty C. D., Torchia J., Yang W. M., Brard G., Ngo S. D., Davie J. R., et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 40.Underhill C., Qutob M. S., Yee S. P., Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 41.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature (London) 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 42.Seigneurin-Berny D., Verdel A., Curtet S., Lemercier C., Garin J., Rousseaux S., Khochbin S. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hook S. S., Orian A., Cowley S. M., Eisenman R. N. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13425–13430. doi: 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westendorf J. J., Zaidi S. K., Cascino J. E., Kahler R., van Wijnen A. J., Lian J. B., Yoshida M., Stein G. S., Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prevot D., Morel A. P., Voeltzel T., Rostan M. C., Rimokh R., Magaud J. P., Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J. Biol. Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]