Abstract
Here, we describe the early events in the disease pathogenesis of Alexander disease. This is a rare and usually fatal neurodegenerative disorder whose pathological hallmark is the abundance of protein aggregates in astrocytes. These aggregates, termed “Rosenthal fibers,” contain the protein chaperones αB-crystallin and HSP27 as well as glial fibrillary acidic protein (GFAP), an intermediate filament (IF) protein found almost exclusively in astrocytes. Heterozygous, missense GFAP mutations that usually arise spontaneously during spermatogenesis have recently been found in the majority of patients with Alexander disease. In this study, we show that one of the more frequently observed mutations, R416W, significantly perturbs in vitro filament assembly. The filamentous structures formed resemble assembly intermediates but aggregate more strongly. Consistent with the heterozygosity of the mutation, this effect is dominant over wild-type GFAP in coassembly experiments. Transient transfection studies demonstrate that R416W GFAP induces the formation of GFAP-containing cytoplasmic aggregates in a wide range of different cell types, including astrocytes. The aggregates have several important features in common with Rosenthal fibers, including the association of αB-crystallin and HSP27. This association occurs simultaneously with the formation of protein aggregates containing R416W GFAP and is also specific, since HSP70 does not partition with them. Monoclonal antibodies specific for R416W GFAP reveal, for the first time for any IF-based disease, the presence of the mutant protein in the characteristic histopathological feature of the disease, namely Rosenthal fibers. Collectively, these data confirm that the effects of the R416W GFAP are dominant, changing the assembly process in a way that encourages aberrant filament-filament interactions that then lead to protein aggregation and chaperone sequestration as early events in Alexander disease.
Alexander disease (MIM 203450) is a rare and often fatal neurological disorder, first described by W. S. Alexander.1 On the basis of age at onset, the disorder has been divided into three subtypes: infantile, juvenile, and adult.2 The infantile form, with onset between birth and age ∼2 years, is the most common type and is characterized by extensive loss of white matter.3–5 A striking neuropathological feature of all forms of Alexander disease is the presence of Rosenthal fibers, unique cytoplasmic inclusions within astrocytes that contain the major astrocytic intermediate filament (IF) protein glial fibrillary acidic protein (GFAP) and the chaperones αB-crystallin and HSP27.6–8 Although the GFAP within Rosenthal fibers appears disorganized, astrocytes in Alexander disease also possess GFAP filaments with conventional 10-nm morphology.
Recently, missense point mutations in GFAP have been identified as a genetic basis for Alexander disease.9 To date, all known mutations have been heterozygous, indicating that the mutant form of the protein is dominant over the wild type. This is consistent with the finding of autosomal dominant mutations in 26 other IF genes that are linked to human disease,10,11 summarized in the online Intermediate Filament Disease Mutation Database. The list of known mutations in GFAP now includes 32 nucleotide changes that affect 24 aa spread throughout the entire sequence12 (see also the Alexander Disease Web site). The mutations usually arise spontaneously during spermatogenesis,13 with familial cases being quite rare because of the high morbidity associated with the disease. The mutation studied in this report, R416W, is one of the four mutations reported in familial cases and is also found in sporadic cases.12
Like other IF family members, GFAP has a characteristic domain structure comprising a central α-helical rod domain flanked by non–α-helical N-terminal “head” and C-terminal “tail” domains.14 The rod domain contains characteristic heptad repeats of hydrophobic residues, which are the underlying basis for the coiled-coil dimer in the filament, and the highly conserved LNDR and TYRKLEGGE motifs that are present at the start and the end of this central rod domain.10 Both of these motifs are highly conserved throughout the whole IF family,15 and those mutations in Alexander disease and other genetic IF protein disorders found within them usually correlate with the severest forms of the diseases.10,11 The crystallization of regions containing these two motifs from vimentin, a closely related type III IF protein16,17 that coassembles with GFAP, has provided the atomic structure of these particular highly conserved motifs. Our knowledge of the important higher order interactions within the filament, however, is still limited to low-resolution studies18–20; therefore, the full structural impact of most of these rod mutations has not yet been detailed.
One of the other common mutations outside the central rod domain of GFAP that causes Alexander disease is R416W. This mutation occurs in the tail domain within the RDG motif, which is conserved among all GFAP proteins from multiple species, as well as the related type III IF proteins vimentin and desmin. Here, we describe the effects of this mutation on GFAP assembly and use this R416W GFAP mutant to identify the early events in the development of Alexander disease.
Material and Methods
Plasmid Construction and Site-Directed Mutagenesis
Total RNA was extracted from human astrocytoma U373MG cells with the RNeasy kit (Qiagen). The complete human GFAP cDNA was amplified by RT-PCR, with the SuperScript RT-PCR system (Invitrogen), with use of oligonucleotides 5′-CATATGGAGAGGAGACGCAT-3′ and 5′-TCACATCACATCCTTGTGCT-3′ as forward and reverse primers, respectively. The amplified PCR product was cloned into the pGEM-T Easy vector (Promega), to generate pGEM-T Easy-WTGFAP, and the entire sequence was confirmed against the GenBank database entry for GFAP (accession number J04569). The R416W mutation was introduced by QuickChange site-directed mutagenesis (Stratagene) with use of the pGEM-T Easy-WTGFAP vector as a template. The following mutagenic oligonucleotides that contained the desired C→T mutation at np 1246 were synthesized: 5′-GAAGACCGTGGAGATGTGGGATGGAGAGGTCAT-3′ and 5′-ATGACCTCTCCATCCCACATCTCCACGGTCTTC-3′. The amplified PCR product containing the R416W mutation was cloned into the pGEM-T Easy vector, and the mutation sequence was confirmed by DNA sequencing. For expression in cultured mammalian cells, both the wild-type and R416W GFAP cDNA in the pGEM-T Easy vector were subcloned into the pcDNA3.1(−) vector (Invitrogen) with use of the XbaI and EcoRI restriction sites. The GFAP mutant R239C was also subcloned into pCDNA3.1 from the pRSVi vector21 with use of the restriction enzyme HindIII. For expression in bacteria, wild-type and R416W GFAP cDNA samples were subcloned into the pET23b vector (Novagen) with use of the NdeI and EcoRI restriction sites.
Expression and Purification of Recombinant GFAPs
For bacterial expression of proteins, pET 23b vector containing either wild-type or mutant GFAP cDNA samples were transformed into Escherichia coli strain BL21(DE3) pLysS (Novagen). After transformation, cultures were grown in Luria Bertani medium supplemented with appropriate antibiotics to OD600 of 0.5–0.6, and protein expression was induced by the addition of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 3 h. Overexpressed GFAP formed inclusion bodies, which were prepared as described elsewhere.22 The final pellets, consisting predominantly of GFAP, were solubilized in extraction buffer (8 M urea, 20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) at room temperature for 3 h, and any insoluble material was removed by centrifugation, at 100,000 g, in a benchtop Optima MAX Ultracentrifuge with use of an MLA-80 rotor (Beckman Coulter). GFAP was further purified by ion-exchange chromatography with use of a Merck-Hitachi Biochromatography system equipped with a Fractogel-EMD TMAE 650S column (Merck) pre-equilibrated in the column buffer (6 M urea, 10 mM Tris-HCl [pH 8.0], 5 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF). GFAP was eluted from the column with a linear gradient of 0–0.5 M NaCl in the same buffer over 1 h at a flow rate of 1 ml/min. The GFAP-enriched fractions were pooled, concentrated, and applied to a Fractogel EMD COO- 650S column (Merck) and were pre-equilibrated with column buffer (6 M urea, 20 mM sodium formate [pH 4.0], 5 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF). After washing with buffer B, GFAP was eluted with a linear gradient of 0–0.5 M NaCl in the same buffer. Column fractions were analyzed by SDS-PAGE, and those containing purified GFAP were collected and stored at −80°C. Protein concentrations were determined by bicinchonic acid assay (BCA reagent [Perbio Science]) with use of BSA as standard.
In Vitro Assembly and Sedimentation Assay
Purified GFAP was diluted to 0.3 mg/ml in 6 M urea in a buffer of 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, 1 mM EGTA, and 1 mM DTT and was dialyzed stepwise against 3 M urea in the same buffer for 4–6 h at room temperature and then against the same buffer without urea overnight at 4°C. Filament assembly was completed by dialyzing against assembly buffer (20 mM imidazole-HCl [pH 6.8], 100 mM NaCl, and 1 mM DTT) for 12–16 h at room temperature. The efficiency of in vitro assembly was assessed by sedimentation assay as described elsewhere.23 In brief, the assembly mixture was layered onto a 0.85-M sucrose cushion in assembly buffer and was centrifuged at 80,000 g for 30 min. To investigate the effect of mutations on filament-filament interactions, assembled filaments were subjected to low-speed centrifugation at 3,000 g for 10 min in a bench-top centrifuge (Eppendorf). The supernatant and pellet fractions were analyzed by SDS-PAGE24 and were visualized by Coomassie blue staining. In some instances, the proportion of GFAP distributed between pellet and supernatant fractions was measured using an image analyzer (LAS-1000plus [FujiFilm]). Coomassie blue signals for individual bands were quantified using the Image Gauge software (v. 4.0) (FujiFilm).
Electron Microscopy
GFAP was diluted in assembly buffer to 100 μg/ml and was negatively stained with 1% (w/v) uranyl acetate (Agar Scientific). Samples on carbon-coated copper grids were examined with a Phillips 400T transmission electron microscope, with use of an accelerating voltage of 80 kV. Images were acquired at a magnification of 17,000× on Kodak 4489 film and then were digitized at 1,200×1,200–pixel resolution before being processed further in Adobe Photoshop 7 (Adobe Systems).
Cell Cultures and Transient Transfection
Human breast cancer epithelial MCF7 cells were obtained from the European Collection of Cell Cultures (Sigma). Human adrenal cortex carcinoma SW/cl.1 and SW13/cl.2 cells were kindly provided by Dr. Robert Evans (University of Colorado Health Sciences Center, Denver). The human astrocytoma cell line U343MG was a gift from Dr. Rutka (Toronto), and the cells were grown in αMEM (Invitrogen). These cells express vimentin and GFAP as well as HSP27 and αB-crystallin. Primary mouse astrocytes from wild-type and vimentin/GFAP double-knock littermates were a generous gift of Dr. Milos Pekny (Göteborg, Sweden). They were prepared and grown as described elsewhere.25 Unless otherwise stated, cells were grown in Dulbecco's modified Eagle medium supplemented with 10% (v/v) fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Sigma) and were maintained at 37°C in a humidified incubator of 95% (v/v) air and 5% (v/v) CO2.
For transient transfection studies, cells grown on 13-mm coverslips at a density of 40%–50% confluency were transfected with pcDNA3.1(−) expressing either wild-type or R416W GFAP, with use of GeneJuice transfection reagent (Novagen) according to the manufacturer’s protocol. In some experiments, the R239C GFAP21 and myc-tagged ubiquitin (His6-myc-Ubiquitin)26 were used. Cells were analyzed by double-label immunofluorescence microscopy 48 h after transfection.
Antibody Production
Mouse monoclonal antibodies were made that specifically recognized human but not wild-type R416W GFAP. The immunogen was a peptide dodecamer centered on the mutation site, KTVEMWDGEVIK (Genemed Synthesis), which was linked to keyhole limpet hemocyanin. Monoclonal antibodies were produced by the UAB Epitope Recognition Core, which also performed an initial ELISA screen against purified recombinant wild-type and R416W mutant GFAP. The corresponding wild-type peptide, KTVEMRDGEVIK, failed to produce monoclonal antibodies specific to the wild-type sequence.
Immunostaining of Cells and Tissues and Immmunofluoresence Microscopy
Immunocytochemistry of cultured cells was performed on coverslips washed twice with PBS, and the cells were fixed in either ice-cold methanol/acetone (1:1 [v/v]) for 20 min or in 4% (w/v) paraformaldehyde/PBS for 10 min. In the case of paraformaldehyde fixation, cells were subsequently permeabilized with 0.5% NP-40 in PBS for 10 min. After being washed twice with PBS containing 0.02% (w/v) sodium azide and 0.02% (w/v) BSA (PBS/BSA/azide), cells were blocked with 10% (v/v) goat serum in PBS/BSA/azide for 20 min and then were incubated with primary antibodies at room temperature for 1 h. The primary antibodies used in this study were mouse monoclonal anti-GFAP (G-A-5, 1:500 [Sigma]), rabbit polyclonal GFAP antibodies (Z0334, 1:500 [Dako]), monoclonal anti-human GFAP (SMI-21, 1:500 [Sternberger Monoclonals]), monoclonal anti-keratin 18 (LE41 [kindly provided by Prof. Birgit Lane, University of Dundee, Dundee, United Kingdom), monoclonal anti-R416W GFAP (19.2 and 1A3, described below, 1:500), rabbit polyclonal anti-GFAP (clone 3270, 1:200),27 polyclonal anti-vimentin (clone 3052, 1:200),28 and the myc-epitope monoclonal antibody (Clone 9E11, 1:10).29 After cells were washed with PBS/BSA/azide, the primary antibodies were detected using Alexa 488 (1:400 [Molecular Probes]) or Alexa 594 (1:600 [Molecular Probes]) conjugated secondary antibodies. All antibodies were diluted in PBS/BSA/azide buffer. The glass coverslips were mounted on slides with the fluorescent protecting agent Citifluor (Citifluor Labs) and were observed with a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss). Optical sections were set to ∼1.0 μm. Images were processed and prepared for figures with Adobe Photoshop 7 (Adobe Systems). Quantitation of the GFAP filament phenotypes was by visual assessment of the cells and by scoring cells for the presence or absence of GFAP-containing aggregates. Approximately 100–150 transfected cells were assessed, and each experiment was repeated at least three times.
Immunohistochemistry from normal human and Alexander disease brain sections was performed on 6 μm–thick paraffin sections kindly provided by Drs. Jim Goldman and Goumei Tang (Columbia Medical School, New York). Internal review board approval was obtained from Columbia Medical School for these studies. Archival material for the infantile R416W Alexander disease case used in this study was described elsewhere.30 Primary antibodies were rabbit anti-cow GFAP (Z0334, 1:5,000 [Dako]), and mouse anti-R416W GFAP (19.2, described below, 1:2,000). Secondary antibody for the peroxidase method was peroxidase-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch) (1:2,000), with staining visualized using 3,3′-diaminobenzidine tetrachloride (DAB, metal-enhanced Substrate Kit [Pierce]). Secondary fluorescent antibodies are described above. Some sections were counterstained with Hoechst 33258 (Sigma) to reveal nuclei.
Ultrastructural Analysis by Immunoelectron Microscopy
MCF7 cells grown on 10-cm2 petri dishes (Greiner Bio-One) were transfected with either wild-type or R416W GFAP for 48 h. Cells were then fixed directly in 80 mM cacodylate buffer (pH 7.2) containing 1.25% (v/v) glutaraldehyde and 1% (w/v) paraformaldehyde for 30 min at room temperature. Cells were scraped off the dish by a rubber policeman, were pelleted by low-speed centrifugation, and were washed three times with cacodylate buffer. The cells were then postfixed with 1% (w/v) osmium tetroxide in cacodylate buffer. After several washes with distilled water, cells were subjected to a series of graded ethanol dehydration, followed by overnight incubation in 1:1 propylene oxide:epoxy resin (Durcupan [Sigma]). After two changes of 100% fresh resin, cell pellets were transferred to BEEM capsules (Agar Scientific) and were polymerized in fresh resin overnight at 60°C. Ultrathin sections were generated using a Leica Ultracut ultramicrotome and were collected on pioloform and carbon-coated nickel grids (Agar Scientific). The grid specimens were then etched with 1% periodic acid, and osmium was removed by 2% (w/v) sodium periodate before incubation with blocking solution consisting of 0.5% (w/v) fish skin gelatin (Sigma) in PBS. Subsequently, sections were incubated with polyclonal anti-GFAP antibodies (clone 3270) diluted 1:20 in blocking solution for 90 min, were washed three times in PBS, and then were incubated with protein A conjugated with 5-nm gold particles (British BioCell International) for 2 h. After several washes in distilled water, specimens were stained with saturated aqueous uranyl acetate (3% [w/v]) for 30 min, followed by staining with lead citrate for 30 min.31 Stained samples were subsequently examined on an FEI Tecnai 12 transmission electron microscope (FEI).
Preparation of Cytoskeletal Fractions and Immunoblotting Analysis
Cells grown on 10-cm2 petri dishes were transfected with control vector (pcDNA3.1) or vectors containing either wild-type or R416W GFAP cDNA. At 48 h after transfection, cells were lysed using two different extraction buffers, designed to test the resistance of GFAP filaments and aggregates to extraction. In the mild extraction protocol, cells were lysed on ice for 15 min in 1 ml mild extraction buffer (MEB; 20 mM Tris-HCl [pH 7.6], 140 mM NaCl, 5 mM EDTA, 1 mM EGTA, 0.5% [v/v] NP-40 supplemented with Complete protease inhibitor cocktail [Roche Diagnostics], and 1 mM PMSF). In the more severe extraction protocol, cells were lysed in 1 ml of a harsher extraction buffer (HEB) containing deoxycholate (20 mM Tris-HCl [pH 7.6], 140 mM NaCl, 5 mM EDTA, 1 mM EGTA, 1% [v/v] NP-40, 0.5% [w/v] sodium deoxycholate supplemented with Complete protease inhibitor cocktail [Roche Diagnostics], and 1 mM PMSF). Cell lysates were collected, were homogenized in a Dounce homogenizer (Wheaton), and were centrifuged at 16,000 g for 15 min at 4°C. The pellet was resuspended in pelleting buffer (20 mM Tris-HCl [pH 8.0], 10 mM MgCl2, and 1 mM PMSF) containing 250 U/ml benzonase nuclease (Novagen) and was incubated for 30 min at room temperature. After repelleting, the final pellets were washed in PBS containing 1 mM PMSF and then were resuspended in Laemmli’s sample buffer,24 in a volume that was equivalent to the supernatant. Supernatant and pellet fractions were then boiled for 5 min in Laemmli’s sample buffer, and equal volumes were analyzed by SDS-PAGE and immunoblotting. Actin was used as a loading control for these samples.
Immunoblotting was performed using the semidry blotting method, according to the manufacturer’s specifications (Bio-Rad Laboratories). After the blotting, protein-transfer efficiency was assessed by Ponceau S (Sigma) staining of the nitrocellulose membrane, followed by destaining in Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.4], and 150 mM NaCl). Membranes were blocked for 2 h in blocking buffer containing 5% (w/v) BSA in either TTBS (TBS containing 0.2% [v/v] Tween 20) or PBST (5% [w/v] nonfat milk and 0.1% [v/v] Tween-20 in PBS) and were incubated for 2 h with mouse monoclonal anti-GFAP (GA-5), monoclonal anti-human GFAP (SMI-21), monoclonal anti-actin (AC-40 [Sigma]), or rabbit polyclonal anti-GFAP antibodies (3270) diluted by 1:5,000 in blocking buffer. In some experiments, the mouse monoclonal antibodies to the c-myc epitope (9E10, diluted by 1:100 in blocking buffer) were used to detect tagged ubiquitin after transfection. After several washes with TTBS, the membrane was incubated for 1 h with horseradish peroxidase–conjugated secondary antibodies (Dako) diluted by 1:2,000 in blocking buffer, followed by washing with TBS for 30 min. Antibody labeling was detected by enhanced chemiluminescence (ECL Plus [Amersham Biosciences]) with use of a luminescent image analyzer (LAS-1000plus [FujiFilm]). The strength of signal was quantified using the Image Gauge software (v. 4.0) (FujiFilm).
Immunoblotting of brain samples was performed using anonymous, frozen tissues kindly provided by Drs. Jim Goldman and Goumei Tang (New York). The tissues were Dounce homogenized on ice in 10 mM Tris-HCl (pH 7.4), 2 mM β-mercaptoethanol, 0.1 M NaCl, 5 mM EDTA, and 1× protease inhibitor cocktail (Sigma) at a 10:1 (v/w) buffer:tissue ratio. The homogenate was centrifuged at 80,000 g for 1 h at 4°C, and the pellet was dissolved in ∼15 volumes of the above buffer containing 2% (w/v) SDS. After determination of protein concentrations with the BCA reagent (Pierce), triplicate sets of 500-ng aliquots of each extract were run on a 10% (w/v) SDS-polyacrylamide gel, with 20-ng samples of purified recombinant wild-type and R416W GFAP. After transfer to Hybond ECL membranes (Amersham Pharmacia Biotech), the blots were probed with either anti-human GFAP monoclonal antibody SMI-21 (Sternberger Monoclonal) (1:2,000) or anti-R416W GFAP monoclonal antibodies 1A3 or 19.2 (1:1,000 dilution). Signals were detected by ECL (Amersham) and were visualized using a ChemImager 4400 (Alpha Innotech).
Results
Effect of R416W Mutation on In Vitro GFAP Assembly
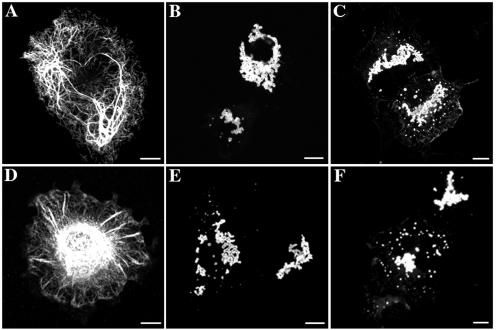
In vitro assembly studies were performed to determine how the R416W mutation affects the structural properties of GFAP filaments. Purified recombinant wild-type and R416W GFAP, produced in E. coli with a pET-based expression system, were assembled in vitro by dialysis-based assembly and were visualized by negative staining with uranyl acetate followed by electron microscopy. Whereas wild-type GFAP assembled into typical 10-nm filaments (fig. 1A), R416W GFAP on its own or in combination with mixtures of wild-type GFAP failed to form extended filaments (fig. 1B–1E). Instead, it formed short rod-like structures that looked like short filament pieces that had failed to elongate and compact properly into 10-nm filaments (fig. 1E). These had a strong tendency to laterally associate into aggregates (fig. 1D). Because the R416W mutation is heterozygous in patients with Alexander disease, we also assessed the assembly behavior of R416W mutant GFAP in the presence of increasing proportions of wild-type GFAP. At both 50:50 (fig. 1C) and 75:25 (fig. 1B) proportions of wild-type:R416W GFAP, normal filament assembly was disrupted, and aggregates similar to those made by R416W GFAP alone (fig. 1D) were formed instead. These data indicate that, in vitro, the R416W mutant GFAP acts in a dominant manner over the wild-type protein. These data suggested that the assembled R416W GFAP filaments were prone to aggregation; therefore, we performed a low-speed centrifugation assay designed to monitor the extent of filament-filament interactions in the whole filament population.32 With use of this assay, only approximately one-third of the wild-type GFAP was found in the supernatant fraction (fig. 1F; e.g., lanes 1 and 2). In contrast, when assembled on its own, nearly all of the R416W GFAP sedimented into the pellet fraction (fig. 1F; e.g., lanes 8 and 7). High-speed centrifugation assay confirmed that the wild-type GFAP had assembled efficiently, because >90% of the protein was sedimented under standard centrifugation conditions23,27 (data not shown). Decreasing the proportion of mutant GFAP in mixtures with wild-type GFAP (fig. 1F; lanes 3–6) failed to markedly increase the proportion of soluble material. These data suggest that the R416W GFAP mutation promotes more interfilament interactions and that this effect is again dominant over the wild-type protein.
Figure 1. .
The dominant effect of R416W GFAP, revealed by in vitro assembly studies. Purified GFAP at a concentration of 0.3 mg/ml was assembled in vitro by stepwise dialysis into assembly buffer, as described in the “Material and Methods” section. Assembled filaments were negatively stained and were visualized by transmission electron microscopy. Under these assembly conditions, wild-type GFAP assembled into typical 10-nm filaments with length of several microns (A), whereas R416W GFAP alone and in different proportions with wild-type protein formed short filamentous intermediates that had a strong tendency to aggregate (B–E). It is difficult to see the structural detail of the aggregates formed by R416W GFAP (D) and mixtures thereof (B and C) when negatively stained with uranyl acetate. Sometimes, less aggregated material can be found, and then, at higher magnification (E), the filamentous structures that comprise the aggregates are clearly seen. Mixing wild-type GFAP in either 75:25 (B) or 50:50 (C) proportions with R416W GFAP failed to rescue intermediate filament formation, and similar aggregates were formed (B and C). A low-speed sedimentation assay was used to assess the extent of this aggregation. F, Wild-type (WT) (lanes 1 and 2) and R416W GFAP (lanes 7 and 8) were assembled, either individually or in mixtures of 75:25 (lanes 3 and 4) or 50:50 (lanes 5 and 6) WT:R416W GFAP. After assembly, the samples were subjected to low-speed centrifugation, and the resulting supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and were visualized by Coomassie blue staining. Whereas only one-third of assembled wild-type GFAP was sedimented (lane 1), almost all the R416W mutant was found in the pellet fraction (lane 8). Mixing wild-type GFAP with the R416W mutant in different proportions did not dramatically increase the GFAP signal in either the supernatant fraction of the 50:50 mixture (lane 5) or the 75:25 mixture (lane 3). These data show that the effects of R416W GFAP on in vitro filament assembly is dominant over the wild-type protein. Bars = 1 μm, except in panel E, where it is 0.1μm.
Effect of R416W Mutation on GFAP Network Formation in Cells Lacking Endogenous GFAP
Transient transfection assays were used to investigate the effects of the R416W mutation on the formation of GFAP networks in a range of cell lines that have different IF compositions. To determine the effect of the R416W mutation on de novo IF network formation, we selected as the host cells the human adrenocarcinoma-derived SW13/cl.2 cell line, which does not contain any endogenous cytoplasmic IFs.33 When expressed in SW13/cl.2 cells, wild-type GFAP formed filaments (fig. 2A), in agreement with previous studies.34,35 In contrast, R416W GFAP failed to assemble into filaments but instead formed clusters of cytoplasmic aggregates (fig. 2B). Similar results were observed when wild-type (fig. 2D) and R416W plasmids (fig. 2E) were transiently transfected into primary astrocytes derived from mice lacking GFAP and vimentin.25 For comparison, R239C GFAP was also transfected into both these cell lines (fig. 2C and 2F), and, like R416W GFAP, it also formed numerous cytoplasmic aggregates when the cytomegalovirus promoter was used to drive mutant GFAP expression. Previous studies have already established that the R239C mutation in GFAP makes the filaments more stable and changes the organization of GFAP networks in transfected cells. Taken together, these data suggest that the R416W mutation, like the R239C mutation, affects the ability of GFAP to self-assemble into an extended filament network in IF-free primary cells and that this deficiency is independent of the cellular background.
Figure 2. .
Effect of R416W and R239C mutations on the de novo GFAP IF network formation in IF-free cells. SW13/cl.2 (A–C) and primary astrocytes derived from GFAP/vimentin-null mice (D-F) were transiently transfected with either wild-type (A and D), R416W (B and E), or R239C (C and F) GFAP. At 48 h after transfection, the distribution of GFAP was assessed by confocal immunofluorescence microscopy with use of the rabbit polyclonal anti-GFAP antibody. When expressed in SW13/cl.2 cells, wild-type GFAP assembled into bundled filaments that extended throughout the cytoplasm (A). In contrast, cells transfected with either R416W (B) or R239C (C) GFAP-expression plasmids exhibited only GFAP-positive aggregates. In the IF-free mouse astrocytes, wild-type GFAP assembled into extended filaments at the cell periphery with some perinuclear accumulations (D), whereas R416W mutant GFAP formed punctuate aggregates scattered throughout the cytoplasm without any detectable filaments (E). Expression of R239C GFAP also induced numerous GFAP aggregates in the cytoplasm (F). For both R416W and R239C GFAP, all the transfected cells had aggregates. Bars = 10 μm.
Recent studies have revealed that two or more IF proteins are required for the formation of most normal IF networks in vivo, including those formed by the type III IF proteins GFAP, desmin, and vimentin. For instance, desmin requires paranemin to produce an extended IF network in nonmuscle-derived cell lines.35 Vimentin requires GFAP to form filaments in astrocytes,36 whereas, in other cell types, vimentin coassembles with nestin.37 Whereas GFAP is able to self-assemble into IFs within astrocytes lacking vimentin, vimentin is required for the correct spacing of the assembled filaments.36 Therefore, it is important to assess the assembly behavior of GFAP in cells containing this key assembly partner.
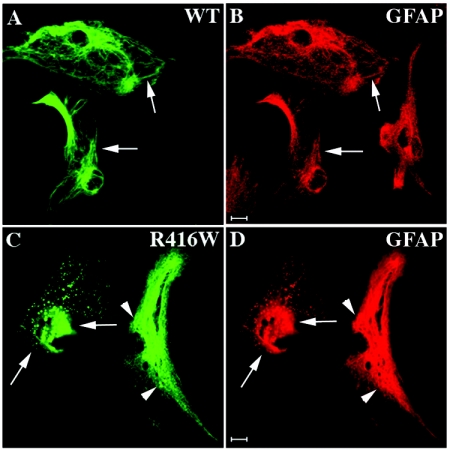
For these experiments, SW13/cl.1 cells that express vimentin IFs were selected for transient transfection studies. In this cell line, wild-type GFAP assembled into filamentous networks (fig. 3A) that colocalized with the endogenous vimentin IFs (fig. 3B). In contrast, most of the cells transfected with R416W GFAP contained large aggregates (fig. 3C; arrows) with small clumps at the cell periphery (fig. 3C; arrowheads). These aggregates disrupted the endogenous vimentin IF networks, usually causing them to collapse into a large perinuclear aggregate (Fig. 3D; arrows).
Figure 3. .
The network-forming abilities of the wild-type and R416W GFAP in SW13/cl.1 cells. SW13/cl.1 cells transiently transfected with either wild-type (A and B) or R416W (C and D) GFAP were fixed at 48 h after transfection and were processed for double-label confocal immunofluorescence microscopy. GFAP immunofluoresence is shown in the green channel (A and C), whereas the counterstaining for vimentin is in the red channel (B and D). Notice that wild-type GFAP (A) incorporated into endogenous vimentin (Vim) (B) networks, whereas this is not the case for R416W GFAP (C). Whereas some transfected cells exhibited one large inclusion with small aggregates at the cell periphery (arrowheads in C), other transfected cells displayed bundled filaments (arrows in C) that coaligned with the endogenous vimentin (arrows in D). Bars = 10 μm.
The finding that R416W GFAP caused the collapse of the endogenous vimentin IF network led us to examine its effects on keratin networks, IF proteins that do not coassemble with GFAP. For these experiments, we used a human breast cancer epithelial MCF7 cell line that expresses only keratin IF proteins (K8, K18, and K19).38 When transfected into this cell line, wild-type GFAP assembled into filaments that tended to bundle in most of the transfected cells (fig. 4A). The GFAP filament networks were found to partially coalign with the keratin IF networks (fig. 4B; arrows), which is consistent with previous observations.22,27 In contrast, nearly all MCF cells expressing R416W mutant GFAP formed aggregates (fig. 4C; arrows) that often collapsed the endogenous keratin IF networks (fig. 4D; arrows). These results suggest that the R416W mutation not only impaired the ability of GFAP to form normal IF network but also revealed a dominant effect on endogenous keratin filament networks in the absence of any obvious coassembly.
Figure 4. .
Expression of R416W mutant in MCF7 cells resulted in the formation of GFAP aggregates. MCF7 cells transfected with either wild-type (A and B) or R416W GFAP (C and D) were processed at 48 h after transfection for confocal double-label immunofluorescence microscopy with use of antibodies against GFAP (A and C) and keratin (B and D). When expressed in these cells, wild-type GFAP formed extended filaments as well as perinuclear filament bundles (A) that partially colocalized with keratin IF networks (arrows in A and B). In contrast, transfected cells expressing R416W GFAP exhibited large aggregates (C) that also cocollapsed the endogenous keratin IF networks (arrows in C and D). Bars = 10 μm.
The relative expression levels and solubility of the wild-type and the R416W GFAP were determined by immunoblotting of extracts from MCF7 cells prepared using a mild lysis buffer protocol. Analysis of both supernatant and pellet fractions revealed no endogenous GFAP expressed in nontransfected MCF7 cells (fig. 5A; lanes 1 and 2). In contrast, cells transfected with either wild-type (fig. 5A; lanes 3 and 4) or R416W (fig. 5A; lanes 5 and 6) GFAP generated proteins of the expected size at comparable levels, suggesting that aggregate formation is likely due to the mutation per se rather than elevated expression levels. Although a small fraction of wild-type GFAP remained in the supernatant fraction (fig. 5A; lane 3), R416W GFAP was found exclusively in the pellet fraction (fig. 5A; lane 6), consistent with its sequestration into cytoplasmic aggregates. Equal loading for the various supernatant and pellet fractions was verified using an anti-actin antibody (fig. 5B).
Figure 5. .
Analysis of wild-type and R416W GFAP expression in transfected MCF7 cells by immunoblotting. MCF7 cells were transfected with either wild-type (lanes 3 and 4) or R416W GFAP (lanes 5 and 6). Untransfected cells were used as a control (lanes 1 and 2). At 48 h after transfection, cells were collected, lysed with MEB, and centrifuged at 18,000 g for 15 min at 4°C. The resulting supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE, followed by immunoblotting using anti-GFAP (A) and anti-actin (B) antibodies. The blots were developed by the ECL system. Notice that, after transfection into MCF7 cells, both wild-type and mutant GFAP expressed at comparable levels, although proteolyzed GFAP fragments with slightly higher electrophoretic mobilities were also detected. Most of the wild-type GFAP was detected in the pellet fraction (A, lane 4) with a small proportion that remained soluble (A, lane 3), whereas the R416W GFAP was found exclusively in the pellet fraction (A, lane 6). Equal loading of each supernatant and pellet fractions was confirmed by probing with anti-actin antibody (B).
Analysis of R416W GFAP Aggregates in MCF7 Cells by Electron Microscopy
The high transfection efficiency of both wild-type and R416W GFAP in MCF7 cells allowed us to further analyze the ultrastructural organization of GFAP by immunoelectron microscopy. MCF7 cells transfected with plasmids expressing either the wild-type or the R416W GFAP were processed for immunogold labeling with anti-GFAP antibodies followed by protein A conjugated with 5-nm gold particles. In cells expressing wild-type GFAP, fibrous regions consisting of 10-nm filaments were observed (fig. 6A and 6B). These filaments were organized into bundles that traversed the cytoplasm (fig. 6A). At a higher magnification, individual filaments were decorated with gold particles (fig. 6B; arrows), confirming the presence of GFAP in these filaments. In contrast, cells expressing R416W GFAP displayed electron-dense aggregates, distributed throughout the cytoplasm (fig. 6C; arrows), that were often accompanied by IFs (fig. 6C; asterisks). At a higher magnification, these electron-dense aggregates appear as amorphous membrane-free structures composed of aggregated GFAP, as evidenced by the presence of gold particles (fig. 6D; arrows).
Figure 6. .
Ultrastructural analysis of wild-type and R416W GFAP in MCF7 cells by immunoelectron microscopy. MCF7 cells transfected with either wild-type (A and B) or R416W (C and D) GFAP were processed at 48 h after transfection for immunogold labeling, as described in the “Material and Methods” section. Immunogold labeling of ultrathin sections was stained and visualized by a transmission electron microscope. Wild-type GFAP assembled into filaments that were organized into parallel bundles (A). In contrast, cells expressing the R416W mutant formed membrane-free irregular-shaped structures composed of electron-dense aggregates at the perinuclear region (C), often in association with IFs (asterisks [*] in C). Panels B and D are higher magnification views of the boxed areas of panels A and C, respectively, showing that both the filaments (B) and aggregates (D) were decorated with 5-nm gold particles (arrows in B and D), confirming the identity of GFAP.
Effects of R416W GFAP on Endogenous GFAP Networks in Primary Astrocytes
The assembly behavior of R416W GFAP was also examined in mouse primary astrocytes, to test the effects of endogenous wild-type GFAP and vimentin IFs on the aggregation process. The distribution of transfected GFAP in relation to the endogenous GFAP was visualized by double-label immunofluorescence microscopy with use of monoclonal antibody SMI-21 (fig. 7A and 7C), which specifically recognizes human GFAP, and polyclonal anti-panGFAP antibody, which recognizes both the endogenous mouse GFAP and the transfected human GFAP (fig. 7B and 7D). Mouse astrocytes transiently transfected with wild-type GFAP mainly formed filaments (fig. 7A) that colocalized with the endogenous GFAP network (fig. 7B; arrows). In contrast, the effects of R416W GFAP expression in astrocytes were similar to those observed for the vimentin-containing SW13/cl.1 cells. Most of the transfected cells contained cytoplasmic aggregates with smaller particles at the cell periphery (fig. 7C), which colocalized with collapsed endogenous GFAP networks (fig. 7C and 7D; arrows). In some transfected cells, however, expressed R416W GFAP was incorporated into the endogenous GFAP networks without any apparent changes (fig. 7C and 7D; cell on the right). Careful examination revealed that there were small aggregates intermingled with the filaments (fig. 7D; arrowheads), which were immunopositive for the human R416W GFAP, indicative of perhaps an early change in the organization of the GFAP filaments preceding the eventual collapse of the network.
Figure 7. .
Transient expression of wild-type or R416W GFAP in primary mouse astrocytes. Primary mouse astrocytes were transfected with either human wild-type (A and B) or R416W (C and D) GFAP. At 48 h after transfection, cells were processed for double-label immunofluoresence microscopy with use of anti-human GFAP monoclonal antibody (SMI-21) and anti-panGFAP polyclonal antibodies (3270). When expressed in mouse primary astrocytes, wild-type GFAP formed filaments (arrows in A) that colocalized with the endogenous mouse GFAP (arrows in B). The expression of human R416W GFAP resulted in both filamentous (arrowheads in C) and aggregate staining patterns (arrows in C), which also costained with the endogenous mouse GFAP (arrows and arrowheads, respectively, in D). Bars = 10 μm.
Detection of R416W GFAP in Alexander Disease Brain with Mutant-Specific Monoclonal Antibodies
The amino acid sequences of mouse and human GFAP are highly homologous, with 91% identity and 95% similarity,39 but species differences could influence the assembly properties of the mutant human R416W GFAP in mouse astrocytes. To follow the fate of the human R416W GFAP in the presence of the endogenous wild-type human GFAP, we sought to develop mouse monoclonal antibodies that specifically recognized human R416W GFAP but not wild-type GFAP, and vice versa (see the “Material and Methods” section for details). The immunogen specific for the wild type failed to elicit any GFAP monoclonal antibodies. The R416W GFAP peptide, however, yielded two monoclonal antibodies, 19.2 and 1A3, that recognized R416W GFAP but not wild-type GFAP, as judged by immunoblots of purified, recombinant proteins (fig. 8A). In addition, neither of these monoclonal antibodies crossreacted with a lysate from a normal brain or with a lysate from a brain containing another GFAP mutation commonly associated with Alexander disease, R239C, and both produced signals from lysates of R416W brain that were nearly identical to those of the control SMI-21 monoclonal antibody (fig. 8A). Suitability for immunohistochemistry was demonstrated by immunostaining SW13/cl2 cells transfected with vectors expressing either the mutant R416W or wild-type protein (data not shown).
Figure 8. .
Characterization of R416W GFAP-specific antibodies and demonstration of its presence in Rosenthal fibers. A, Immunoblots, performed as described in the “Material and Methods” section, with use of purified, recombinant human wild-type (hGF-WT) and R416W (hGF-R416W) GFAP and lysates from brain samples taken from either control human (WT) or patients with Alexander disease that harbor either an R239C mutation (R239C) or an R416W mutation (R416W) in GFAP. The general anti-human GFAP monoclonal antibody (SMI-21) reacts with all samples, whereas the anti-R416W monoclonal antibodies (1A3 and 19.2) produce signals from R416W-containing samples only, with the pattern for the R416W patient lysate identical to that of SMI-21. The identity of the immunopositive bands above and below the prominent GFAP-positive band are as yet unknown. The lower bands most likely correspond to degradation products, since these are normally seen in control brain samples. The upper bands are common to both the R239C and R416W samples, suggesting these are a common feature of Alexander disease pathology, but, as yet, the reason for their slower electrophoretic mobility is unknown. Panels B and C are striatum in a control (normal) brain stained with standard polyclonal GFAP antibody (B) and by R416W monoclonal antibody 19.2 (C). Note that the R416W antibody does not crossreact with normal human brain tissue. Panels D–F are brain sections from a patient with the R416W GFAP mutation that are stained with the monoclonal 19.2 antibody and then are visualized by either peroxidase- (D) or rhodamine-tagged secondary antibodies (F) or are stained with the rabbit polyclonal GFAP antibody (Dako) and then are detected with FITC-tagged secondary antibodies (E). Nuclei are counterstained with Hoechst 33258 (E and F [see the “Material and Methods” section for procedure details]) to assist comparison of the panels E and F. Numerous Rosenthal fibers are stained around their periphery (arrows in E and F), a feature often reported for these aggregates (e.g., the work of Tomokane et al.6). Normal-looking GFAP filaments are also stained by the R416W-specific mAb (arrowheads in F) and can be detected by the diffuse staining in other parts of the section.
The R416W GFAP–specific monoclonal antibodies then allowed us to determine whether R416W GFAP incorporated into Rosenthal fibers, normal-appearing filaments, or both. Staining of normal, control brain was first tested as a negative control. Control brain was readily stained by a standard, GFAP polyclonal antibody (fig. 8B), but no signal was produced with the R416W antibody (fig. 8C), confirming the immunoblot results that the R416W antibody does not crossreact with normal human brain GFAP. On the other hand, the R416W antibody strongly stained brain from a patient with Alexander disease who harbored an R416W mutation, with use of either peroxidase (fig. 8D) or fluorescent (fig. 8F) detection methods. This staining was apparent both along the periphery of the Rosenthal fibers and in normal-appearing filaments (fig. 8F) and largely colocalized with staining of total GFAP (fig. 8E). These data confirm the specificity of the R416W antibodies produced. Importantly, they also demonstrate, for the first time, that the mutant protein is stably expressed in patients with the R416W mutation and that it is incorporated into both filaments and Rosenthal fibers.
Assembly Properties of Mutant GFAP Expressed in Human Cells Containing Endogenous GFAP Filament Networks
These R416W GFAP–specific monoclonal antibodies also permitted us to follow the fate of R416W GFAP when transiently expressed in human astrocytoma U343MG cells. This cell system would be expected to better mimic the scenario of R416W GFAP being expressed in a human astrocyte cell background and removes potential species conflicts. In this model system, R416W GFAP induced the formation of GFAP-rich aggregates (fig. 9A and 9B; arrows). These data show that R416W GFAP is capable of disrupting the endogenous networks of wild-type GFAP filaments within the context of a human astrocytoma cell line and once again demonstrate the dominant negative potential of the mutant R416W GFAP on the endogenous IFs.
Figure 9. .
The similarity between Rosenthal fibers and GFAP aggregates formed in transfected human astrocytoma cells. U343MG cells were transiently transfected with R416W GFAP and were routinely stained with rabbit polyclonal antibodies (3270) to GFAP (A, C, E, and G) and then were double stained with mouse monoclonal antibodies specific to R416W GFAP (B), αB-crystallin (αB-cry) (D), or HSP27 (F). Notice that the GFAP containing aggregates are also positive for both αB-crystallin and HSP27 (arrows in C–F). To demonstrate the presence of ubiquitin in the GFAP aggregates, cells were cotransfected with His6-myc ubiquitin as well as R416W GFAP and then were stained with rabbit polyclonal antibodies to GFAP (G) and the mouse monoclonal antibodies that recognize the myc epitope (H), showing that the GFAP aggregates contain ubiquitin. Bars = 10 μm. I, Wild-type and R416W GFAP (R416W) were transiently expressed in the human astrocyte cell line U343MG, and supernatant (S) and pellet (P) fractions were prepared from these culture and were compared with mock transfected cells. Cell fractionation used HEB, which almost completely solubilized wild-type GFAP. R416W GFAP, on the other hand, remained in the pellet fraction. Immunoblots of the cell fractions were probed with antibodies to GFAP, αB-crystallin, HSP27, HSP70, and finally actin, which was used as a loading control. Notice that, when cells were transfected with R416W GFAP, a significant proportion of the HSP27 and αB-crystallin but not HSP70 remained in the pellet fraction along with the R416W GFAP. Both the sHSPs and R416W GFAP were more resistant to extraction compared with these proteins in the wild-type GFAP transfected cells.
Similarities between the R416W-Induced Aggregates in Transfected Cells and Rosenthal Fibers
In previous studies of Alexander disease pathology, several other proteins were found to associate with Rosenthal fibers, including the small heat shock proteins (sHSPs) αB-crystallin6 and HSP27.40 We investigated whether these proteins would associate with the aggregates of R416W GFAP formed in the U343MG astrocytoma cells (fig. 9C–9F). Both αB-crystallin (fig. 9D; arrows) and HSP27 (fig. 9F; arrows) colocalized with the GFAP-containing aggregates in these cells (fig. 9C and 9E, respectively; arrows). Rosenthal fibers are also ubiquitinated.6 That the GFAP aggregates in transfected cells are also ubiquitinated was demonstrated by cotransfection of His6-myc tagged ubiquitin26 along with R416W GFAP into the human astrocytoma cells (fig. 9G and 9H; arrows). Thus, the aggregates formed by R416W GFAP in the transiently transfected human astrocytoma cells have many features reported elsewhere and expected for Rosenthal fibers.41
To obtain biochemical evidence of the similarities between the Rosenthal fibers in patients and the R416W GFAP aggregates formed in the transfected U343MG cells, we extracted the cells and monitored the solubility of the GFAP and the associated protein chaperones (fig. 9I). With use of an extraction buffer containing deoxycholate, wild-type GFAP was almost completely extracted from both the untransfected (fig. 9I; Mock) and wild-type GFAP transfected cells (fig. 9I; WT), conditions that also extracted αB-crystallin, HSP27, and HSP70 (fig. 9I; Mock and WT). In contrast, R416W GFAP remained entirely in the pellet fraction of the extracted cells that had been transiently transfected with R416W GFAP (fig. 9I; R416W). Interestingly, when the immunoblots of the supernatant and pellet fractions were also probed with antibodies to αB-crystallin (fig. 9I; αB-cry), HSP27 (fig. 9I; HSP27), and HSP70 (fig. 9I; HSP70), a significant proportion of both αB-crystallin and HSP27—but not HSP70—remained in the pellet fractions of R416W GFAP–transfected cells (fig. 9I; R416W). These data show that a proportion of both sHSPs is associated with the insoluble R416W GFAP. Since HSP7027 was completely extracted from the R416W GFAP–transfected cells under these conditions (fig. 9I; HSP70), the association of the sHSPs with R416W GFAP is specific and not a general property for all protein chaperones.
Discussion
The R416W Mutation in GFAP: A Dominant Mutation That Affects Interfilament Interactions
In this study, we investigated the properties of the common Alexander disease–causing R416W GFAP mutation to obtain insights into the initial stages of the disease process. We demonstrated that this R416W GFAP mutation dramatically alters assembly, both in vitro and in transfected cells (figs. 1, 2, 4, 7, and 9), and does so in a dominant manner. The mutant protein can also disturb endogenous IF networks in cultured cells, including those of vimentin (fig. 3D), keratin (fig. 4), and GFAP (figs. 7 and 9A). The effects of R416W GFAP are different in several respects from those of R239C, the only other Alexander disease–causing mutation that has been studied in detail.21 When assembled in vitro, the R239C mutant formed IFs that appeared indistinguishable from wild-type filaments,21 whereas the R416W GFAP formed short filament intermediates that associated laterally (fig. 1). Despite this difference, we show here that both the R239C and the R416W GFAP form aggregates when expressed in either the vimentin-negative SW13/cl.2 cell line or the primary mouse astrocytes lacking both vimentin and GFAP (fig. 2), confirming some of the results in a previous study.21 When R239C GFAP was transiently expressed in primary rat astrocytes, however, its assembly properties were not radically different from those of transiently expressed wild-type GFAP.21 In contrast, we found that the R416W mutant invariably formed aggregates when transiently overexpressed in several different cell lines, including primary mouse astrocytes (figs. 2 and 7) and human astrocytoma cells (fig. 9B). Despite these differences, both mutations were found to increase the resistance of GFAP to extraction (figs. 5 and 9I).21 Taken together, these data indicate that, whereas the immediate structural consequences of different GFAP mutations are likely to differ, as has been found for other intermediate filament proteins,10,42,43 there will likely be common consequences that lead to Alexander disease.
The RDG-Containing Motif and Its Role in Filament Assembly
The R416W mutation lies within the RDG motif that is conserved within the tail domain of nearly all of the type III IF proteins. Previous studies of GFAP had concluded that the RDG motif was important to filament assembly, but only in the context of the rest of the C-terminal domain.44 Our assembly data, presented here, show that changing the precise sequence of this motif to WDG does have dramatic effects on GFAP assembly (figs. 1 and 2). Change of the equivalent arginine (R449) to proline in vimentin also severely disrupted the in vitro assembly and network formation in transiently transfected cells.45 These combined data suggest that this arginine residue in the RDG motif within the C-terminal domain is very important for the assembly IF proteins with the motif.
Previous studies proposed that the C-terminal domain associates intramolecularly with the C-terminal end of the rod domain.46 Crystallization of the helix IIB rod domain suggests that the two α helices fold away from the dimer axis,17 which is consistent with the hypothesis that the C-terminal domain could form a surface-exposed loop structure and prevent inappropriate subunit interactions in the self-assembly process.46 This offers an explanation of the altered width of vimentin filaments assembled from tail-truncated and deleted forms of vimentin47 and the changed in vitro assembly characteristics of the R416W GFAP we report here (fig. 1).
Our studies also suggest that the RDG motif will contribute to interfilament interactions as well as subunit organization within the IF. We have demonstrated the increased tendency of R416W GFAP to aggregate in vitro (fig. 1) as well as in transfected cells (figs. 2, 4, 7, and 9). Studies of keratins also show that the C-terminal domain is very important in promoting filament-filament interactions.48–50 Together with our data presented here, a consensus is thus beginning to emerge that C-terminal sequences of cytoplasmic IFs help regulate both intra- and interfilament associations.
The Mutant R416W GFAP: A Component of Rosenthal Fibers
The presence of Rosenthal fibers has been documented in two cases of Alexander disease caused by the R416W GFAP mutation.6,9,51 For one of these cases, we show, for the first time for any Alexander disease case, that the mutant GFAP is present as the defining histopathological feature (fig. 8D and 8F). This is also the first time for any IF-based human disease that the link between the presence of the mutated protein and a characteristic histopathogical IF aggregate has been rigorously shown. In a case of epidermolysis bullosa simplex, a keratin-blistering disease, the loss of an epitope was used to confirm the genotype of patients,52 but this reagent was unable to demonstrate the presence of the mutant keratin 5 in the aggregates found in keratinocytes. Another study described the generation of a polyclonal antibody specific for a keratin 8 variant (R340H), which may predispose carriers to liver disease,53 but these antibodies were used only for immunblotting studies. Even in the case of Mallory bodies formed as a result of alcoholic hepatitis, where antibodies were developed that selectively stained the Mallory bodies and not the surrounding keratin filaments, the antibodies still detected wild-type keratins.54 Our antibody reagents are therefore the first to specifically identify a mutant missense IF protein in the presence of its wild-type counterpart. These antibodies allowed us to demonstrate that the mutant protein is a component of Rosenthal fibers in human patient brain tissue (fig. 8D and 8F) and also in GFAP-containing aggregates of transfected human astrocytoma cells (fig. 9B).
Association of αB-Crystallin and HSP27 with R416W GFAP Aggregates
We also discovered that the protein chaperones αB-crystallin and HSP27 specifically associate with the GFAP aggregates that are formed in transfected cells that express R416W GFAP. Both chaperones are components of Rosenthal fibers found in patients, including those with the R416W mutation.6,40 These chaperones normally minimize filament-filament associations,27 and their overexpression can dissolve accumulations of wild-type GFAP filaments.55 Their presence, however, in both brain astrocytes of patients with Alexander disease and in cultured cells does not prevent the changes in GFAP-filament solubility, filament aggregation, or the formation of Rosenthal fibers by R416W GFAP. The association of HSP27 and αB-crystallin with R416W GFAP and resistance to extraction is likely due to the altered filament properties induced by this mutation. Although other stresses have been shown to stimulate the association of both HSP27 and αB-crystallin with GFAP filaments,27,40 those studies involved wild-type proteins. Our data show that it is specifically the presence of the R416W GFAP, and not wild-type GFAP, that both stabilizes GFAP and leads to the sequestration of sHSPs (figs. 9C–9F and 9I).
The sequestration of HSP27 and αB-crystallin could potentially compromise the astrocyte stress response and therefore contribute to the initiation of Alexander disease. For example, it has already been shown that either the reduction of HSP27 or the loss of αB-crystallin compromises cytoskeletal integrity and function56 or induces muscular atrophy,57 respectively. HSP27 is a key protein in protecting cells, including neurons,58 against apoptosis by inhibiting caspase activity59 and preserving mitochondrial function.60 HSP27 has also been implicated in regulating the stress response through the ubiquitin-mediated proteosomal regulation of the key transcription factor, NFκB.61 Importantly, HSP27 protects cells against other protein aggregation–based diseases caused by huntingtin62 and α-synuclein.63 Such studies identify diminution of sHSP levels as a potential Achilles’ heel in the cellular response to protein aggregate–based diseases that require either up-regulation58 or overexpression63 to ameliorate the disease phenotype. These observations suggest that the early sequestration of sHSPs into GFAP aggregates (this study) and Rosenthal fibers6 is a key event at the onset of Alexander disease. Figure 10 incorporates these ideas into a diagram of the events that we believe to be important in the early stages of Alexander disease.
Figure 10. .
GFAP aggregation caused by the R416W mutation induces sHSP association and the association of ubiquitin as early events in the etiology of Alexander disease. The presence of the R416W GFAP mutation decreases the solubility of the GFAP filaments, probably by altering the filament-filament interactions in a manner that encourages aggregation. This is accompanied by the sequestration of the sHSP protein chaperones—αB-crystallin and HSP27 (shaded circles)—and GFAP into aggregates. Both proteins also localize to Rosenthal fibers, which also contain ubiquitin (Ub). The filament aggregates undergo a maturing process, with the additional posttranslational modification of integral components, such as the phosphorylation64 and ubiquitination8 of αB-crystallin to form the Rosenthal fibers. The model is not exclusive to R416W GFAP, since Rosenthal fibers are a characteristic diagnostic feature of Alexander disease.12 Other GFAP mutations differ in the details of the mechanism by which they produce aberrant filament-filament interactions leading to the formation of stabilized aggregates, but, once formed, they then follow a common pathway to Rosenthal fiber formation. Increased GFAP filament stability and the specific association of sHSPs are predicted to be the earliest events in the development of Alexander disease.
Acknowledgments
We thank Dr. R. M. Evans (University of Colorado Health Sciences Center, Denver), for generously providing SW13/cl.1 and SW13/cl.2 cells; Dr. M. Pekny and Lizhen Li (The Arvid Carlsson Institute, Institute of Clinical Neuroscience, Sahlgrenska Academy, Göteborg University, Sweden), for mouse primary astrocytes; Ron Kopito (School of Medicine, Stanford, Stanford), for the His6-myc-ubiquitin expression construct; Dr. J. T. Rutka (Division of Neurosurgery, University of Toronto, Toronto), for U343 cells; Dr. M. A. Accavitti-Loper and the UAB Epitope Recognition Core, for production of the monoclonal antibodies; and Drs. Jim Goldman and Guomei Tang (Department of Pathology, Columbia University, New York), for the control and Alexander disease brain tissues. We also thank John James (CHIPs, School of Life Sciences, University of Dundee, Dundee, United Kingdom) for excellent technical support. This work was supported by National Institute of Neurological Disorders and Stroke grant P01NS42803.
Web Resources
The accession number and URLs for data presented herein are as follows:
- Alexander Disease Web site, http://www.waisman.wisc.edu/alexander/home.htmlx
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for GFAP [accession number J04569])
- Intermediate Filament Disease Mutation Database, http://www.interfil.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Alexander disease)
References
- 1.Alexander WS (1949) Progressive fibrinoid degeneration of fibrillary astrocytes associated with mental retardation in a hydrocephalic infant. Brain 72:373–381 [DOI] [PubMed] [Google Scholar]
- 2.Russo LS Jr, Aron A, Anderson PJ (1976) Alexander’s disease: a report and reappraisal. Neurology 26:607–614 [DOI] [PubMed] [Google Scholar]
- 3.Neal JW, Cave EM, Singhrao SK, Cole G, Wallace SJ (1992) Alexander’s disease in infancy and childhood: a report of two cases. Acta Neuropathol (Berl) 84:322–327 [DOI] [PubMed] [Google Scholar]
- 4.Deprez M, D’Hooghe M, Misson JP, de Leval L, Ceuterick C, Reznik M, Martin JJ, D’Hooge M (1999) Infantile and juvenile presentations of Alexander’s disease: a report of two cases. Acta Neurol Scand 99:158–165 [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez D, Gauthier F, Bertini E, Bugiani M, Brenner M, N’Guyen S, Goizet C, Gelot A, Surtees R, Pedespan J-M, Hernandorena X, Troncoso M, Uziel G, Messing A, Ponsot G, Pham-Dinh D, Dautigny A, Boespflug-Tanguy O (2001) Infantile Alexander disease: spectrum of GFAP mutations and genotype-phenotype correlation. Am J Hum Genet 69:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomokane N, Iwaki T, Tateishi J, Iwaki A, Goldman JE (1991) Rosenthal fibers share epitopes with alpha B-crystallin, glial fibrillary acidic protein, and ubiquitin, but not with vimentin: immunoelectron microscopy with colloidal gold. Am J Pathol 138:875–885 [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AB, Bettica A (1989) On-grid immunogold labeling of glial intermediate filaments in epoxy-embedded tissue. Am J Anat 185:335–341 10.1002/aja.1001850228 [DOI] [PubMed] [Google Scholar]
- 8.Head MW, Corbin E, Goldman JE (1993) Overexpression and abnormal modification of the stress proteins alpha B-crystallin and HSP27 in Alexander disease. Am J Pathol 143:1743–1753 [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner M, Johnson AB, Boespflug-Tanguy O, Rodriguez D, Goldman JE, Messing A (2001) Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat Genet 27:117–120 10.1038/83679 [DOI] [PubMed] [Google Scholar]
- 10.Omary MB, Coulombe PA, McLean WH (2004) Intermediate filament proteins and their associated diseases. N Engl J Med 351:2087–2100 10.1056/NEJMra040319 [DOI] [PubMed] [Google Scholar]
- 11.McLean WH, Smith FJ, Cassidy AJ (2005) Insights into genotype-phenotype correlation in pachyonychia congenita from the human intermediate filament mutation database. J Investig Dermatol Symp Proc 10:31–36 10.1111/j.1087-0024.2005.10205.x [DOI] [PubMed] [Google Scholar]
- 12.Li R, Johnson AB, Salomons G, Goldman JE, Naidu S, Quinlan R, Cree B, Ruyle SZ, Banwell B, D’Hooghe M, Siebert JR, Rolf CM, Cox H, Reddy A, Gutierrez-Solana LG, Collins A, Weller RO, Messing A, van der Knaap MS, Brenner M (2005) Glial fibrillary acidic protein mutations in infantile, juvenile, and adult forms of Alexander disease. Ann Neurol 57:310–326 10.1002/ana.20406 [DOI] [PubMed] [Google Scholar]
- 13.Li R, Johnson AB, Salomons GS, van der Knaap MS, Rodriguez D, Boespflug-Tanguy O, Gorospe JR, Goldman JE, Messing A, Brenner M (2006) Propensity for paternal inheritance of de novo mutations in Alexander disease. Hum Genet 119:137–144 10.1007/s00439-005-0116-7 [DOI] [PubMed] [Google Scholar]
- 14.Herrmann H, Aebi U (2000) Intermediate filaments and their associates: multi-talented structural elements specifying cytoarchitecture and cytodynamics. Curr Opin Cell Biol 12:79–90 10.1016/S0955-0674(99)00060-5 [DOI] [PubMed] [Google Scholar]
- 15.Herrmann H, Aebi U (2004) Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem 73:749–789 10.1146/annurev.biochem.73.011303.073823 [DOI] [PubMed] [Google Scholar]
- 16.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P (2002) Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J 21:1255–1266 10.1093/emboj/21.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strelkov SV, Herrmann H, Aebi U (2003) Molecular architecture of intermediate filaments. Bioessays 25:243–251 10.1002/bies.10246 [DOI] [PubMed] [Google Scholar]
- 18.Wu KC, Bryan JT, Morasso MI, Jang SI, Lee JH, Yang JM, Marekov LN, Parry DA, Steinert PM (2000) Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol Biol Cell 11:3539–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada S, Wirtz D, Coulombe PA (2002) Pairwise assembly determines the intrinsic potential for self-organization and mechanical properties of keratin filaments. Mol Biol Cell 13:382–391 10.1091/mbc.01-10-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernot KM, Lee CH, Coulombe PA (2005) A small surface hydrophobic stripe in the coiled-coil domain of type I keratins mediates tetramer stability. J Cell Biol 168:965–974 10.1083/jcb.200408116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao VC, Tian R, Long H, Der Perng M, Brenner M, Quinlan RA, Goldman JE (2005) Alexander-disease mutation of GFAP causes filament disorganization and decreased solubility of GFAP. J Cell Sci 118:2057–2065 10.1242/jcs.02339 [DOI] [PubMed] [Google Scholar]
- 22.Ralton JE, Lu X, Hutcheson AM, Quinlan RA (1994) Identification of two N-terminal non-alpha-helical domain motifs important in the assembly of glial fibrillary acidic protein. J Cell Sci 107:1935–1948 [DOI] [PubMed] [Google Scholar]
- 23.Nicholl ID, Quinlan RA (1994) Chaperone activity of alpha-crystallins modulates intermediate filament assembly. EMBO J 13:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 25.Ding M, Eliasson C, Betsholtz C, Hamberger A, Pekny M (1998) Altered taurine release following hypotonic stress in astrocytes from mice deficient for GFAP and vimentin. Brain Res Mol Brain Res 62:77–81 10.1016/S0169-328X(98)00240-X [DOI] [PubMed] [Google Scholar]
- 26.Ward CL, Omura S, Kopito RR (1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121–127 10.1016/0092-8674(95)90240-6 [DOI] [PubMed] [Google Scholar]
- 27.Perng MD, Cairns L, van den IJsse P, Prescott A, Hutcheson AM, Quinlan RA (1999) Intermediate filament interactions can be altered by HSP27 and alphaB-crystallin. J Cell Sci 112:2099–2112 [DOI] [PubMed] [Google Scholar]
- 28.Sandilands A, Prescott AR, Carter JM, Hutcheson AM, Quinlan RA, Richards J, FitzGerald PG (1995) Vimentin and CP49/filensin form distinct networks in the lens which are independently modulated during lens fibre cell differentiation. J Cell Sci 108:1397–1406 [DOI] [PubMed] [Google Scholar]
- 29.Evan GI, Lewis GK, Ramsay G, Bishop JM (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5:3610–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwaki T, Wisniewski T, Iwaki A, Corbin E, Tomokane N, Tateishi J, Goldman JE (1992) Accumulation of alpha B-crystallin in central nervous system glia and neurons in pathologic conditions. Am J Pathol 140:345–356 [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard TD, Cooper JA (1982) Methods to characterize actin filament networks. Methods Enzymol 85:211–233 [DOI] [PubMed] [Google Scholar]
- 33.Hedberg KK, Chen LB (1986) Absence of intermediate filaments in a human adrenal cortex carcinoma-derived cell line. Exp Cell Res 163:509–517 10.1016/0014-4827(86)90081-9 [DOI] [PubMed] [Google Scholar]
- 34.Chen WJ, Liem RK (1994) The endless story of the glial fibrillary acidic protein. J Cell Sci 107:2299–2311 [DOI] [PubMed] [Google Scholar]
- 35.Schweitzer SC, Klymkowsky MW, Bellin RM, Robson RM, Capetanaki Y, Evans RM (2001) Paranemin and the organization of desmin filament networks. J Cell Sci 114:1079–1089 [DOI] [PubMed] [Google Scholar]
- 36.Eliasson C, Sahlgren C, Berthold CH, Stakeberg J, Celis JE, Betsholtz C, Eriksson JE, Pekny M (1999) Intermediate filament protein partnership in astrocytes. J Biol Chem 274:23996–24006 10.1074/jbc.274.34.23996 [DOI] [PubMed] [Google Scholar]
- 37.Steinert PM, Chou YH, Prahlad V, Parry DA, Marekov LN, Wu KC, Jang SI, Goldman RD (1999) A high molecular weight intermediate filament-associated protein in BHK-21 cells is nestin, a type VI intermediate filament protein: limited co-assembly in vitro to form heteropolymers with type III vimentin and type IV alpha-internexin. J Biol Chem 274:9881–9890 10.1074/jbc.274.14.9881 [DOI] [PubMed] [Google Scholar]
- 38.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R (1982) The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 31:11–24 10.1016/0092-8674(82)90400-7 [DOI] [PubMed] [Google Scholar]
- 39.Brenner M, Lampel K, Nakatani Y, Mill J, Banner C, Mearow K, Dohadwala M, Lipsky R, Freese E (1990) Characterization of human cDNA and genomic clones for glial fibrillary acidic protein. Brain Res Mol Brain Res 7:277–286 10.1016/0169-328X(90)90078-R [DOI] [PubMed] [Google Scholar]
- 40.Iwaki T, Iwaki A, Tateishi J, Sakaki Y, Goldman JE (1993) Alpha-b-crystallin and 27-kd heat-shock protein are regulated by stress conditions in the central-nervous-system and accumulate in rosenthal fibers. Am J Path 143:487–495 [PMC free article] [PubMed] [Google Scholar]
- 41.Iwaki A, Iwaki T, Goldman JE, Ogomori K, Tateishi J, Sakaki Y (1992) Accumulation of alpha B-crystallin in brains of patients with Alexander’s disease is not due to an abnormality of the 5′-flanking and coding sequence of the genomic DNA. Neurosci Lett 140:89–92 10.1016/0304-3940(92)90689-5 [DOI] [PubMed] [Google Scholar]
- 42.Herrmann H, Hesse M, Reichenzeller M, Aebi U, Magin TM (2003) Functional complexity of intermediate filament cytoskeletons: from structure to assembly to gene ablation. Int Rev Cytol 223:83–175 [DOI] [PubMed] [Google Scholar]
- 43.Bar H, Mucke N, Kostareva A, Sjoberg G, Aebi U, Herrmann H (2005) Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc Natl Acad Sci USA 102:15099–15104 10.1073/pnas.0504568102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen WJ, Liem RK (1994) Reexpression of glial fibrillary acidic protein rescues the ability of astrocytoma cells to form processes in response to neurons. J Cell Biol 127:813–823 10.1083/jcb.127.3.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCormick MB, Kouklis P, Syder A, Fuchs E (1993) The roles of the rod end and the tail in vimentin IF assembly and IF network formation. J Cell Biol 122:395–407 10.1083/jcb.122.2.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouklis PD, Papamarcaki T, Merdes A, Georgatos SD (1991) A potential role for the COOH-terminal domain in the lateral packing of type III intermediate filaments. J Cell Biol 114:773–786 10.1083/jcb.114.4.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U (1996) Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol 264:933–953 10.1006/jmbi.1996.0688 [DOI] [PubMed] [Google Scholar]
- 48.Bousquet O, Ma L, Yamada S, Gu C, Idei T, Takahashi K, Wirtz D, Coulombe PA (2001) The nonhelical tail domain of keratin 14 promotes filament bundling and enhances the mechanical properties of keratin intermediate filaments in vitro. J Cell Biol 155:747–754 10.1083/jcb.200104063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu LH, Coulombe PA (2005) Defining the properties of the nonhelical tail domain in type II keratin 5: insight from a bullous disease-causing mutation. Mol Biol Cell 16:1427–1438 10.1091/mbc.E04-06-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada S, Wirtz D, Coulombe PA (2003) The mechanical properties of simple epithelial keratins 8 and 18: discriminating between interfacial and bulk elasticities. J Struct Biol 143:45–55 10.1016/S1047-8477(03)00101-1 [DOI] [PubMed] [Google Scholar]
- 51.Reichard EA, Ball WS Jr, Bove KE (1996) Alexander disease: a case report and review of the literature. Pediatr Pathol Lab Med 16:327–343 10.1080/107710496175804 [DOI] [PubMed] [Google Scholar]
- 52.Lane EB, Rugg EL, Navsaria H, Leigh IM, Heagerty AH, Ishida-Yamamoto A, Eady RA (1992) A mutation in the conserved helix termination peptide of keratin 5 in hereditary skin blistering. Nature 356:244–246 10.1038/356244a0 [DOI] [PubMed] [Google Scholar]
- 53.Ku NO, Lim JK, Krams SM, Esquivel CO, Keeffe EB, Wright TL, Parry DA, Omary MB (2005) Keratins as susceptibility genes for end-stage liver disease. Gastroenterology 129:885–893 10.1053/j.gastro.2005.06.065 [DOI] [PubMed] [Google Scholar]
- 54.Hazan R, Denk H, Franke WW, Lackinger E, Schiller DL (1986) Change of cytokeratin organization during development of Mallory bodies as revealed by a monoclonal antibody. Lab Invest 54:543–553 [PubMed] [Google Scholar]
- 55.Koyama Y, Goldman JE (1999) Formation of GFAP cytoplasmic inclusions in astrocytes and their disaggregation by alphaB-crystallin. Am J Pathol 154:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mairesse N, Horman S, Mosselmans R, Galand P (1996) Antisense inhibition of the 27 kDa heat shock protein production affects growth rate and cytoskeletal organization in MCF-7 cells. Cell Biol Int 20:205–212 10.1006/cbir.1996.0025 [DOI] [PubMed] [Google Scholar]
- 57.Brady JP, Garland DL, Green DE, Tamm ER, Giblin FJ, Wawrousek EF (2001) AlphaB-crystallin in lens development and muscle integrity: a gene knockout approach. Invest Ophthalmol Vis Sci 42:2924–2934 [PubMed] [Google Scholar]
- 58.Benn SC, Perrelet D, Kato AC, Scholz J, Decosterd I, Mannion RJ, Bakowska JC, Woolf CJ (2002) Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron 36:45–56 10.1016/S0896-6273(02)00941-8 [DOI] [PubMed] [Google Scholar]
- 59.Kamradt MC, Chen F, Sam S, Cryns VL (2002) The small heat shock protein alpha B-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J Biol Chem 277:38731–38736 10.1074/jbc.M201770200 [DOI] [PubMed] [Google Scholar]
- 60.Paul C, Manero F, Gonin S, Kretz-Remy C, Virot S, Arrigo AP (2002) Hsp27 as a negative regulator of cytochrome C release. Mol Cell Biol 22:816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parcellier A, Schmitt E, Gurbuxani S, Seigneurin-Berny D, Pance A, Chantome A, Plenchette S, Khochbin S, Solary E, Garrido C (2003) HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol Cell Biol 23:5790–5802 10.1128/MCB.23.16.5790-5802.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo AP, Rubinsztein DC (2002) Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet 11:1137–1151 10.1093/hmg/11.9.1137 [DOI] [PubMed] [Google Scholar]
- 63.Zourlidou A, Payne Smith MD, Latchman DS (2004) HSP27 but not HSP70 has a potent protective effect against alpha-synuclein-induced cell death in mammalian neuronal cells. J Neurochem 88:1439–1448 [DOI] [PubMed] [Google Scholar]
- 64.Mann E, McDermott MJ, Goldman J, Chiesa R, Spector A (1991) Phosphorylation of alpha-crystallin B in Alexander’s disease brain. FEBS Lett 294:133–136 10.1016/0014-5793(91)81359-G [DOI] [PubMed] [Google Scholar]