Abstract
γ-Glutamyl hydrolase (GGH) catalyzes degradation of the active polyglutamates of natural folates and the antifolate methotrexate (MTX). We found that GGH activity is directly related to GGH messenger RNA expression in acute lymphoblastic leukemia (ALL) cells of patients with a wild-type germline GGH genotype. We identified two CpG islands (CpG1 and CpG2) in the region extending from the GGH promoter through the first exon and into intron 1 and showed that methylation of both CpG islands in the GGH promoter (seen in leukemia cells from ∼15% of patients with nonhyperdiploid B-lineage ALL) is associated with significantly reduced GGH mRNA expression and catalytic activity and with significantly higher accumulation of MTX polyglutamates (MTXPG4–7) in ALL cells. Furthermore, methylation of CpG1 was leukemia-cell specific and had a pronounced effect on GGH expression, whereas methylation of CpG2 was common in leukemia cells and normal leukocytes but did not significantly alter GGH expression. These findings indicate that GGH activity in human leukemia cells is regulated by epigenetic changes, in addition to previously recognized genetic polymorphisms and karyotypic abnormalities, which collectively determine interindividual differences in GGH activity and influence MTXPG accumulation in leukemia cells.
Intracellular folates are required as cofactors in DNA synthesis, repair, and methylation, and they are essential for normal cellular growth and replication.1,2 Antifolates, such as methotrexate (MTX), are competitive inhibitors of folate-dependent enzymes and are widely used in the treatment of many human cancers, including acute lymphoblastic leukemia (ALL). Folates and most antifolates exist intracellularly as more-active polyglutamated derivatives, which are retained longer in cells because they are generally not substrates of export transporters.1 Hydrolytic removal of γ-linked polyglutamates, including folylpoly-γ-glutamates and antifolylpoly-γ-glutamates, is catalyzed by the lysosomal peptidase γ-glutamyl hydrolase (GGH [MIM 601509]).3
MTX is a major component of the curative treatment of childhood ALL,4 and the extent of accumulation of its active polyglutamates (MTXPG) determines its cytotoxicity and influences treatment response in childhood ALL.5 GGH converts long-chain MTXPG into shorter-chain MTXPG and ultimately into MTX, which can be effluxed from cells.6,7 In children with nonhyperdiploid B-lineage ALL (BNHD-ALL), GGH activity in leukemia cells is inversely correlated with long-chain MTXPG accumulation.8 Hyperdiploid B-lineage ALL (BHD-ALL) cells with trisomy of chromosome 8 that contains a wild-type GGH allele have significantly higher GGH activity and accumulate less MTXPG than do those with disomy 8.9
GGH activity and MTXPG accumulation in leukemia cells are also affected by a substrate-specific functional genetic polymorphism in GGH, 452C→T (T127I, rs11545078).8 This SNP alters GGH’s surface conformation at the substrate-binding cleft and reduces its binding affinity to long-chain MTXPG. Among patients with the same ALL subtype (e.g., BNHD-ALL, T-lineage ALL, or BHD-ALL without an additional chromosome 8), 452C→T was found at high frequency among patients with low GGH activity, and it was not found in patients with high GGH activity.8 However, there remains substantial interindividual variability in GGH activity that is not explained by genetic polymorphism or karyotypic abnormalities.
Cancer has been linked to both genetic and epigenetic changes.10 Epigenetic modifications, mainly DNA methylation at CpG dinucleotides, affect the regulation of gene transcription without altering the gene’s sequence. Short sequences rich in CpG dinucleotides (i.e., CpG islands) are often found in the 5′ promoter region of genes and can extend into the first exon and sometimes into intron 1.10,11 Aberrant methylation of CpG islands is a common feature of cancer cells,12,13 and methylated CpG islands have been consistently detected in primary human acute leukemia samples, sometimes with consequent silencing of gene expression.13 In the current study, we examined epigenetic modifications of GGH in leukemia cells and normal leukocytes from pediatric patients with ALL and identified two CpG islands in the promoter region of GGH. CpG island 1 (CpG1) was methylated in leukemia cells but not in normal leukocytes, whereas CpG island 2 (CpG2) was methylated in both normal leukocytes and leukemia cells. Methylation of CpG1 alone in leukemia cells was associated with modest down-regulation of GGH expression, whereas methylation in both CpG1 and CpG2 was associated with significantly reduced GGH mRNA expression and GGH catalytic activity. Our findings establish that epigenetic mechanisms alter GGH activity and MTXPG accumulation in human leukemia cells via methylation of CpG islands in the human GGH promoter region.
Subjects and Methods
Patients and Pharmacologic Analyses
We studied 93 children (aged ⩽21 years) who had newly diagnosed ALL and were enrolled in St. Jude Total Therapy XV protocols. We also studied six children (aged ⩽21 years) who had newly diagnosed acute myeloid leukemia (AML), had no structural aberrations on chromosome 8, and were enrolled in the St. Jude AML-97 protocol. Of the 93 patients with ALL, 46 had BNHD-ALL and 47 had either BHD-ALL (n=31) or T-lineage ALL (n=16). We had sufficient ALL cells to determine both GGH catalytic activity and mRNA expression in 40 of 46 patients with BNHD-ALL. Of these 40 patients, 34 had a wild-type GGH genotype for the SNP at position 452 (452CC), and 6 had a variant GGH allele (452CT or 452TT genotype). Among the 31 patients with BHD-ALL, 12 had trisomy of chromosome 8 in their leukemia cells. The treatments and research protocols were approved by the Institutional Review Board of St. Jude Children’s Research Hospital, and informed consent was obtained from patients or their guardians (or both) before enrollment. Patient assent was also obtained when appropriate (from those aged ⩾14 years). The diagnoses of ALL and AML were based on morphologic and molecular analyses described elsewhere,14,15 and complete chromosomal analysis was performed on leukemia cells from each patient, according to the International System for Human Cytogenetic Nomenclature (ISCN 1995).16
Leukemia cells were isolated by applying a Ficoll-Hypaque gradient to bone marrow aspirates obtained at diagnosis (median 97% blast cells), and GGH activity and MTXPG concentration were measured as described elsewhere.8 Normal leukocytes were isolated from peripheral blood samples obtained after the successful completion of remission-induction therapy (on days 45–48 after the start of treatment).
Gene Expression Analysis and Germline Genotyping
Genomic DNA and RNA were extracted from leukemia cells and normal leukocytes by use of TriReagent (MRC). GGH germline genotype was determined in 611 children with ALL by use of leukocyte DNA as described elsewhere.8
By methods described elsewhere,17 total RNA was processed and hybridized to the HG-U133A oligonucleotide microarray (Affymetrix) according to the manufacturer’s protocol. We used the default settings of the Affymetrix Microarray Suite software, version 5 (MAS 5.0), to calculate scaled gene-expression values. GGH expression was assessed by evaluating the Affymetrix expression signal resulting from hybridization with probe set 203560_at. GGH expression in leukemia cells was also measured by RT-PCR (table 1) in a subset of patients.
Table 1. .
Primer Sequences
| Primer Type and Name | Sequence (5′→3′) |
| Primer for RT-PCR: | |
| GGHf | TTTTGAAAGGCGGCGGGAG |
| GGHr | TGAAGAATCAGAGCCAGGCAC |
| GAPDHf | CTCTCTGCTCCTCCTGTTCGACA |
| GAPDHr | TTGTGCTCTTGCTGGGGCTG |
| Primer for CpG islands amplification: | |
| CpG1f | TTGGTTTTTATAGATGGAAATTTTTATT |
| CpG1r | AAAACTAAACTAACCAACCCAAATC |
| CpG2f | TTTGGGTTGGTTAGTTTAGTTTTG |
| CpG2r | AAACCTATAAAAAATACAATCC |
| Primer for methylation-specific PCR: | |
| CpG1-Mf | GCGTGTTCGTTTTAAAGTAGAC |
| CpG1-Mr | GAAAATAATAACAACGTTCGCT |
| CpG1-Uf | GGGTGTGTTTGTTTTAAAGTAGAT |
| CpG1-Ur | TCCAAAAATAATAACAACATTCACT |
| CpG2-Mf | GTTTTTAGAGTTTTCGTTTAGGC |
| CpG2-Mr | TATAAAAAATACAATCCCGACG |
| CpG2-Uf | TTTTAGAGTTTTTGTTTAGGTGG |
| CpG2-Ur | TTTTAGAGTTTTTGTTTAGGTGG |
Promoter Methylation Analysis
MethPrimer18 was used to predict promoter CpG islands and to design primers for bisulfite sequencing and nested methylation-specific PCR (NMSP) (table 1). Sodium bisulfite modification was performed using the CpGenome DNA Modification Kit (Chemicon International) according to the manufacturer’s protocol, with minor modifications to optimize performance. In brief, genomic DNA (100 ng for bisulfite sequencing and 40 ng for NMSP) was denatured by incubation with NaOH (final concentration 0.2 M) for 20 min at 50°C. Sodium bisulfite solution (pH 5.0), freshly prepared, was added, and the mixture was incubated at 50°C for 14 h. The modified DNA was then treated with NaOH (final concentration 0.3 M) for 5 min at room temperature, was precipitated with ethanol, and then was resuspended in 25 μl TE buffer (10 mM Tris [pH 8] and 1 mM EDTA).
GGH promoter CpG1 and CpG2 were amplified using 3 μl bisulfite-modified genomic DNA and 1× AmpliTaq Gold master mix (Applied Biosystems) at an annealing temperature of 55°C, with 35 cycles for bisulfite sequencing and 20–25 cycles for NMSP. For bisulfite sequencing, amplified CpG1 and CpG2 were subjected to TA cloning (Invitrogen), and >10 clones were selected for cycle sequencing. For NMSP, 1-μl aliquots of amplified fragments were used in second-round PCR with 1× True Allele PCR mix (Applied Biosystems) for 2 min at 50°C and 10 min at 95°C followed by 25–40 cycles for 15 s at 95°C and 1 min at 60°C. Genomic DNA isolated from a healthy individual whose GGH promoter was unmethylated was used as a negative control, and genomic DNA treated with SssI methylase (at 37°C for 16 h and 65°C for 20 min) was used as a positive (methylated) control.
Promoter Constructs and Luciferase Assays
Wild-type fragments of the entire GGH promoter (−1002 to −23), CpG1 (−1002 to −419) and CpG2 in the promoter region (−418 to −23), were cloned from human Nalm6 ALL cells by use of the TOPO TA cloning kit (Invitrogen). Reconstructed GGH promoter (CpG1 and CpG2) was subcloned into the pGL3-Basic vector (Promega) from NheI to HindIII sites. CpG1, CpG2, or the entire GGH promoter was removed from the vector by digestion with the appropriate restriction enzyme (NheI plus BglII, BglII plus HindIII, or NheI plus HindIII) and was incubated with 1 U/μg SssI methylase (New England Biolabs) and 200 nM S-adenosylmethionine at 37°C for 16 h and then at 65°C for 20 min. Methylated CpG island fragments and the remaining portions of the constructs were religated after in vitro methylation. The ligation efficiency was monitored by gel electrophoresis, and the ligated products were extracted by gel purification (Qiagen). To confirm complete methylation, reconstructed GGH promoter was restriction digested with NheI and HindIII followed by BstUI digestion at 60°C for 1 h.
Nalm6 cells (2×106 cells/ml) were transfected with 2 μg of firefly luciferase promoter-reporter plasmid containing methylated CpG1 alone, methylated CpG2 alone, or methylated and unmethylated entire promoter fragment, with 0.1 μg of phRL-TK (Promega), by use of Lipofectamine 2000 (Invitrogen). Activities of firefly and Renilla luciferases were measured 48 h after transfection by use of the Dual-Glo luciferase assay system (Promega). A pGL3-Basic vector without inserts and subcloned complete GGH promoter were used as negative and positive controls, respectively. Luciferase activity was calculated as the ratio of firefly to Renilla luciferase activities. Relative luciferase activity in the experimental sample was expressed as a percentage of that in the positive control.
Statistical Analysis
The objective of this study called for comparisons of continuous phenotypes across two or more groups formed by methylation status or genotype. The exact Kruskal-Wallis test was used to compare the general differences in phenotype across more than two groups; the exact Wilcoxon-Mann-Whitney test was used to compare two genotype groups. Multivariate analysis of variance (MANOVA) model was used to compare the effect of methylation on expression and activity jointly. All computations were performed using SAS (The SAS Institute).
Results
GGH Activity in ALL Cells
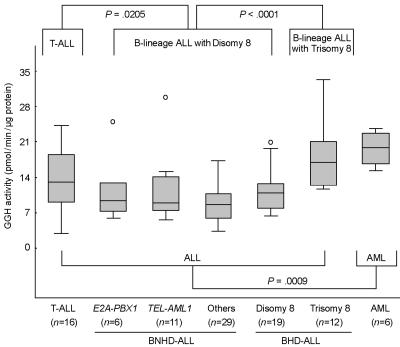
We measured GGH activity in ALL cells isolated from diagnostic bone marrow aspirates of children with newly diagnosed ALL, and we found that GGH activity was significantly higher in T-lineage ALL cells (n=93; P=.0205) and in B-lineage ALL cells with trisomy 8 (n=12; P<.0001) than in B-lineage ALL cells with disomy 8 (n=65) (fig. 1). In patients with B-lineage ALL, GGH activity was not significantly associated with karyotypic abnormalities other than trisomy 8, such as presence of the TEL-AML1 or E2A-PBX1 gene fusion, or other numerical aberrations (ploidy differences). There was no significant difference in GGH expression between T-lineage ALL (n=16) and B-lineage ALL cells with disomy 8 (n=65; P=.79), a finding consistent with our previous results,19 which indicates that other posttranslation factor(s) might affect GGH activity in T-lineage ALL cells. Primary AML cells had significantly higher GGH activity than ALL cells (P=.0021) (fig. 1), consistent with a previous report.20
Figure 1. .
Comparison of GGH activity in acute leukemia cells with different genetic and lineage subtypes. GGH activity was measured in leukemia cells isolated from diagnostic bone marrow aspirates from pediatric patients with ALL (n=93) or primary AML (n=6). Patients with ALL were grouped according to ALL lineage, DNA ploidy, number of chromosome 8, and whether the leukemia cells contain the E2A-PBX1 or TEL-AML1 gene fusion. Differences between pairs were assessed using the exact Wilcoxon-Mann-Whitney test. Boxes represent the 25%–75% quartiles, lines in the boxes represent the median level of GGH activity, whiskers represent the nonoutlier range, and circles represent the outliers.
Relationship between GGH Activity and mRNA Expression in Leukemia Cells
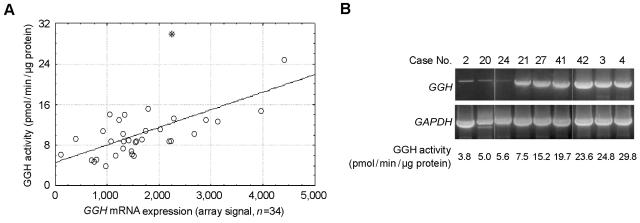
Because ALL lineage (T- vs. B-lineage), a GGH SNP (452C→T), and chromosome number (trisomy 8) influence GGH activity in ALL cells, we limited the current studies to patients with BNHD-ALL (⩽50 chromosomes), a wild-type germline GGH genotype (452CC), and two copies of chromosome 8 (this group represents ∼48% of B-lineage ALL cases9). In these patients (n=34), GGH activity in leukemia cells was highly variable (coefficient of variation 51.5%; activity range 3.8–29.8 pmol/min/μg protein), and this variability was significantly explained by the level of GGH mRNA expression (P<.0001; R2=0.49) (fig. 2).
Figure 2. .
Relationship between GGH expression and GGH activity. GGH activity was measured in leukemia cells obtained at diagnosis from 34 children with BNHD-ALL. These patients had wild-type GGH and disomy of chromosome 8. A, GGH expression, as detected by Affymetrix gene expression array, was significantly correlated with GGH activity. Excluding the outlier indicated by the asterisk (*) (n=33), we obtained P<.0001 and R2=0.49; including the outlier (n=34), we obtained P=.0002 and R2=0.36. B, Verification of GGH expression by RT-PCR with primers that amplified the entire GGH coding region.
To determine whether SNPs in the GGH promoter region and 3′ UTR affect GGH expression and activity, we sequenced the GGH promoter region and exon 9 in patients with BNHD-ALL who had high or low GGH activity (n=9). Excluding the 3′ UTR SNP 1102A→G,8 we found the GGH promoter SNPs −1187G→A (dbSNP accession number rs11988534), −679C→T (rs3758147), −649G→A (rs2736676), −605C→G (rs28365060), −424C→A (rs719236), −401C→T (rs3758149), −354G→T (rs719235), and −124T→C (rs11545076) (numbered relative to the A of the translation start codon at nt +1) in ALL cells with high GGH activity and in cases with low GGH activity. However, in 611 children with ALL, the 3′ UTR SNP 1102A→G was in high linkage disequilibrium with 452C→T (in whites [n=459], in African Americans [n=102], and in all other patients [n=50], P<.001 for each test, calculated using Haplotype procedure [SAS/Genetics]). Using primers that amplified the entire GGH coding region (table 1), we did not find any evidence of GGH splice variation in ALL cells (fig. 2B).
GGH Promoter CpG Islands and Their Effect on GGH mRNA Expression
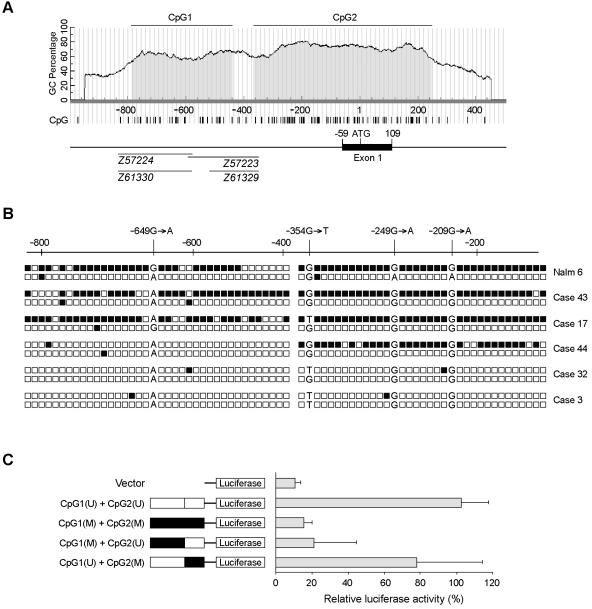
We identified two CpG islands within the 5-kb region around exon 1 of the human GGH gene, including the 2-kb 5′ upstream regulatory region. These two CpG islands, at nt −790 to −442 (CpG1) and −369 to 245 (CpG2) (fig. 3A), have a G+C content >60% and a CpG density per 100 bp >70%. CpG1 is confined to the GGH promoter region, whereas CpG2 extends from the GGH promoter region21 through the first exon and into intron 1. A search of the GenBank database revealed four “H.sapiens CpG island DNA genomic Mse1 fragments” (accession numbers Z57223, Z57224, Z61329, and Z61330) in the region of GGH CpG1 (fig. 3A).
Figure 3. .
CpG islands in the GGH promoter region. A, Two CpG islands in the GGH 5′ promoter region extend into intron 1. The CpG1 and CpG2 regions (shaded areas) are indicated relative to the A of the translation start codon, defined as nt +1. B, Sequence of the GGH promoter region (nt −905 to −118) of each allele in the Nalm6 cell line and primary leukemia cells from five cases of ALL, determined by bisulfite sequencing. GGH promoter methylation was confirmed by NMSP in all five cases (data not shown). Each box represents a CpG dinucleotide; methylated CpG sites are blackened; unmethylated CpG sites are unblackened. The polymorphisms at nt −649, −249, and −209 were used to define allelic methylation. C, Regulation of transcriptional activity of GGH promoter by selective in vitro methylation of CpG islands in the GGH promoter region. The left panel shows a schematic diagram of the individually methylated CpG islands within GGH promoter constructs linked to a luciferase reporter. Methylated CpG island fragments are blackened; unmethylated fragments are unblackened.
To determine the distribution of methylation in the GGH promoter CpG islands, we analyzed the GGH promoter by using bisulfite-modified genomic DNA isolated from bone marrow leukemia cells and from the human ALL cell line Nalm6. We observed hypermethylation of both CpG1 and CpG2 in GGH of Nalm6 cells (fig. 3B) but found that methylation was restricted to one GGH allele in these cells (the G allele for SNP −649G→A). Consistent with this finding is that Nalm6 cells have relatively low GGH activity (6.9 pmol/h/μg protein).7 We also observed methylation in CpG1 and CpG2 in leukemia cells from two patients (cases 17 and 43) whose GGH mRNA expression array signals (707 and 831, respectively) were in the lowest quartile among the patients studied. In these two cases, the GGH promoter was methylated in the A allele for SNP −649G→A. None of the three patients with high GGH expression levels in their leukemia cells (array expression signals were between 3,968 and 4,421) had methylation in CpG1, but we observed methylation in CpG2 alone in one patient (case 44).
To further assess the role of CpG island methylation in regulating GGH expression, we reconstructed GGH promoters with one or both CpG islands methylated, and we measured promoter activity by a promoter luciferase assay, as described in the “Subjects and Methods” section. Methylation of CpG1 alone reduced transcription by 88.4% (relative luciferase activity 21.4%±23.3%), whereas methylation of CpG2 alone reduced transcription by only 26.9% (relative luciferase activity 77.9%±36.2%) (fig. 3C). Promoter activity was essentially eliminated by methylation of the entire GGH promoter region (CpG1 and CpG2) (fig. 3C).
GGH Promoter Methylation and Its Regulation of GGH Expression in Leukemia Cells
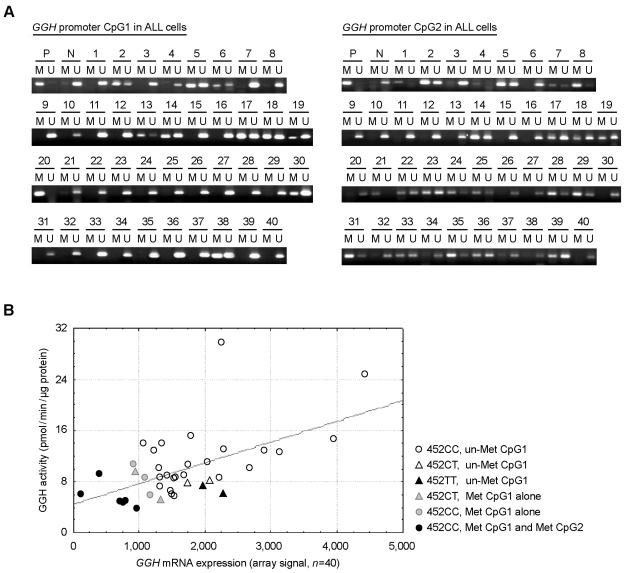
Using NMSP, we analyzed the GGH promoter in leukemia cells isolated from diagnostic bone marrow aspirates obtained from 40 patients with BNHD-ALL. GGH mRNA expression in these same leukemia cells was measured by Affymetrix microarray analysis. We detected methylated GGH CpG1 in leukemia cells from 11 patients (28%), including 6 (15%) with methylation in both CpG1 and CpG2 (fig. 4A). Methylated CpG2 alone was detected in leukemia cells from 16 patients (40%). Leukemia cells in which both CpG1 and CpG2 were methylated showed significantly lower GGH expression (2.1-fold reduction) and catalytic activity (2-fold reduction) (n=40; P=.003, by one-way MANOVA) when compared with leukemia cells having only one or no CpG island methylated (fig. 4B). There was no correlation between methylation of CpG2 alone and GGH expression or GGH activity (data not shown). Compared with leukemia cells without GGH promoter methylation or with methylation of CpG2 alone, those with methylated CpG1 alone expressed significantly less GGH (1.5-fold reduction) (n=34; P=.0003, by exact Wilcoxon-Mann-Whitney test), but the difference in GGH catalytic activity did not reach statistical significance (n=34; P=.16, by exact Wilcoxon-Mann-Whitney test).
Figure 4. .
GGH promoter methylation status in ALL cells. A, NMSP analysis of the GGH promoter in leukemia cells from 40 patients with BNHD-ALL. Two sets of primers were used to analyze the methylation status of GGH promoter CpG1 and CpG2. M = methylated PCR products; N = negative (unmethylated) control; P = positive (methylated) control; U = unmethylated PCR products. B, Hypermethylation of both CpG islands in the GGH promoter was associated with down-regulated GGH expression. GGH activity, RNA expression (Affymetrix array signal), and promoter methylation were measured in leukemia cells obtained at diagnosis from 40 children with BNHD-ALL. The germline genotype of GGH at nt 452 (C→T) was assessed by examining genomic DNA isolated from normal peripheral leukocytes from the same patients. Met = methylated; un-Met = unmethylated.
Leukemia Cell–Specific Methylation in GGH CpG1
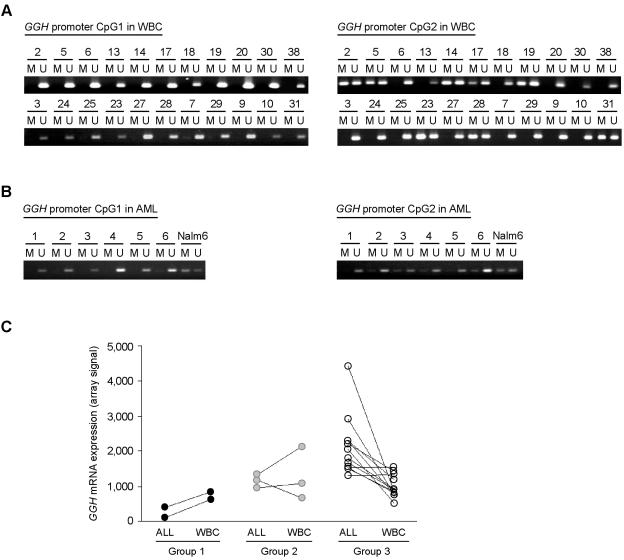
We analyzed GGH promoter methylation in paired normal peripheral blood leukocytes and leukemia cells obtained from 22 patients, including 11 whose leukemia cells contained methylated CpG1. None had methylated CpG1 in their normal leukocytes (fig. 5A). In contrast, CpG2 methylation was found in normal leukocytes and in leukemia cells (CpG2 methylation was found in normal leukocytes of 36.4% of patients and in leukemia cells of 55% of patients) (figs. 4A and 5A).
Figure 5. .
GGH promoter methylation status in normal leukocytes and AML cells. A, NMSP analysis of the GGH promoter in normal peripheral leukocytes (white blood cells [WBC]) isolated from 22 children with ALL. M = methylated CpG island; U = unmethylated CpG island. B, NMSP analysis of the GGH promoter in leukemia cells isolated from six children with primary AML. C, Comparison of GGH expression in leukemia cells (ALL) and normal leukocytes (WBC) from patients with ALL (n=16). Group 1 includes patients whose leukemia cells contained methylated CpG1 and CpG2; group 2, patients whose leukemia cells contained methylated CpG1 but not methylated CpG2; and group 3, patients in whose leukemia cells CpG1 was not methylated. CpG1 was not methylated in the normal leukocytes (WBC) of all patients.
We also analyzed GGH promoter methylation status in leukemia cells isolated from diagnostic bone marrow aspirates from six children with primary AML. GGH CpG2 was methylated in the AML cells of three patients, but none of the six patients had methylated CpG1 in their AML cells (fig. 5B).
Comparison of GGH Expression in Leukemia Cells and Normal Leukocytes from Patients with ALL
To determine the effect of GGH promoter methylation on GGH expression in normal leukocytes and leukemia cells, we measured GGH expression levels in normal leukocytes and leukemia cells from 17 children with BNHD-ALL by using Affymetrix expression arrays. In 10 of 12 patients whose leukemia cells did not contain methylated CpG1 (group 3, fig. 5C), GGH was more highly expressed (by a factor of 1.6–5.4) in leukemia cells than in the normal leukocytes; in the remaining 2 patients, GGH expression levels were similar in normal leukocytes and leukemia cells. In contrast, in patients in whose leukemia cells both CpG1 and CpG2 were methylated (group 1, fig. 5C), GGH was more highly expressed (by a factor of 2.1–5.3) in normal leukocytes than in leukemia cells. Among patients with only CpG1 methylated in their leukemia cells (group 2, fig. 5C), there was not a consistent difference in GGH expression between leukemia cells and normal leukocytes. GGH expression was not up-regulated in patients whose normal leukocytes lacked CpG2 methylation (cases 7, 24, 25, and 29), consistent with in vitro results showing that methylation in CpG2 alone does not affect GGH expression. These findings indicate that epigenetic modification of the entire GGH promoter occurs in leukemia cells but not in normal leukocytes and further demonstrate that methylation in both CpG islands is associated with the greatest reduction in GGH expression. (We calculated the ratio of GGH expression in ALL cells to that in normal leukocytes for each patient and used this ratio in a three-group comparison: n=17; P=.002, by exact Kruskal-Wallis test.)
Genetic Factors Altering GGH Activity in Leukemia Cells
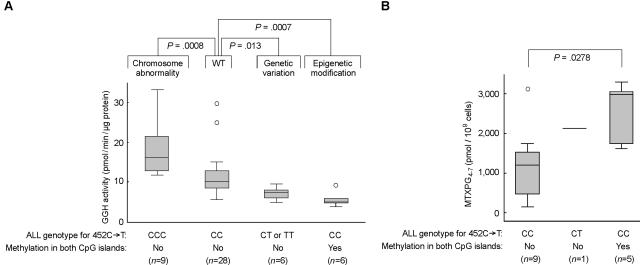
As depicted in figure 6, three distinct genetic factors were found to influence interindividual differences in GGH activity in leukemia cells: chromosomal abnormalities (trisomy 8), genetic polymorphism (SNP 452C→T), and epigenetic modification (promoter methylation). These factors result in significant differences in GGH activity in leukemia cells from different subgroups of patients exhibiting these genetic and epigenetic differences (n=49; P<.0001, by exact Kruskal-Wallis test).
Figure 6. .
GGH phenotype, GGH genotype, and MTXPG accumulation. A, Genetic and epigenetic factors affecting GGH activity in leukemia cells from patients with ALL. Overall comparison was performed using the exact Kruskal-Wallis test (n=49; P<.0001). Pairwise comparison was performed using the exact Wilcoxon-Mann-Whitney test. WT = wild type. B, Comparison of GGH activity and MTXPG accumulation in BNHD-ALL cells without the E2A-PBX1 or TEL-AML1 gene fusion. Boxes represent the 25%–75% quartiles, lines in the boxes represent the median level, whiskers represent the nonoutlier range, and circles represent the outliers.
Because MTXPG accumulation in ALL cells is significantly influenced by ALL lineage and DNA ploidy and by chromosomal translocations creating either the TEL-AML1 or the E2A-PBX1 gene fusion,19 we compared GGH activity and MTXPG accumulation in BNHD-ALL cells not carrying the TEL-AML1 or E2A-PBX1 gene fusion, a subtype that arises in ∼36% of cases of childhood ALL.22 In these cells, methylation of the two CpG islands in the GGH promoter was associated with significantly reduced GGH mRNA expression, reduced GGH catalytic activity, and increased MTXPG accumulation after in vivo treatment with 1 g/m2 intravenous MTX19 (fig. 6).
Discussion
Because MTXPG accumulation in leukemia cells is a determinant of the in vivo antileukemic effects of MTX,4,5,23 we and others have focused on elucidating mechanisms underlying the highly variable accumulation of MTXPG in leukemia cells.7,24,25 We have already documented significantly differential expression of the genes encoding reduced folate carrier (SLC19A1 [MIM 600424]); breast cancer resistance protein (ABCG2 [MIM 603756]); ATP-binding cassette, subfamily C, member 1 (ABCC1 [MIM 158343]); and folylpolyglutamate synthetase (FPGS [MIM 136510]) as a mechanism underlying differences in MTXPG accumulation among some subtypes of ALL.19 We have also shown that the catalytic activity of GGH in ALL cells exhibits substantial interindividual differences that are influenced by the number of copies of chromosome 8 containing a wild-type GGH allele9 and that GGH activity is inversely related to accumulation of long-chain MTXPG in BNHD-ALL cells.8 However, before now, we did not know what mechanism is responsible for differences in GGH mRNA expression and catalytic activity in ALL cells of patients with a diploid wild-type GGH genotype. Here, we report epigenetic mechanisms controlling GGH expression and catalytic activity in B-lineage ALL cells containing two wild-type GGH alleles. We found significant concordance between GGH activity and GGH mRNA expression in leukemia cells from these patients. Furthermore, we observed methylation spanning the entire GGH promoter region in leukemia cells of ∼15% of pediatric patients with ALL and its concordance with significantly reduced GGH mRNA expression and catalytic activity. Moreover, we showed that this was associated with higher intracellular MTXPG concentrations after in vivo treatment with MTX.
DNA methylation is an important mechanism regulating gene expression, and it is required for normal mammalian development.26–28 Alteration in DNA methylation is a hallmark of cancer10,29,30 and may be observed as a loss of global methylation or as a gain of methylation of selected CpG islands, either of which can result in alteration of the expression of hundreds of genes.12,13,31 We identified two CpG islands in the region extending from the GGH promoter through the first exon and into intron 1. To perform population screening for GGH promoter methylation status, we designed an NMSP that requires as little as 40 ng of genomic DNA from each patient sample. We confirmed GGH promoter methylation status in primary leukemia cells and a BNHD-ALL cell line by sequencing bisulfite-treated samples. We found that methylation of CpG1 has a much greater effect on GGH expression than does methylation of CpG2 and that methylation of both CpG islands has the most pronounced effect on GGH expression.
Allele-specific methylation has been observed in CpG islands of imprinted genes.27,32 In the present study, however, methylation of CpG1 was specific to ALL cells and occurred in the leukemia cells of 28% of children with ALL, but it was not found in normal leukocytes from the same patients. Methylation in CpG1 was also not detected in leukemia cells from children with primary AML, a form of leukemia that is less sensitive to MTX therapy. Furthermore, we identified four “H.sapiens CpG island DNA genomic Mse1 fragments” in GenBank that are homologous to GGH CpG1. These CpG island genomic DNA MseI fragments are GC-rich chromosomal fragments that are not methylated in primary, normal blood cells but are methylated after in vitro treatment with methylase.33 A library of CpG island genomic DNA MseI fragments has been used to identify hypermethylated CpG islands in cancer cells,34 but this has not been reported for GGH. Consistent with our data is that no evidence exists that GGH is an imprinted gene,27,32 and no imprinted genes have been reported in the locus around the human GGH coding region. Although GGH promoter methylation was restricted to the G allele for SNP −649G→A in Nalm6 cells, methylation of the A allele was observed in primary ALL cells from two of five patients (fig. 3B).
The GGH promoter contains multiple transcriptional start sites,21 all in the CpG2 region. However, we found that methylation in GGH CpG1 has a more pronounced effect on GGH expression (down-regulation) than does methylation in CpG2. Methylation in CpG2 was common in leukemia cells and normal leukocytes from patients with ALL, but it occurred at a somewhat higher frequency in the leukemia cells. However, there was no relationship between CpG2 methylation status and GGH expression in ALL cells, and an absence of CpG2 methylation in normal cells was not associated with higher GGH expression. This finding is consistent with earlier reports that a high level of methylation around a transcriptional start site does not always block transcription, especially for genes with multiple transcriptional start sites.35
Hypermethylation of CpG islands in cancer exhibits heterogeneity both within the same tumor types and among different tumor types.36,37 Methylated CpG islands are often, but not always, associated with transcriptional silencing.38,39 A recent study suggested that aberrant methylation of CpG islands in malignancy might be less frequent than expected, because only a small set of promoters was differentially methylated in normal and transformed human cells.39 Our study has revealed that aberrant methylation in cancer cells can occur in specific CpG islands (e.g., GGH CpG1) but not in other CpG islands (e.g., GGH CpG2) within the same gene promoter region. Furthermore, the present findings indicate that cancer-specific CpG-island promoter methylation can preferentially target CpG islands that have an effect on gene expression (i.e., CpG1 was methylated only in cancer cells, whereas CpG2 was methylated in cancer and normal cells). Therefore, a global assessment of methylation profile—for example, by use of restriction landmark genomic scanning40 or an oligonucleotide microarray-based methylation analysis41—would need to be performed in a manner that permits one to determine whether methylation in specific CpG islands (certain promoter regions) of a given gene is associated with altered gene transcription or cancer-cell phenotype (e.g., enzyme activity). An array based on a CpG island library created by isolation of genomic fragments that contain CpG dinucleotide regions that are poorly methylated in normal tissue has been used as a genomewide method of detecting cancer-specific CpG island methylation.33,42 Consistent with our findings, this CpG island library contains the CpG1 region but not the CpG2 region from the GGH promoter region.
Mechanisms by which CpG islands are hypermethylated in cancer have not been fully elucidated. CpG island methylation in cancer cells may be blocked by the binding of the transcription factor Sp1,43,44 and it has been suggested that a boundary sequence separating methylated and unmethylated promoter regions exists.45,46 Recent studies have indicated that DNA methylation in cancer cells can be induced by micro-RNAs.47,48 De novo methylation in cancer cells has been suggested to start from a methylation “hotspot” consisting of several nonadjacent CpG sites. These hotspots are methylated in both normal and cancer cells, and they serve as a starting point for CpG island hypermethylation in cancer cells.35 Because we found that methylation frequently occurs in GGH CpG2 in both leukemia cells and normal leukocytes and that both CpG1 and CpG2 are methylated on a single allele, it is conceivable that methylation in CpG2 serves as the seeding point from which de novo methylation of the entire GGH promoter occurs in leukemia cells. However, the finding that the leukemia cells of several patients had GGH promoter methylation of only CpG1 suggests that leukemia cell–specific methylation of CpG1 arises by another mechanism. Further study is needed to determine whether methylation of a particular allele is a selective process. Our findings that none of the GGH promoter SNPs examined was associated with altered GGH mRNA expression and that either GGH allele (G or A at nt −649) can be methylated in primary ALL cells indicate that GGH promoter SNPs are not the driving force for single-allele methylation.
Epigenetic silencing of genes that determine tumor invasiveness, growth patterns of tumors, and tumor-cell apoptosis12,29 may also affect the expression of drug-metabolizing enzymes, thereby providing an additional genetic mechanism in pharmacogenomics. Trisomy 8 that contains wild-type GGH could be used as a first parameter to define a subgroup of ALL with higher GGH activity than that of the ALL subgroup with disomy 8.9 Among B-lineage ALL cells containing disomy 8, epigenetic modification (methylation in both CpG1 and CpG2) had a more pronounced effect on GGH activity than did the 452C→T SNP (fig. 4B). ALL cells in which both CpG1 and CpG2 were methylated showed more than twofold lower GGH expression and GGH activity than did all the others. We have shown elsewhere that the GGH SNP 452C→T changes the protein surface conformation at the end of substrate-binding cleft and reduces binding affinity with long-chain MTXPG.8 This SNP significantly lowers but does not abolish catalytic activity and has a much less pronounced effect on GGH activity than does hypermethylation of the GGH promoter.
Because numerous DNA changes, such as chromosomal aberration and epigenetic modification, are often cancer-cell specific, germline genotyping does not always fully assess quantitative differences in cancer genomes or cancer-cell phenotypes. Unequivocal elucidation of cancer-cell pharmacogenomics may therefore require consideration of the special features of the cancer-cell genome.9,49 Indeed, we show that GGH activity in human leukemia cells is determined by epigenetic changes as well as germline genetic polymorphisms and karyotypic abnormalities in leukemia cells, which together account for a substantial proportion of interindividual differences in GGH activity and MTXPG disposition in human ALL cells (fig. 6). The current findings are important because they show that multiple genomic mechanisms can control the expression and function of a single gene in cancer cells and thereby determine the pharmacogenomics of anticancer agents.
Our results also indicate that genetic and epigenetic changes can have differing or additive effects on pharmacogenetics of an anticancer drug in cancer cells. In this study, we report substantial interindividual variability in GGH activity and MTXPG disposition in human ALL cells and that this variability is determined by multiple genetic and epigenetic mechanisms. It has been reported that undetectable GGH mRNA expression and GGH protein are associated with good prognosis of pulmonary neuroendocrine tumors50 and that high accumulation of MTXPG in ALL cells is associated with better treatment response.5,23 It will be important to extend these studies to larger cohorts of patients, in which the effects of these epigenetic changes on MTX activity and treatment outcome can be more fully elucidated. The current studies are an important foundation for future studies to define the utility of these genetic, karyotypic, and epigenetic factors for individualizing MTX therapy for ALL on the basis of pharmacogenomics.
Acknowledgments
We gratefully acknowledge the technical support of Paxton Baker, Natasha Lenchik, YaQin Chu, and Nancy Duran; the statistical computation by Wei Liu; the contributions of Nancy Kornegay and WenJian Yang in establishing our research databases; and Dr. Meyling Cheok for the Affymetrix gene expression analyses. We thank Dr. Janet R. Davies for the help in editing the manuscript. This work was supported in part by National Institutes of Health (NIH) grants CA36401 (R37), CA78224 (R01), CA51001 (R01), GM61393 (U01), and CA21765 (Cancer Center Support Grant); by the NIH Pharmacogenetics Research Network; by an FM Kirby Clinical Research Professorship from the American Cancer Society; and by the American Lebanese Syrian Associated Charities.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/ (for GGH promoter SNPs [accession numbers rs11988534, rs3758147, rs2736676, rs28365060, rs719236, rs3758149, rs719235, and rs11545076])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for GGH sequences [accession numbers NM_003878, NT_008183.1, and AF147081] and H.sapiens CpG island DNA genomic Mse1 fragments [accession numbers Z57223, Z57224, Z61329, and Z61330])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GGH, SLC19A1, ABCG2, ABCC1, and FPGS)
References
- 1.Zhao R, Goldman ID (2003) Resistance to antifolates. Oncogene 22:7431–7457 10.1038/sj.onc.1206946 [DOI] [PubMed] [Google Scholar]
- 2.Lightfoot TJ, Roman E (2004) Causes of childhood leukaemia and lymphoma. Toxicol Appl Pharmacol 199:104–117 10.1016/j.taap.2003.12.032 [DOI] [PubMed] [Google Scholar]
- 3.Galivan J, Ryan TJ, Chave K, Rhee M, Yao R, Yin D (2000) Glutamyl hydrolase: pharmacological role and enzymatic characterization. Pharmacol Ther 85:207–215 10.1016/S0163-7258(99)00063-7 [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Evans WE (2006) Treatment of acute lymphoblastic leukemia. N Engl J Med 354:166–178 10.1056/NEJMra052603 [DOI] [PubMed] [Google Scholar]
- 5.Masson E, Relling MV, Synold TW, Liu Q, Schuetz JD, Sandlund JT, Pui CH, Evans WE (1996) Accumulation of methotrexate polyglutamates in lymphoblasts is a determinant of antileukemic effects in vivo: a rationale for high-dose methotrexate. J Clin Invest 97:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galivan J, Ryan T, Rhee M, Yao R, Chave K (1999) Glutamyl hydrolase: properties and pharmacologic impact. Semin Oncol 26:33–37 [PubMed] [Google Scholar]
- 7.Panetta JC, Wall A, Pui CH, Relling MV, Evans WE (2002) Methotrexate intracellular disposition in acute lymphoblastic leukemia: a mathematical model of γ-glutamyl hydrolase activity. Clin Cancer Res 8:2423–2429 [PubMed] [Google Scholar]
- 8.Cheng Q, Wu B, Kager L, Panetta JC, Zheng J, Pui CH, Relling MV, Evans WE (2004) A substrate specific functional polymorphism of human γ-glutamyl hydrolase alters catalytic activity and methotrexate polyglutamate accumulation in acute lymphoblastic leukaemia cells. Pharmacogenetics 14:557–567 10.1097/01.fpc.0000114761.78957.7e [DOI] [PubMed] [Google Scholar]
- 9.Cheng Q, Yang W, Raimondi SC, Pui CH, Relling MV, Evans WE (2005) Karyotypic abnormalities create discordance of germline genotype and cancer cell phenotypes. Nat Genet 37:878–882 10.1038/ng1612 [DOI] [PubMed] [Google Scholar]
- 10.Plass C (2002) Cancer epigenomics. Hum Mol Genet 11:2479–2488 10.1093/hmg/11.20.2479 [DOI] [PubMed] [Google Scholar]
- 11.Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321:209–213 10.1038/321209a0 [DOI] [PubMed] [Google Scholar]
- 12.Issa JP (2004) CpG island methylator phenotype in cancer. Nat Rev Cancer 4:988–993 10.1038/nrc1507 [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428 [DOI] [PubMed] [Google Scholar]
- 14.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR (2002) Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1:133–143 10.1016/S1535-6108(02)00032-6 [DOI] [PubMed] [Google Scholar]
- 15.Crews KR, Gandhi V, Srivastava DK, Razzouk BI, Tong X, Behm FG, Plunkett W, Raimondi SC, Pui CH, Rubnitz JE, Stewart CF, Ribeiro RC (2002) Interim comparison of a continuous infusion versus a short daily infusion of cytarabine given in combination with cladribine for pediatric acute myeloid leukemia. J Clin Oncol 20:4217–4224 10.1200/JCO.2002.10.006 [DOI] [PubMed] [Google Scholar]
- 16.Mitelman F (ed) (1995) ISCN 1995: an international system for human cytogenetic nomenclature. Karger, Basel [Google Scholar]
- 17.Cheok MH, Yang W, Pui CH, Downing JR, Cheng C, Naeve CW, Relling MV, Evans WE (2003) Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet 34:85–90 10.1038/ng1151 [DOI] [PubMed] [Google Scholar]
- 18.Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- 19.Kager L, Cheok M, Yang W, Zaza G, Cheng Q, Panetta JC, Pui CH, Downing JR, Relling MV, Evans WE (2005) Folate pathway gene expression differs in subtypes of acute lymphoblastic leukemia and influences methotrexate pharmacodynamics. J Clin Invest 115:110–117 10.1172/JCI200522477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rots MG, Pieters R, Peters GJ, Noordhuis P, Van Zantwijk CH, Kaspers GJ, Hahlen K, Creutzig U, Veerman AJ, Jansen G (1999) Role of folylpolyglutamate synthetase and folylpolyglutamate hydrolase in methotrexate accumulation and polyglutamylation in childhood leukemia. Blood 93:1677–1683 [PubMed] [Google Scholar]
- 21.Yin D, Chave KJ, Macaluso CR, Galivan J, Yao R (1999) Structural organization of the human γ-glutamyl hydrolase gene. Gene 238:463–470 10.1016/S0378-1119(99)00362-5 [DOI] [PubMed] [Google Scholar]
- 22.Pui CH, Relling MV, Downing JR (2004) Acute lymphoblastic leukemia. N Engl J Med 350:1535–1548 10.1056/NEJMra023001 [DOI] [PubMed] [Google Scholar]
- 23.Whitehead VM, Rosenblatt DS, Vuchich MJ, Shuster JJ, Witte A, Beaulieu D (1990) Accumulation of methotrexate and methotrexate polyglutamates in lymphoblasts at diagnosis of childhood acute lymphoblastic leukemia: a pilot prognostic factor analysis. Blood 76:44–49 [PubMed] [Google Scholar]
- 24.Synold TW, Relling MV, Boyett JM, Rivera GK, Sandlund JT, Mahmoud H, Crist WM, Pui CH, Evans WE (1994) Blast cell methotrexate-polyglutamate accumulation in vivo differs by lineage, ploidy, and methotrexate dose in acute lymphoblastic leukemia. J Clin Invest 94:1996–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead VM, Payment C, Cooley L, Lauer SJ, Mahoney DH, Shuster JJ, Vuchich MJ, Bernstein ML, Look AT, Pullen DJ, Camitta B (2001) The association of the TEL-AML1 chromosomal translocation with the accumulation of methotrexate polyglutamates in lymphoblasts and with ploidy in childhood B-progenitor cell acute lymphoblastic leukemia: a Pediatric Oncology Group study. Leukemia 15:1081–1088 10.1038/sj.leu.2402165 [DOI] [PubMed] [Google Scholar]
- 26.Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 27.Reik W, Walter J (2001) Genomic imprinting: parental influence on the genome. Nat Rev Genet 2:21–32 10.1038/35047554 [DOI] [PubMed] [Google Scholar]
- 28.Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–926 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- 29.Laird PW (2005) Cancer epigenetics. Hum Mol Genet Spec No 1 14:R65–R76 10.1093/hmg/ddi113 [DOI] [PubMed] [Google Scholar]
- 30.Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4:143–153 [DOI] [PubMed] [Google Scholar]
- 31.Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054 10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- 32.Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6:597–610 10.1038/nrm1228 [DOI] [PubMed] [Google Scholar]
- 33.Cross SH, Charlton JA, Nan X, Bird AP (1994) Purification of CpG islands using a methylated DNA binding column. Nat Genet 6:236–244 10.1038/ng0394-236 [DOI] [PubMed] [Google Scholar]
- 34.Huang TH, Perry MR, Laux DE (1999) Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet 8:459–470 10.1093/hmg/8.3.459 [DOI] [PubMed] [Google Scholar]
- 35.Pao MM, Tsutsumi M, Liang G, Uzvolgyi E, Gonzales FA, Jones PA (2001) The endothelin receptor B (EDNRB) promoter displays heterogeneous, site specific methylation patterns in normal and tumor cells. Hum Mol Genet 10:903–910 10.1093/hmg/10.9.903 [DOI] [PubMed] [Google Scholar]
- 36.Jones PA, Gonzalgo ML (1997) Altered DNA methylation and genome instability: a new pathway to cancer? Proc Natl Acad Sci USA 94:2103–2105 10.1073/pnas.94.6.2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su HH, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C (2000) Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nat Genet 24:132–138 10.1038/72785 [DOI] [PubMed] [Google Scholar]
- 38.Banelli B, Gelvi I, Di VA, Scaruffi P, Casciano I, Allemanni G, Bonassi S, Tonini GP, Romani M (2005) Distinct CpG methylation profiles characterize different clinical groups of neuroblastic tumors. Oncogene 24:5619–5628 10.1038/sj.onc.1208722 [DOI] [PubMed] [Google Scholar]
- 39.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- 40.Kawai J, Hirose K, Fushiki S, Hirotsune S, Ozawa N, Hara A, Hayashizaki Y, Watanabe S (1994) Comparison of DNA methylation patterns among mouse cell lines by restriction landmark genomic scanning. Mol Cell Biol 14:7421–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura N, Oda R, Inaki Y, Suzuki O (2004) Attachment of oligonucleotide probes to poly carbodiimide-coated glass for microarray applications. Nucleic Acids Res 32:e68 10.1093/nar/gnh057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heisler LE, Torti D, Boutros PC, Watson J, Chan C, Winegarden N, Takahashi M, Yau P, Huang TH, Farnham PJ, Jurisica I, Woodgett JR, Bremner R, Penn LZ, Der SD (2005) CpG island microarray probe sequences derived from a physical library are representative of CpG islands annotated on the human genome. Nucleic Acids Res 33:2952–2961 10.1093/nar/gki582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H (1994) Sp1 elements protect a CpG island from de novo methylation. Nature 371:435–438 10.1038/371435a0 [DOI] [PubMed] [Google Scholar]
- 44.Mummaneni P, Yates P, Simpson J, Rose J, Turker MS (1998) The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res 26:5163–5169 10.1093/nar/26.22.5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melki JR, Vincent PC, Clark SJ (1999) Cancer-specific region of hypermethylation identified within the HIC1 putative tumour suppressor gene in acute myeloid leukaemia. Leukemia 13:877–883 10.1038/sj/leu/2401401 [DOI] [PubMed] [Google Scholar]
- 46.Millar DS, Paul CL, Molloy PL, Clark SJ (2000) A distinct sequence (ATAAA)n separates methylated and unmethylated domains at the 5′-end of the GSTP1 CpG island. J Biol Chem 275:24893–24899 10.1074/jbc.M906538199 [DOI] [PubMed] [Google Scholar]
- 47.Morris KV, Chan SW, Jacobsen SE, Looney DJ (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305:1289–1292 10.1126/science.1101372 [DOI] [PubMed] [Google Scholar]
- 48.Kawasaki H, Taira K (2004) Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431:211–217 10.1038/nature02889 [DOI] [PubMed] [Google Scholar]
- 49.Cheng Q, Evans WE (2005) Cancer pharmacogenomics may require both qualitative and quantitative approaches. Cell Cycle 4:1506–1509 [DOI] [PubMed] [Google Scholar]
- 50.He P, Varticovski L, Bowman ED, Fukuoka J, Welsh JA, Miura K, Jen J, Gabrielson E, Brambilla E, Travis WD, Harris CC (2004) Identification of carboxypeptidase E and γ-glutamyl hydrolase as biomarkers for pulmonary neuroendocrine tumors by cDNA microarray. Hum Pathol 35:1196–1209 10.1016/j.humpath.2004.06.014 [DOI] [PubMed] [Google Scholar]