To the Editor: In the July 2005 and March 2006 issues of the Journal, Schneider et al.1 and Zinn et al.,2 respectively, reported mapping studies of SHOX (MIM 312865) deletions in patients with Léri-Weill dyschondrosteosis (LWD [MIM 127300]). In their study, Schneider et al.1 reported that the majority (73%) of patients with LWD who had SHOX deletions shared a 3′ deletion breakpoint hotspot located downstream of SHOX. Zinn et al.2 identified a different 3′ breakpoint hotspot located several hundred kilobases farther downstream in 86% of Hispanic patients, whereas the recombination hotspot described by Schneider et al.1 was not observed.
We characterized the SHOX deletion limits in a cohort of 48 European patients with LWD (n=47) and Langer mesomelic dysplasia (LMD [MIM 249700]) (n=1). SHOX deletions were originally detected by multiplex ligation probe amplification (MLPA) (MRC Holland) or microsatellite analysis (DXYS10092, DXYS201, DYS290, DXYS10093, DXYS233, and DXYS234) and subsequently were finely mapped using a dense panel of microsatellites and SNPs.3 Four newly identified microsatellites (Tandem Repeat Finder), located 133, 54, 31, and 19 kb 5′ of SHOX (table 1), and 59 SNPs, 12 of which were previously unreported (table 2), were analyzed.
Table 1. .
Novel Microsatellite Markers[Note]
| Oligonucleotide(5′→3′) |
||||
| Markera | Sense | Antisense | Size Range (bp) |
Heterozygosity |
| DXYS10136 | CTGAACTCAGAATCGGGACC | CCCAGGAGCCCAGGAGATTGA | 303–323 | .85 |
| DXYS10137 | CCCAGGCCCTGTTTACGCTTCG | TATCCTCACAACTGCGTCTTCC | 180–210 | .90 |
| DXYS10138 | GTACATAGATGGCAGATAGATG | CTGCATGTATACACACTGTAAT | 197–221 | .73 |
| DXYS10139 | AGCCCCAACCCTCCATGATACTGA | GCAAAGGCATCTGTTTAAGTAACG | 131–203 | .24 |
Note.— PCR conditions for the amplification of microsatellites DXYS10136, DXYS10137, DXYS10138, and DXYS10139 were as follows: 1× Qiagen Hotstart Taq buffer, 0.5 units of Hotstart Taq polymerase, 2.0 mM MgCl2, 200 μM dNTPs (50 μM each), and 400 nM of each primer. Cycling conditions for DXYS10137, DXYS10138, and DXYS10139 were as follows: initial denaturation at 94°C for 15 min; 35 cycles for 30 s at 94°C; 30 s at 49°C, 58°C, and 55°C, respectively; and 40 s at 72°C, with a final extension for 8 min at 72°C. For the amplification of DXYS10136, we used a touchdown protocol for easier allele calling. Cycling conditions were as follows: initial denaturation at 94°C for 15 min, 16 cycles for 30 s at 94°C, 30 s at 58°C–66°C ramp (−0.5°C per cycle), and 40 s at 72°C, followed by 16 cycles for 30 s at 94°C, 30 s at 58°C, and 40 s at 72°C. The program terminated with a final extension for 8 min at 72°C.
Further details can be found at the GDB Human Genome Database.
Table 2. .
Oligonucleotide Sequences and PCR Conditions of the PAR1 Amplicons That Contain the 18 Analyzed SNPs[Note]
| Oligonucleotide(5′→3′) |
||||||
| dbSNP ID | Our Amplicon ID |
SNP | Sense | Antisense | Annealing Temperature (°C) |
Amplicon Size (bp) |
| rs5988284 | S27 | G/A | GTCATGGCCGGATCCT | ACTTGTTACCACGAGCCCG | 52 | 422 |
| rs6644380 | G/A | |||||
| rs5946512 | G/A | |||||
| rs17148729 | S21 | A/C | GGATTCCTGGTGGTTGCTAT | GCAGTAATCACATTAGGGTAAATAA | 55 | 997 |
| ss49845888 | G/A | |||||
| ss49845889 | T/A | |||||
| ss49845890 | S22 | C/T | TGGGGACAAATTATTCATTGGATTC | CTTGACCTTGTGATCTGCCCTCTTC | 55 | 986 |
| ss49845891 | G/A | |||||
| ss49845892 | G/A | |||||
| ss49845893 | T/C | |||||
| ss49845894 | G/A | |||||
| ss49845895 | S23 | T/C | CCACCACCATGCCTAGCTGA | ATGAGAGACAGGTTTCCACTGT | 55 | 377 |
| ss49845896 | S25 | C/T | CTCCCCATTGCGGTTGCACGAATTT | CCGTATGCACAATTCATGGGCG | 56 | 1,182 |
| rs6644384 | A/G | |||||
| rs6644385 | C/T | |||||
| ss49845885 | S20 | T/C | CAATACCAATCTTGCTTCAACCCAC | GGGATCAACAGACACTAATACGCA | 55 | 720 |
| ss49845886 | G/A | |||||
| ss49845887 | C/T | |||||
Note.— The 12 previously unreported SNPs are shown in bold.
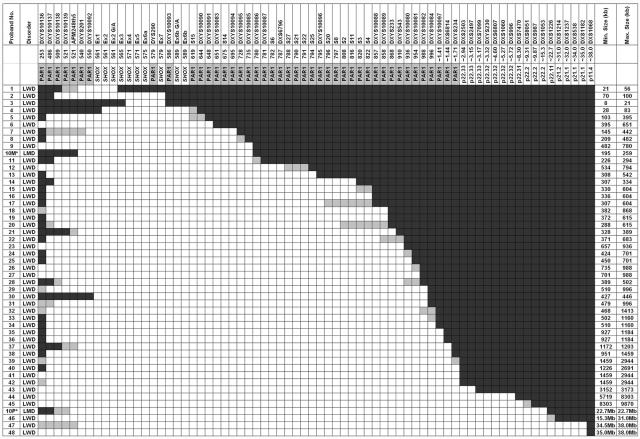
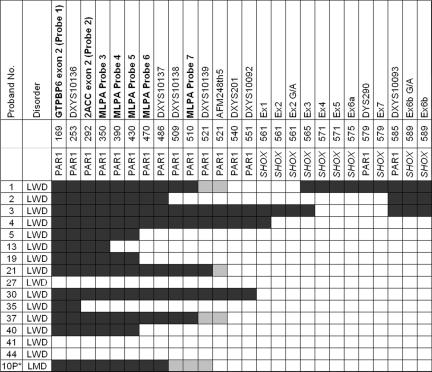
In our study, the SHOX-encompassing deletions were highly heterogeneous for both extension and breakpoint localization, with no recombination hotspot observed (fig. 1). Deletion extensions varied from at least 8 kb to 38 Mb. The majority of deletions (45 of 49) encompassed the entire SHOX gene, whereas 4 were partial deletions (deletions 1–4) (fig. 1). The smallest deletion (in proband 3) included exons 4–6a (8–21 kb) and was detected only by MLPA5 (fig. 1). In seven probands (14%), deletions extended into the Xp-specific region (figs. 1 and 2). The majority (61%) of deletions were interstitial, whereas the remaining 39% encompassed all analyzed telomeric markers, thus possibly extending to the telomere. Because of the lower microsatellite density in the telomeric pseudoautosomal region 1 (PAR1), the 5′ breakpoints of 16 subjects were reevaluated using the commercial SHOX MLPA kit complemented with seven additional synthetic MLPA probes (table 3 and fig. 3). Only three patients—29, 30, and 31—possibly share a 3′ breakpoint between DXYS10082 and DXYS10084, and only probands 35 and 36 possibly share the same 5′ and 3′ breakpoint.
Figure 1. .
Characterization of PAR1 deletions in 47 patients with LWD and 1 patient with LMD, through use of a panel of microsatellites and SNPs (not to scale). Blackened areas indicate the presence of two copies of the marker or SNP, unblackened areas indicate the presence of a deletion, and shaded areas indicate the noninformative areas where the breakpoints are located. Distances are according to Ensembl Genome Browser coordinates; markers DXYS10136–DXYS234 are stated in kilobases, whereas markers DXYS10097–DXS1068 are stated in megabases. Deletion sizes are in kilobases unless otherwise stated. Proband 10 (indicated by an asterisk [*]), the subject with LMD, has been reported elsewhere; the proband inherited a familial deletion from her mother (10M), and a de novo deletion of the paternal allele (10P) was also included in the cohort.4 AFM248th5 is an STS marker.
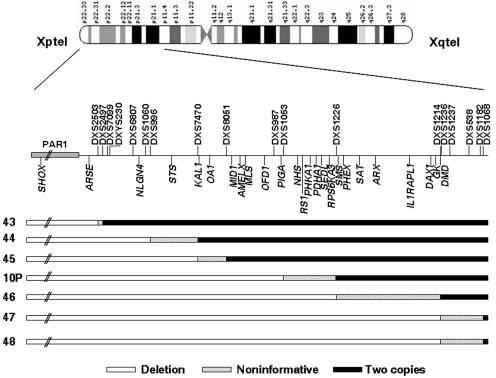
Figure 2. .
Detailed schematic representation of the 3′ deletion limits of probands 10P and 43–48, which extend beyond the PAR1 into X-specific regions. These samples correspond to one male (proband 43), five sporadic females (44, 45, 10P, 47, and 48), and proband 46, a familial case from a family in which only affected females were observed. Blackened areas indicate the presence of two copies of the marker or SNP, unblackened areas indicate the presence of a deletion, and shaded areas indicate the noninformative areas where the breakpoints are located. Localization of the microsatellite markers and the genes located within the deletions are indicated by vertical lines above and below the line, respectively.
Table 3. .
Novel Synthetic MLPA Probes Incorporated into the MRC Holland SHOX Kit
| Nomenclature | Approximate Distance from Telomere (kb) |
Sequence at Ligation Site |
| Probe 1 (GTPBP6 exon 2) | 169 | AAGATCAGGAAGGCCTTGGACAGG-CTTCGCAAGAAGAGGCACCTGCTC |
| Probe 2 (PPP2R3B exon 2) | 292 | CGCAGGACTCCGTCAACGTGGATG-CCGTCATCAGCAAGATCGAGAGCA |
| Probe 3 | 350 | AGCCAGCATCCGTGGTCTCTCTAT-AGTGGCCTCACGGTCTCCAGCCAG |
| Probe 4 | 390 | TCATCTTGTCTCAGAGACCTCGGA-GAGCTCCCAGAGCCTGGCTGCCAC |
| Probe 5 | 430 | GGTGCTCAGAGCCTCTAGGAGGAT-CCTTTCGGAAAGCAAGTCTGCTGT |
| Probe 6 | 470 | TCAGGGCCCAAGCCAGCGGAAGCG-CTGCGCTCACTAAAGACGCTCCGT |
| Probe 7 | 510 | ATGCTGGCAATATGG CGGTCACCA-ATAGTGTTCATCAACTCCAGAGGG |
Figure 3. .
Fine mapping of 16 PAR1 deletions in patients with LWD and LMD with use of seven new synthetic Xp telomeric MLPA probes, indicated in bold. Distances are not to scale and are according to Ensembl Genome Browser coordinates (in kb). Blackened areas indicate the presence of two copies of the MLPA probe, marker, or SNP; unblackened areas indicate the presence of a deletion; and shaded areas indicate the noninformative areas where the breakpoints are located. “10P*” indicates the de novo deletion of the paternal allele.
The results from this study are in contrast to previous reports of the presence of 3′ deletion breakpoint hotspots in patients with LWD.1,2 Schneider et al.1 reported a common proximal breakpoint in 20 of 27 patients with LWD. In a subset of six patients with LWD, this common breakpoint was refined to an ∼5-kb interval 666–671 kb from Xpter, corresponding to a point between DXYS10083 and S14. In contrast, no breakpoint hotspots were observed in our study; moreover, none of our 48 subjects had a breakpoint within this interval. This discrepancy is unlikely to be explained by population differences between studies, since all subjects were of European origin. The presence of multiple 5′ breakpoints in both studies also excludes the possibility of a founder effect. Differences in the methodology used for deletion mapping may explain the discrepancy between the studies. Whereas Schneider et al.1 initially used microsatellites to identify the deletion limits, subsequent mapping was performed by FISH. They analyzed a number of PAR1 cosmids, by “walking” away from SHOX until they identified a probe that gave two hybridization signals. In 20 subjects, the first nondeleted probe downstream of SHOX was cosmid P0117. No additional probes downstream of P0117 were reported in these 20 subjects. The deletion breakpoint was defined by cosmids 29B11 distally and P0117 proximally. Further mapping of the breakpoint region was achieved by SNP analysis in 6 of these 20 subjects. A breakpoint hotspot was observed within a 5-kb interval. These findings were then extrapolated to all 20 subjects with a common breakpoint detected by FISH. In our opinion, this inference must be taken with care because of the observation reported elsewhere that PAR1 is enriched for repetitive elements and duplications6 and the fact that marker P117, located within cosmid P0117, was mapped to multiple Xp locations.7 Thus, if the cosmid P0117 is misassigned or comprises repetitive sequences, two FISH signals may be observed. This may be misleading, since the presence of the deletion would be masked.
In our study, we employed MLPA and segregation analysis of a large number of microsatellites and SNPs for probands with LWD and their parents. The deletions carried by 42 of 48 patients extended beyond the breakpoint reported by Schneider et al.1 The large number of polymorphic markers analyzed excludes the possibility of allele dropout and nonpaternity. To investigate the discrepancy between our study and that of Schneider et al.,1 we tested a subset of subjects with use of FISH. As expected, all four tested subjects had deletion of cosmid 29B11. We also attempted to analyze cosmid P0117 (a kind gift from Dr. G.-J. van Ommen), but we could not obtain hybridization signals for any of the four subjects. However, none of the cases were deleted for a series of fosmids, which mapped to the same region (613–747 kb) (information available from the authors). Furthermore, no deletion breakpoint hotspots were identified in patients with LWD who had PAR1 deletions located downstream of SHOX5 or among the three largest studies of patients with Xp deletions.8–11
In the second recent study of SHOX deletion mapping of probands with LWD, Zinn et al.2 employed molecular methods to analyze 30 LWD-affected families with SHOX deletions. In 17 of 26 informative cases, the deletion included the DXYS233 marker—that is, extended beyond the 666–671 kb interval defined by Schneider et al.1 Detailed STS mapping was performed on a subset of 11 subjects for whom human-hamster hybrids were constructed. This revealed a second hotspot in 8 of the 11 subjects, between 1.18 and 1.42 Mb, in a region that still contains a number of gaps in the human PAR1 sequence. In contrast to the results of Zinn et al.,2 only three of our subjects had a 3′ breakpoint within this region. However, a founder effect could not be excluded in their study, since five of the six probands also shared 5′ breakpoints.
Although the underlying molecular mechanism remains to be elucidated, nonhomologous end joining may explain the occurrence of the scattered deletion breakpoints observed in the present study. The architectural features of the PAR1 deletion–flanking sequences include a high incidence of Alu and long interspersed nucleotide elements (LINE),12 which has resulted in a recombination fraction ∼20-fold higher per unit of physical distance for the whole PAR1 than for the genome-average rate of 1 cM per Mb.13 Furthermore, even within different segments of the PAR1, the recombination fraction seems to be variable, with a range from a 13–23-fold greater increase at the telomere to a 26–38-fold increase near the pseudoautosomal boundary.14 Thus, one can expect—and, indeed, observe—the recurrent incidence of deletions in this region in LWD.
An important diagnostic point was the observation that a significant proportion of probands (14% [7 of 48]) were found to carry PAR1 deletions that extended into the X-specific region of Xp22.3-Xp21.2 (fig. 2). Male offspring of these females would have a 50% chance of inheriting the Xp terminal deletion and therefore could be either inviable or at risk of presenting with severe clinical complications. In the LWD/idiopathic short stature (ISS) cohort studied by Schneider et al.,1 3 of 27 subjects with LWD and 1 of 6 subjects with ISS also presented with deletions of the entire analyzed PAR1 and with the likely extension into the Xp22.3 region, although the deletion limits were not available.
In conclusion, we detected a high level of genetic heterogeneity of SHOX deletions in a European cohort of 48 patients with LWD/LMD who have a significant proportion of deletions extending beyond the PAR1 boundary. Until recently, molecular analysis of patients with LWD included only the screening of SHOX deletions and insertions, deletions, and point mutations within SHOX. Our recent findings3 and those reported here emphasize the necessity of improving the molecular diagnosis given to LWD-, LMD-, and ISS-affected families by means of including deletion screening of the PAR1 region downstream of SHOX and, in certain cases, of analyzing whether the 3′ boundaries extend into the Xp22.3 region.
Acknowledgments
This work was supported by Ministerio de Ciencia y Tecnología grant SAF2003-02511; Fondo de Investigación Sanitaria grants C03/07, PI051675, and PI021663; and a grant from Fundación Mutua Madrileña. This study was also partly supported by the Fundación Endocrinología y Nutrición, Serono España, and Lilly France. The French patients were included in the module SHOX GeNesis. We thank all the clinicians and patients who participated in the study, and we thank Kevin Baker, who performed the synthetic MLPA work.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNP identification numbers [listed in ], including the new accession numbers ss49845885, ss49845886, ss49845887, ss49845888, ss49845889, ss49845890, ss49845891, ss49845892, ss49845893, ss49845894, ss49845895, and ss49845896)
- Ensembl Genome Browser, http://www.ensembl.org/ (for sequence information of the human X and Y chromosomes)
- GDB Human Genome Database, http://www.gdb.org (for further details about the new microsatellites DXYS10136, DXYS10137, DXYS10138, and DXYS10139)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SHOX, LWD, and LMD)
- Tandem Repeat Finder, http://tandem.bu.edu/ (for identifying microsatellites in sequence data)
References
- 1.Schneider KU, Sabherwal N, Jantz K, Röth R, Muncke N, Blum WF, Cutler GB Jr, Rappold G (2005) Identification of a major recombination hotspot in patients with short stature and SHOX deficiency. Am J Hum Genet 77:89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinn AR, Ramos P, Ross JL (2006) A second recombination hotspot associated with SHOX deletions. Am J Hum Genet 78:523–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benito-Sanz S, Thomas NS, Huber C, Gorbenko del Blanco D, Aza-Carmona D, Crolla JA, Maloney V, Rappold G, Argente J, Campos-Barros A, Cormier-Daire V, Heath KE (2005) A novel class of pseudoautosomal region 1 deletions downstream of SHOX is associated with Léri-Weill dyschondrosteosis. Am J Hum Genet 77:533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas NS, Maloney V, Bass P, Mulik V, Wellesley D, Castle B (2004) SHOX mutations in a family and a fetus with Langer mesomelic dwarfism. Am J Med Genet A 128:179–184 10.1002/ajmg.a.30095 [DOI] [PubMed] [Google Scholar]
- 5.Schouten JP, McElgunn CJ, Waaijer R, Zwinenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30:e57 10.1093/nar/gnf056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wapenaar MC, Petit C, Basler E, Ballabio A, Henke A, Rappold GA, van Paassen HMB, Blonden LAJ, van Ommen GJB (1992) Physical mapping of 14 new DNA markers isolated from the human distal Xp region. Genomics 13:167–175 10.1016/0888-7543(92)90217-G [DOI] [PubMed] [Google Scholar]
- 8.James RS, Coppin B, Dalton P, Dennis NR, Mitchell C, Sharp AJ, Skuse DH, Thomas NS, Jacobs PA (1998) A study of females with deletions of the short arm of the X chromosome. Hum Genet 102:507–516 10.1007/s004390050733 [DOI] [PubMed] [Google Scholar]
- 9.Ross JL, Roeltgen D, Kushner H, Wei F, Zinn AR (2000) The Turner syndrome–associated neurocognitive phenotype maps to distal Xp. Am J Hum Genet 67:672–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata T, Muroya K, Matsuo N, Shinohara O, Yorifuji T, Nishi Y, Hasegawa Y, Horikawa R, Tachibana K (2001) Turner syndrome and Xp deletions: clinical and molecular studies in 47 patients. J Clin Endocrinol Metab 86:5498–5508 10.1210/jc.86.11.5498 [DOI] [PubMed] [Google Scholar]
- 11.Kosho T, Muroya K, Nagai T, Fujimoto M, Yokoya S, Sakamoto H, Hirano T, Terasaki H, Ohashi H, Nishimura G, Sato S, Matsuo N, Ogata T (1999) Skeletal features and growth patterns in 14 patients with haploinsufficiency of SHOX: implications for the development of Turner syndrome. J Clin Endo Metab 84:4613–4621 10.1210/jc.84.12.4613 [DOI] [PubMed] [Google Scholar]
- 12.Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- 13.May CA, Shone AC, Kalydjieva L, Sajantila A, Jeffreys AJ (2002) Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat Genet 31:272–275 10.1038/ng918 [DOI] [PubMed] [Google Scholar]
- 14.Lien S, Szyda J, Schechinger B, Rappold G, Arnheim N (2000) Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am J Hum Genet 66:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]