Abstract
The human mitochondrial 12S ribosomal RNA (rRNA) A1555G mutation has been associated with aminoglycoside-induced and nonsyndromic deafness in many families worldwide. Our previous investigation revealed that the A1555G mutation is a primary factor underlying the development of deafness but is not sufficient to produce a deafness phenotype. However, it has been proposed that nuclear-modifier genes modulate the phenotypic manifestation of the A1555G mutation. Here, we identified the nuclear-modifier gene TRMU, which encodes a highly conserved mitochondrial protein related to transfer RNA (tRNA) modification. Genotyping analysis of TRMU in 613 subjects from 1 Arab-Israeli kindred, 210 European (Italian pedigrees and Spanish pedigrees) families, and 31 Chinese pedigrees carrying the A1555G or the C1494T mutation revealed a missense mutation (G28T) altering an invariant amino acid residue (A10S) in the evolutionarily conserved N-terminal region of the TRMU protein. Interestingly, all 18 Arab-Israeli/Italian-Spanish matrilineal relatives carrying both the TRMU A10S and 12S rRNA A1555G mutations exhibited prelingual profound deafness. Functional analysis showed that this mutation did not affect importation of TRMU precursors into mitochondria. However, the homozygous A10S mutation leads to a marked failure in mitochondrial tRNA metabolisms, specifically reducing the steady-state levels of mitochondrial tRNA. As a consequence, these defects contribute to the impairment of mitochondrial-protein synthesis. Resultant biochemical defects aggravate the mitochondrial dysfunction associated with the A1555G mutation, exceeding the threshold for expressing the deafness phenotype. These findings indicate that the mutated TRMU, acting as a modifier factor, modulates the phenotypic manifestation of the deafness-associated 12S rRNA mutations.
Mutations in mtDNA have been found to be one of the most important causes of sensorineural hearing loss.1–2 In particular, the homoplasmic A1555G and C1494T mutations in the highly conserved decoding site of the mitochondrial 12S rRNA (MIM 561000) have been associated with both aminoglycoside-induced and nonsyndromic deafness [MIM 580000) in many families worldwide.3–8 Matrilineal relatives within and among families carrying the A1555G mutation exhibited a wide range of penetrance and expressivity, including severity and age at onset of deafness.4–6 Functional characterization demonstrated more-severe biochemical defects in the mutant lymphoblastoid cell lines derived from symptomatic individuals of an Arab-Israeli family carrying the A1555G mutation than from those of cell lines derived from asymptomatic individuals in the same family.9 However, under a constant nuclear background, a nearly identical degree of mitochondrial dysfunction was observed in cybrid cell lines derived from symptomatic and asymptomatic individuals from this family.10 These findings strongly indicate that the A1555G mutation is a primary factor underlying the development of deafness but is itself insufficient to produce a deafness phenotype. Thus, other modifier factors, such as aminoglycosides or nuclear-modifier genes, modulate the phenotypic manifestation of the A1555G or the C1494T mutation.3,8–11 The product of nuclear-modifier gene(s), which may functionally interact with the mutated 12S rRNA, influences the phenotypic manifestation of the A1555G mutation by enhancing or suppressing the effect of the mutation.9
Extensive genomewide linkage studies of Arab-Israeli and European (Italian and Spanish) families revealed that the phenotypic expression of the A1555G mutation is, in fact, influenced by a complex inheritance of multiple nuclear-encoded modifier genes.12 Despite statistical support for the linkages of several putative modifier loci, including one locus localized to chromosome 8p23.1,13,14 no mutations in these modifier genes have been identified. Thus, the involvement of multiple factors and a relatively rare disorder make it very difficult to identify such nuclear-modifier genes by use of conventional genetic approaches, such as genomewide linkage analysis. Recently, we proposed an interesting model for nuclear-mtDNA interaction for the phenotypic manifestation of the A1555G or the C1494T mutation.15–17 In the yeast Saccharomyces cerevisiae, the mutant alleles of MTO1, MSS1, or MTO2 that encode mitochondrial proteins manifest a respiratory-deficient phenotype only when coupled with the mitochondrial 15S rRNA C1409G mutation17–19 corresponding to human 12S rRNA C1494T mutation.3,8 This strongly suggests that Mss1p, Mto1p, or Mto2p affects the phenotypic expression of the C1409G mutation, by functionally interacting with the region of C1409G in mitochondrial 15S rRNA. In Escherichia coli, the products of mnmE (homolog of MSS1),20 gidA (homolog of MTO1),21 and trmU (homolog of MTO2)22 have been shown to be involved in the biosynthesis of the hypermodified nucleoside 5-methyl-aminomethy-2-thio-uridine (mnm5s2U34).23 This modified nucleotide, found in the wobble position of several bacterial and human mitochondrial tRNAs (mt tRNAs) specific for glutamate, lysine, and glutamine, has a pivotal role in the structure and function of tRNAs, including structural stabilization, aminoacylation, and codon recognition at the decoding site of small rRNA.21,23,24
Of these, TRMU has been shown to be responsible for the 2-thiolation of mnm5s2U34 in tRNALys, tRNAGlu, and tRNAGln in bacteria,22,25 yeast, and human mitochondria.24 Recently, we demonstrated that isolated human TRMU cDNA can complement the respiratory-deficient phenotype of yeast mto2 cells carrying the 15S rRNA C1409G mutation.17 Furthermore, we showed that, in families carrying the A1555G mutation, there was highly suggestive linkage and linkage disequilibrium between microsatellite markers adjacent to TRMU and the presence of deafness.26 These findings strongly suggest that TRMU is a candidate nuclear-modifier gene for the phenotypic manifestation of the deafness-associated 12S rRNA A1555G or C1494T mutation. To assess the contribution that allelic variant(s) in this gene makes toward the phenotypic expression of the A1555G mutation, we performed mutational analysis of TRMU in 613 subjects with nonsyndromic deafness from 1 Arab-Israeli kindred, 210 European families, and 31 Chinese pedigrees. Functional significance of the TRMU allelic variant has been evaluated by examination of import and processing of TRMU, mt tRNA modification and stability, and protein synthesis, through use of lymphoblastoid mutant cell lines derived from Arab-Israeli controls and the Arab-Israeli family members carrying the A1555G mutation.
Material and Methods
Families and Subjects
DNA samples used for this investigation were from members of families who carried the mtDNA A1555G mutation and exhibited nonsyndromic hearing loss: 67 members of a large Arab-Israeli kindred,12 100 hearing-impaired members of 30 Chinese pedigrees,5,6,27 and 420 deaf individuals from 210 pedigrees of Italian-Spanish background.14,28 In addition, DNA samples from 26 hearing-impaired members of a large Chinese family carrying the mtDNA C1494T mutation8 were also used for the mutation analysis. The control DNA samples were obtained from the following panels: 137 individuals of Jewish background with normal hearing, 100 hearing-impaired white subjects lacking the mtDNA A1555G mutation, and 142 unaffected individuals of Han Chinese ancestry. Informed consent, blood samples, and clinical evaluations were obtained from all participating family members, under protocols approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
Cell Lines and Culture Conditions
Human cell lines 143BTK− and HeLa were used for the extraction of RNA, mitochondrial protein import, and in vivo mitochondrial protein–labeling experiments. Both 143BTK− and HeLa cells were grown in regular Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Six human immortalized lymphoblastoid cell lines derived from members of the Arab-Israeli family (V-4, V-8, IV-5, IV-9, V-1, and V-3) and two genetically unrelated Arab-Israeli control individuals (#2 and #3)12 were grown in RPMI1640 medium (Invitrogen) supplemented with 10% FBS.
Mutational Analysis of the TRMU Gene
Nine pairs of primers for PCR-amplifying exons and their flanking sequences, including splicing-donor and acceptor-consensus sequences of TRMU, were used for this analysis. The forward and reverse primers for PCR amplification and sequence analysis are shown in table 1.
Table 1. .
Oligonucleotide Primers for Amplification of Human TRMU Exons
| Primer Sequence(5′→3′) |
||||
| Exon(s) | Forward | Reverse | Annealing Temperature (°C) |
Product Size (bp) |
| 1 | ACAGCGCAGAAGAAGAGCAGT | ACTACACAGGTGGAGGGCGA | 58 | 572 |
| 2 | CTCAGGCACCAAGATGGAAAC | GAGGCCTCTTGCAGTCTTCAG | 56 | 492 |
| 3 | CGTAGTGGCAGAGAATAACACC | ACAGTTGTGACACCATCTCCAA | 53 | 525 |
| 4 | CAGAGTGCTAGGATTACAGGC | GGCAGGGACACTGTTTACTAC | 50 | 725 |
| 5 | GAGTGTTGATGTCTGCCTCTGA | CCTCAGCAAACTCCTCCATCT | 53 | 453 |
| 6 and 7 | TCTAAGGCTCTGGCATCGTGT | GGACGACAGGAACTCTGGTCTAG | 56 | 491 |
| 8 | GATGTGCTCAGGTGCTTGGT | GACCAGCATACAACTCAGCCTA | 54 | 495 |
| 9 and 10 | GTGTGCTGGTAGGACAGTTGTT | GTGACACCAGAGTGGAAATCC | 52 | 830 |
| 11 | TCTCCTGTTCAGCAGCAGCA | ACACAGGTCAGCATCGCAGG | 56 | 858 |
Fragments spanning 11 exons and flanking sequences from 15 matrilineal relatives of this Arab-Israeli family,12 one spouse, and two unrelated controls were PCR amplified, purified, and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer with use of the Big Dye Terminator Cycle sequencing reaction kit (Applied Biosystems). These sequence results were compared with the TRMU genomic sequence (GenBank accession number AF448221).26 Genotyping for the G28T variant in other subjects was PCR amplified for exon 1 and was followed by digestion of the 467-bp segment with the restriction enzyme Bsp1286I. The forward and reverse primers for exon 1 are 5′-ACAGCGCAGAAGAAGAGCAGT-3′ and 5′-ACAACGCCACGACGGACG-3′, respectively. The Bsp1286I-digested products were analyzed on 1.5% agarose gels.
Mitochondrial Import Experiment
pGEM7-TRMU and pGEM7-TRMU-A10S were constructed by inserting full-length wild-type and A10S mutant cDNA into pGEM7Zf(+) (Promega).17 TRMU and mTERF precursors were synthesized in vitro with use of the TNT-coupled reticulocyte lysate system (Promega) and pGEM7-TRMU, pGEM7-TRMU-Ser10, and pmTERF29 constructs in the presence of [35S]methionine. Mitochondrial fractions were isolated from HeLa cells as described elsewhere,30 were washed twice with an incubation buffer (10 mM Tris-HCl, 25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM KH2PO4, 0.05 mM EDTA, and 5 mM MgCl2 [pH 7.4] at 25°C), and were resuspended in the incubation buffer containing 1 mg/ml BSA, 2 mM Na succinate, 1 mM ATP, and 1 mM methionine. Then, 10 μl reticulocyte-lysate synthesized protein was added to 90 μl of mitochondrial suspension, and the reactions were incubated at 37°C for 30 min. Mitochondria were washed twice with ice-chilled incubation buffer, and the final pellets were lysed with twice-concentrated gel sample buffer. In some experiments, 10 μM uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP), 0.5 μg/ml valinomccycin, and 20 μg/ml oligomycin were added to the reaction 5 min before the import initiation, to dissipate the electrochemical potential (ΔΨ). In other assays, mitochondria, after the import incubation, were treated with 100 μg/ml proteinase K in the incubation buffer for 15 min at room temperature, followed by the addition of 0.5 μM phenylmethylsulfonyl fluoride, and were washed once in incubation buffer. Mitochondrial lysates were analyzed by SDS-PAGE, as described elsewhere.29
APM Gel Electrophoresis to Quantify 2-Thiouridine Modification in tRNAs
Total RNA samples from the lymphoblastoid cell lines (1×108) were obtained by use of TRIzol reagent (Invitrogen). mt tRNALys, tRNAGlu, and tRNAGln were specifically labeled with 32P at the 3′ termini, following the method described elsewhere.31 Oligonucleotide probes are as follows: 5′-GTGGTCACTGTAAAGAGGTGTTG-3′ (tRNALys), 5′-GTGGTATTCTCGCACGGACTACAACC-3′ (tRNAGlu), and 5′-GTGGCTAGGACTATGAGAATCGAACC-3′ (tRNAGln). (N-acryloylamino)phenyl mercuric chloride (APM) gel electrophoresis and quantification of 2-thiouridine modification in tRNAs were conducted as detailed elsewhere.32–33
mt tRNA Analysis
Total mtRNA preparations were obtained by using TOTALLY RNA (Ambion) from mitochondria isolated from lymphobloid cell lines (∼4.0×107 cells), as described elsewhere.30 Five μg of total mitochondrial RNA was electrophoresed through a 10% polyacrylamide/7 M urea gel in Tris-borate-EDTA buffer (TBE) (after heating the sample at 65°C for 10 min) and then was electroblotted onto a positively charged nylon membrane (Roche) for the hybridization analysis with specific oligodeoxynucleotide probes. For the detection of tRNASer(UCN), tRNALeu(UUR), tRNALys, tRNAMet, tRNAHis, tRNAGlu, 12S rRNA, and 5S RNA, the following nonradioactive digoxigenin (DIG)-labeled oligodeoxynucleotides specific for each RNA were used: 5′-CAAGCCAACCCCATGGCCTC-3′ (tRNASer[UCN]), 5′-GGTAAATAAGGGGTCGTAAGC-3′ (tRNAHis), 5′-TCACTGTAAAGAGGTGTTGG-3′ (tRNALys), 5′-TGTTAAGAAGAGGAATTGAA-3′ (tRNALeu[UUR]), 5′-TATTCTCGCACGGACTACAA-3′ (tRNAGlu), 5′-TAGTACGGGAAGGGTATAACC-3′ (tRNAMet), 5′-GAAAGGCTAGGACCAAACCTA-3′ (12S rRNA),34 and 5′-GGGTGGTATGGCCGTAGAC-3′ (5S RNA).35 DIG-labeled oligodeoxynucleotides were generated by using DIG Oligonucleotide Tailing Kit (Roche). The hybridization was performed as detailed elsewhere.36 Quantification of density in each band was made as detailed elsewhere.36,37
Analysis of Mitochondrial-Protein Synthesis
Pulse labeling of the cell lines for 30 min with [35S]methionine-[35S]cysteine in methionine-free DMEM in the presence of emetine, electrophoretic analysis of the translation products, and quantification of radioactivity in the whole electrophoretic patterns or in individual well-resolved bands was performed as detailed elsewhere.9,38
Computer Analysis
Statistical analysis was performed by the unpaired, two-tailed Student’s t-test contained in the Microsoft Excel program for Macintosh (v. 5). Correlation analysis was performed using the curve fitting in the CA-Cricket Graph III program for Macintosh (v. 1.5.2).
Results
Mutation Screening of TRMU in Families with Nonsyndromic Deafness
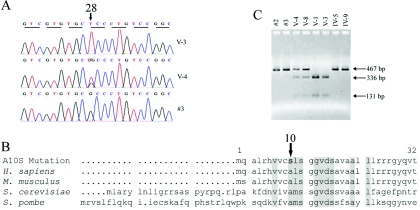
To determine whether TRMU modulates the phenotypic expression of the A1555G or the C1494T mutation, we performed a genotyping analysis of TRMU, using DNA samples derived from three cohorts of subjects carrying the A1555G or the C1494T mutation: (1) 67 members of a large Arab-Israeli kindred,12 (2) 420 hearing-impaired subjects from 210 European pedigrees,13,14,28 and (3) 126 hearing-impaired members of 31 Chinese families.5,6,8,27 No subjects had a history of exposure to aminoglycosides. First, we performed the PCR-amplification and sequence analysis of DNA fragments spanning exons and their flanking sequences of TRMU from 15 matrilineal relatives of a large Arab-Israeli family, one spouse, and two unrelated control individuals with normal hearing who were of Arab descent.12 In this family, most hearing-impaired matrilineal relatives exhibited congenital severe-to-profound hearing loss, a few showed late-onset and/or mild-to-moderate hearing impairment, and other matrilineal relatives had normal hearing. This analysis identified a G→T transversion at position 28 (G28T) (fig. 1A) in exon 1 of some individuals, which caused an alanine→serine replacement (A10S) at position 10. No other sequence changes in this gene were identified among these individuals. As shown in figure 1B, the A10 at the N-terminal sequence of this polypeptide is a highly conserved residue, indicating that this alanine might be important for the function of this protein.
Figure 1. .
Mutation analysis of TRMU. A, Partial sequence chromatograms of TRMU from affected individual V-3 and normal-hearing individual V-4 of the Arab-Israeli family11 and control #3. The arrow indicates the location of the nucleotide changes at position 28. B, RFLP PCR for the G28T mutation in several members of the Arab-Israeli family. C, Sequence alignment of the N-terminal sequence of human TRMU with its homologs. The arrow indicates the position of the A10S mutation.
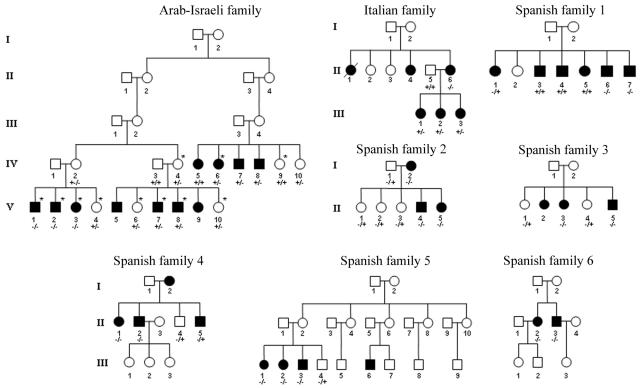
We further analyzed the presence of the G28T mutation in other subjects, by RFLP analysis, since the G28T mutation creates a Bsp1286I site, as shown in figure 1C. In the Arab-Israeli family, as shown in figure 2, three of the deaf children (V-1, V-2, V-3) of asymptomatic mother IV-2 (who harbors the heterozygous G28T mutation) carry the homozygous G28T mutation, whereas their asymptomatic sibling (V-4) is heterozygous for the G28T mutation. Of other symptomatic members, 12 individuals are heterozygous for the G28T mutation, whereas 29 members lack this mutation. Of 18 asymptomatic individuals, 10 are heterozygous for the TRMU G28T mutation, whereas 8 do not carry this mutation. However, the G28T mutation was absent in the 126 hearing-impaired Chinese subjects of 31 pedigrees carrying either the A1555G or the C1494T mutation. Of 420 hearing-impaired subjects of 210 Italian-Spanish pedigrees, 15 subjects—who belong to 1 Italian pedigree and 6 Spanish pedigrees, as shown in figure 2—were homozygous for the G28T mutation. Interestingly, all 15 subjects exhibited prelingual profound hearing loss. Of other Italian-Spanish subjects, 73 were heterozygous for the G28T mutation, and the others lacked this mutation. These data indicate that other putative modifier genes may contribute to the phenotypic manifestation of the A1555G mutation in those hearing-impaired individuals.
Figure 2. .
Eight pedigrees with nonsyndromic deafness. The TRMU G28T mutation was initially identified, by direct sequencing of PCR fragments spanning coding regions, with use of genomic DNA derived from members of an Arab-Israeli kindred as template and subsequently screened by RFLP PCR. Members marked with an asterisk (*) were examined for the TRMU mutation by direct sequencing of PCR fragments spanning coding regions. Hearing-impaired individuals are indicated by blackened symbols. Individuals who harbored homozygous (−/−), heterozygous (+/−), or wild type (+/+) mutations are indicated.
Finally, we screened for the presence of this mutation in 137 Jewish controls, 142 Chinese controls, and 100 deaf whites without the A1555G mutation. The heterozygous G28T mutation was not detected in these Chinese controls but was present in 14 Jewish controls and 12 deaf whites. However, we did not identify the homozygous G28T mutation in these control populations. This translates to ∼10% frequency of this variant in the Jewish and deaf white populations. However, the allele frequency of this variant was ∼25% in both the Arab-Israeli family and the 420 hearing-impaired subjects of 210 Italian-Spanish pedigrees. The higher incidence of this TRMU variant indicates that this variant may be involved in deafness expression in these subjects carrying the A1555G mutation.
Import and Processing of the Human TRMU Precursor Not Affected by the A10S Mutation
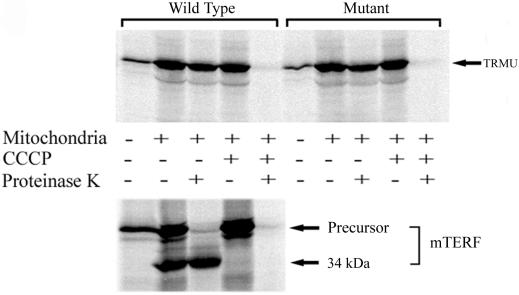
To examine whether the A10S mutation affects TRMU import, import/processing reactions were performed in which the wild-type or mutant TRMU or mTERF putative precursors were labeled in vitro with an amino acid and then were incubated with isolated HeLa cell mitochondria under various conditions. As shown in figure 3, both [35S]methionine-labeled precursors synthesized from wild-type and mutant TRMU constructs, after incubation with mitochondria, produced only one band with the same electrophoretic mobility as did in vitro–synthesized precursors. By contrast, [35S]methionine-labeled precursors of mTERF that contains cleavable mitochondrial targeting presequence yielded, after incubation with the mitochondria, two bands that corresponded to the mTERF precursor and 34-kDa mature forms.29 After the [35S]methionine-labeled precursors were incubated with mitochondria and then were digested with proteinase K, only the 34-kDa mature form and not the precursor of mTERF appeared, whereas there was one band showing the same size as the precursors of wild-type and mutant TRMU. In fact, the imported polypeptides were resistant to proteinase K digestion, whereas nonimported polypeptides were sensitive to proteinase K digestion. In the presence of uncoupler CCCP to block the ΔΨ necessary for mitochondrial-protein import, all [35S]methionine-labeled polypeptides were unable to import into mitochondria; thus, they digested completely with proteinase K. Those results clearly indicated that both wild-type and mutant TRMU were imported into mitochondria but, unlike the majority of nucleus-encoded mitochondrial proteins such as mTERF,29 were not processed after import. Most importantly, efficiencies of the import of wild-type and A10S TRMU into mitochondria are 38.4% and 36.7%, respectively. Thus, there is no obvious difference, in terms of imported efficiency, between wild-type and mutant TRMU. This result strongly indicated that the A10S mutation does not affect TRMU import into mitochondria.
Figure 3. .
Import into mitochondria and processing of human TRMU precursor. SDS-PAGE analysis of the in vitro transcribed/translated products synthesized in the presence of [35S]methionine, with use of cDNA carrying human wild-type and mutant (A10S) TRMU or mTERF as template, analyzed directly or after incubation with isolated HeLa cell mitochondria for 30 min at 37°C under various conditions, detailed in the “Material and Methods” section.
Defect in 2-Thiouridine Modification at Position 34 in mt tRNALys, tRNAGlu, and tRNAGln Caused by the A10S Mutation
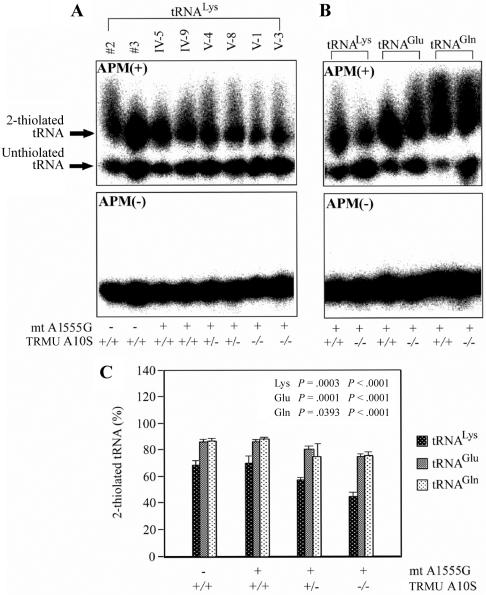
To investigate whether the A10S mutation alters 2-thiouridine modification at position 34 in mt tRNAs, the 2-thiouridylation levels of tRNAs were determined by isolating total RNA from eight lymphoblastoid cell lines, purifying mt tRNAs, qualifying the 2-thiouridine modification by the retardation of electrophoresis mobility in polyacrylamide gel containing 0.05 mg/ml APM,32–33 and hybridizing specific probes for tRNALys, tRNAGlu, and tRNAGln. In this system, the mercuric compound can specifically interact with the tRNAs containing the thiocarbonyl group—such as tRNALys, tRNAGlu, and tRNAGln—thereby retarding tRNA migration.
As can be seen in figure 4, the 2-thiouridylation levels of tRNALys, tRNAGlu, and tRNAGln were reduced significantly in mutant cells carrying the A1555G and the homozygous A10S mutations, compared with control cells. In particular, decreases of 35% and 16% of 2-thiouridylation and tRNALys, respectively, were observed in mutant cells carrying the homozygous and heterozygous A10S mutation, in comparison with wild-type cells. Furthermore, reductions of ∼13% and ∼6% in 2-thiouridylation of tRNAGlu were found in mutant cells carrying the A1555G mutation and the homozygous or heterozygous A10S mutation, respectively, as compared with that of wild-type cells. Similarly, a reduction of ∼13% of 2-thiouridylation of tRNAGln was observed in mutant cells carrying the A10S mutation (both homozygous and heterozygous), as compared with wild-type cells. As expected, the levels of 2-thiouridylation of tRNALys, tRNAGlu, and tRNAGln in mutant cells carrying the A1555G mutation but lacking the A10S mutation are comparable with those in controls. These data strongly suggest that the A10S mutation leads to defects in 2-thiouridylation of mt tRNALys, tRNAGlu, and tRNAGln.
Figure 4. .
Quantification of 2-thiouridine modification in mt tRNALys, tRNAGlu, and tRNAGln. A, The 3′ end-labeled mt tRNALys from each lymphoblastoid cell line, analyzed by 10% polyacrylamide gel electrophoresis with (+) or without (−) APM. The retarded bands of 2-thiolated tRNAs and nonretarded bands of tRNA without thiolation are marked with arrows. B, The 3′ end-labeled mt tRNALys, tRNAGlu, and tRNAGln from mutant lymphoblastoid cell lines carrying the A10S mutation, analyzed by 10% polyacrylamide gel electrophoresis with (+) or without (−) APM. C, Proportion in vivo of the 2-thiouridine modification levels of mt tRNAs. The calculations were based on three independent determinations of each mt tRNA in each cell line. The error bars indicate 2 SEMs; P indicates the significance, according to Student's t test, of the difference between mutant and control values for each mt tRNA.
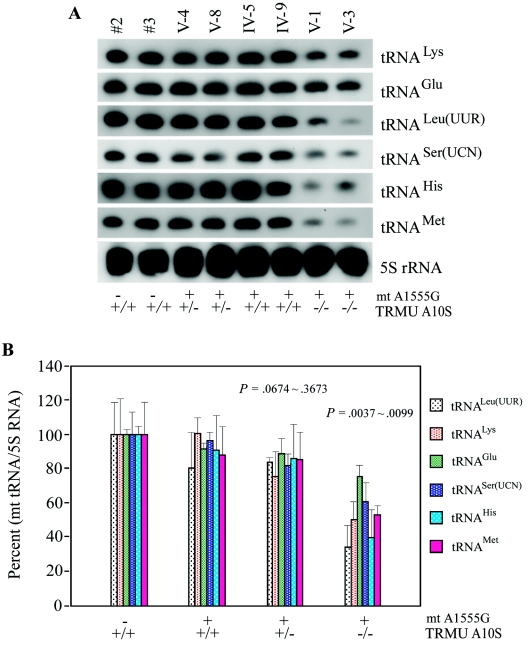
A10S Mutation Leads to a Decrease of Steady-State Level of mt tRNAs
As illustrated in figure 5A, the amounts of tRNALys, tRNAGlu, tRNALeu(UUR), tRNASer(UCN), tRNAMet, and tRNAHis were markedly decreased in mutant cells carrying both the A1555G and the homozygous TRMU A10S mutations, compared with control cells. For comparison, the average levels of each tRNA in the various control cell lines or mutant cell lines were then normalized to the average levels in the same cell lines for the reference 5S RNA, a nuclear-encoded mitochondrial small RNA.35 As shown in figure 5B, the average steady-state levels of those tRNAs were significantly decreased in the mutant cell lines carrying both A1555G and homozygous A10S mutations, relative to the controls (P=.0037–.0099). In particular, the average levels of tRNAs in those mutant cells are 50% in the tRNALys, 75% in tRNAGlu, 34% in the tRNALeu(UUR), 60% in tRNASer(UCN), 40% in tRNAHis, and 53% in tRNAMet of control levels. Strikingly, the variations in the steady-state level of tRNALys and tRNAGlu among the individual control and mutant cells carrying the A1555G and the homozygous A10S mutations were correlated with the levels of 2-thiouridylation of tRNALys (r=0.96; P<.001) and tRNAGlu (r=0.97; P<.001). However, there is no significant reduction in the average levels of mt tRNAs in mutant cells carrying both A1555G and heterozygous A10S mutations relative to controls (P=.0674–.3673): 78% in the tRNALys, 88% in the tRNAGlu, 84% in the tRNALeu(UUR), 82% in the tRNASer(UCN), 86% in the tRNAHis, and 85% in the tRNAMet of control levels. Furthermore, the average levels of those tRNAs in mutant cell lines carrying the A1555G mutation but lacking A10S mutation were comparable to those of control cell lines. In addition, the average levels of each tRNA in the various control cell lines or mutant cell lines were also normalized to the average levels in the same cell lines for the mitochondrial 12S rRNA.9 In fact, the reduction in the average steady-state levels of those tRNAs in the mutant cell lines carrying both A1555G and homozygous A10S mutations relative to the controls were comparable with those detected using 5S RNA as reference marker (data not shown). These data indicate that the homozygous A10S mutation leads to a defect in mt tRNA metabolism, thus causing a marked reduction in the steady-state level of mt tRNAs.
Figure 5. .
A, Northern-blot analysis of mt tRNA. Equal amounts (5 μg) of total mtRNA samples from the various cell lines were electrophoresed through a denaturing polyacrylamide gel, were electroblotted, and were hybridized with DIG-labeled oligonucleotide probes specific for the mt tRNALys, tRNAGlu, tRNALeu(UUR), tRNASer(UCN), tRNAHis, and tRNAMet; the blots were then stripped and rehybridized with DIG-labeled 5S rRNA probe as a control. B, Quantification of the levels of mt tRNA. Average relative tRNALys, tRNAGlu, tRNALeu(UUR), tRNASer(UCN), tRNAHis, and tRNAMet content per cell, was normalized to the average content per cell of 5S RNA in the two control cell lines and in the six mutant cell lines. The values for the latter are expressed as percentages of the average values for the control cell lines. The calculations were based on three independent determinations of each mt tRNA in each cell line. The error bars indicate 2 SEMs; P indicates the significance, according to Student's t test, of the difference between mutant and control values for each mt tRNA.
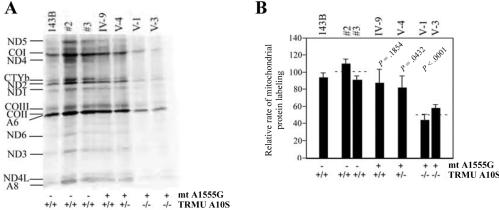
Mitochondrial-Protein Synthesis Defect in the Lymphoblastoid Cell Lines
Figure 6A shows typical electrophoretic patterns of the organelle-specific translation products of the mutant and control lymphoblastoid cell lines. The patterns of the mtDNA-encoded polypeptides of the mutation-carrying lymphoblastoid cell lines were qualitatively identical, in terms of electrophoretic mobility of the various polypeptides, to those of two control lymphoblastoid cell lines and of 143B.TK− (fig. 6A). However, the lymphoblastoid cell lines carrying the mutations showed a clear tendency toward decrease in the total rate of labeling of mitochondrial translation products relative to control cell lines. Figure 6B illustrates a quantification of the results of a large number of labeling experiments and electrophoretic runs, which was performed by densitometric analysis of appropriate exposures of the fluorograms and normalization to the data obtained for the 143B.TK− sample included in each gel. In particular, the decrease in the rate of mitochondrial-protein labeling in cell lines V-1 and V-3, carrying both A1555G and homozygous A10S mutations, relative to the controls had a range of 44%–57%, with an average of 51%. Furthermore, the rate of mitochondrial-protein labeling in cell line V-4, which carried both A1555G and heterozygous A10S mutations, revealed a mild reduction (81%) relative to controls. Moreover, the rate of mitochondrial-protein labeling in mutant cell line IV-9, carrying only A1555G mutation but lacking A10S mutation, was 87% of control cell lines. These data clearly indicate that the homozygous A10S mutation leads to a defect in mitochondrial-protein labeling.
Figure 6. .
Mitochondrial protein–labeling analysis. A, Electrophoretic patterns of the mitochondrial translation products. The lymphoblastoid cell lines and of 143B.TK− cells were labeled for 30 min with [35S]methionine in the presence of 100 μg of emetine per ml, an inhibitor for cytosolic protein synthesis. Samples containing equal amounts of protein (30 μg) were run in SDS/polyacrylamide gradient gels. COI, COII, and COIII, subunits I, II, and III of cytochrome c oxidase; ND1, ND2, ND3, ND4, ND4L, ND5, and ND6, subunits 1, 2, 3, 4, 4L, 5, and 6 of the respiratory-chain NADH dehydrogenase; A6 and A8, subunits 6 and 8 of the H+-ATPase; CYTb, apocytochrome b. B, Quantification of the rates of labeling of the mitochondrial translation products in lymphoblastoid cell lines. The rates of mitochondrial-protein labeling, determined as described elsewhere,9 are expressed as percentages of the value for 143B.TK− in each gel, with error bars indicating 2 SEMs. Three independent labeling experiments and three or four electrophoretic analyses of each labeled preparation were performed on each lymphoblastoid cell line. The horizontal dashed lines represent the average value for each group. P indicates the significance, according to Student's t test, of the difference between mutant and control values for each mt tRNA.
Discussion
In this study, we identified and characterized the nuclear-modifier gene TRMU for the phenotypic expression of deafness-associated mitochondrial 12S rRNA mutations. In fact, TRMU encodes a highly conserved 5-methylaminomethyl-2-thiouridylate-methyltransferase responsible for the biosynthesis of 5-taurinomethyl-2-thiouridine (τm5s2U) of mt tRNALys, tRNAGlu, and tRNAGln in the wobble position.24,39 The τm5s2U is further modified to mnm5s2U34 in the same position of those tRNAs in E. coli and human mitochondria.24,40 This modified nucleotide contributes to the high fidelity of codon recognition and the structural formation and stabilization of functional tRNAs.23 We showed that isolated human TRMU cDNA partially restored the respiratory-deficient phenotype of yeast Mto2 cells carrying the C1409G mutation,17 and there was highly suggestive linkage and linkage disequilibrium between microsatellite markers adjacent to TRMU and the presence of deafness.26 This led us to test whether TRMU acts as a modifier gene that modulates the phenotypic expression of the deafness-associated A1555G or C1494T mutation in mitochondrial 12S rRNA.
Mutation analysis identified a single missense mutation (A10S) in TRMU in some members of the Arab-Israeli family and other European families. The A10S mutation locates at the evolutionarily conserved N-terminal region of the TRMU protein. This mutation occurs in 10% of Jewish and white populations lacking the A1555G mutation, whereas the frequency of this variant was ∼25% in both the Arab-Israeli and the European cohorts carrying the A1555G mutation but not in either the deaf or normal-hearing Chinese cohorts. These findings imply that the A10S mutation, even in a homozygous form, is not sufficient to cause a clinical phenotype. However, all 18 matrilineal relatives carrying both the homozygous TRMU A10S and mtDNA A1555G mutations exhibited prelingual profound deafness. This strongly indicates that the coexistence of the homozygous TRMU A10S variant with the 12S rRNA A1555G mutation led to the expression of deafness in these subjects. However, the observation that other symptomatic individuals in these families carried heterozygous A10S mutation or lacked this mutation suggests that other putative modifier genes may contribute to the phenotypic manifestation of the A1555G mutation. In these cases, the deafness phenotype caused by the interaction between the TRMU A10S mutation and mtDNA A1555G mutation did not have a significant linkage and did not well fit the typical correlation of genotype and phenotype caused by a disease with the single-gene inheritance. Therefore, strong functional evidences are necessary for understanding the pathogenesis of diseases with a complex inheritance of multiple factors.
The A10S mutation, despite localizing at the putative mitochondrial targeting sequence,26 does not affect the import of this human TRMU protein into mitochondria but led to an alteration of TRMU enzymatic function. Indeed, the homozygous A10S mutation caused a significant defect in 2-thio modification in the wobble position of mt tRNALys, tRNAGlu, and tRNAGln. These findings are consistent with the fact that the small interfering RNA down-regulation of human TRMU led to the defect in 2-thiouridylation24 in mt tRNALys, tRNAGlu, and tRNAGln. A failure in 2-thio modification likely results in a large proportion of unmodified mt tRNAs. Those unmodified tRNAs caused by the A10S mutation likely leave the tRNAs more exposed to degradation, thereby lowering the steady-state level of those tRNAs. Indeed, decreases of 50% and ∼25% in the steady-state levels of tRNALys and tRNAGlu, respectively, in those mutant cells is significantly correlated with the degree of decreases in the 2-thiouridylation between tRNALys and tRNAGlu. Furthermore, the steady-state levels of mt tRNALeu(UUR), tRNASer(UCN), tRNAMet, and tRNAHis were also decreased significantly in cells carrying both the homozygous A10S and A1555G mutations. In fact, modifications other than the 2-thio modification occur in the wobble position of those mt tRNAs, such as 5-taurinomethyluridine40 in the tRNALeu(UUR) and 5-formylcytidine41 in the tRNAMet. Thus, the decreased TRMU activity caused by the A10S mutation likely leads to transcriptional/translational defects and thereby reduces the steady-state levels of those tRNAs. Alternatively, these unmodified tRNAs may not function accurately with the altered mitochondrial ribosomes carrying the A1555G mutation and especially affect the efficiency and accuracy of codon-anticodon interaction. As a result, the mutated TRMU, by functionally interacting with the decoding region of small rRNA, particularly in the site of the A1555G mutation, affects the translational efficiency and accuracy of codon-anticodon base pairings in the mitochondrial ribosomes, thereby leading to the impairment in mitochondrial-protein synthesis. In fact, it has been suggested that the wobble modification defect is the primary factor for dispossessing the mutant tRNALys of its cognate codon–binding affinity, forcing the mutant tRNALys(UUU) to become translationally inactive, which subsequently results in mitochondrial dysfunction.42 Thus, a failure in tRNA metabolism, caused by mutated TRMU, most probably leads to a reduction in the overall rate of mitochondrial translation but is not sufficient to produce a clinical phenotype, as in the case of the yeast mto2 mutant strains.17
The A1555G and the C1494T mutations locate at the aminoacyl-tRNA decoding site (A site) of the mitochondrial ribosome where the codon-anticodon interaction occurs43,44 and aminoglycoside antibiotics interact.45–47 These nucleotides are adjacent to the A-site tRNA-binding bases, including A1408, A1492, and A1493 of bacterial 16S rRNA.48 Therefore, the A1555G or the C1494T mutation may result in a local conformational change in the A-site of 12S rRNA, thereby affecting the efficiency and accuracy of mitochondrial translation. Hypermodified tRNAs, synthesized by the participation of TRMU, MTO1, and GTPBP3, are less efficient for the decoding of the codon ending in G than of the codon ending in A of small rRNA.17,49 Indeed, the A1555G or the C1494T mutation produces an ∼30% or an ∼40% decrease in the rate of mitochondrial translation, respectively.9,10,50 However, an ∼50% decrease in the rate of mitochondrial translation responsible for significant respiratory defects was proposed as the threshold level that produces the deafness phenotype associated with the A1555G or the C1494T mutation.9,50 Furthermore, the new G-C or U-A base-pairing, created by the A1555G or the C1494T mutation, in the A site of 12S rRNA facilitates the binding of aminoglycosides,47,51 thus causing sensitivity to those drugs.3,8,9,11,50 The exposure to aminoglycosides yielded an additional 30% decrease in the rate of mitochondrial-protein synthesis in cells carrying the A1555G mutation.11 In fact, aminoglycosides are concentrated in the perilymph and endolymph of the inner ear but are rapidly cleared in other tissues or organs.52 Therefore, a 50%–60% decrease in the rate of mitochondrial translation, caused by the combination of the A1555G mutation with aminoglycosides, leads to cell dysfunction or death of the auditory system, thereby inducing or worsening hearing loss.
Unmodified tRNAs, caused by the TRMU A10S mutation, may be much less efficient in binding to the A-site nucleotides of mitochondrial ribosome,53 thus leading to a defect in translation, as in the case of E. coli tRNALys(UUU). Those unmodified tRNA caused by the A10S mutation may interact very faultily with mitochondrial ribosomes carrying the A1555G or the C1494T mutation, thereby worsening defects in mitochondrial translation associated with the A1555G or the C1494T mutation. In S. cerevisiae, the 15S rRNA C1409G mutation, in conjunction with the mto2 mutation, led to nearly complete loss of mitochondrial-protein synthesis.17 Here, a failure in tRNA metabolism, caused by the homozygous TRMU A10S mutation, accounted for a >20% decrease in the rate of mitochondrial-protein synthesis. Indeed, the ∼50% decrease in the rate of mitochondrial translation observed in cells derived from symptomatic individuals V-1 and V-3 of the Arab-Israeli family was the consequence of a faulty interaction between unmodified tRNAs caused by the TRMU A10S mutation and mitochondrial ribosomes carrying the 12S rRNA A1555G mutation. Defects in the mitochondrial translation consequently led to a respiratory phenotype and a decline in ATP production below the threshold level required for normal cell function in the auditory organs, including cochlea, thus producing the deafness phenotype.
In summary, our study has identified the nuclear-modifier gene TRMU for the phenotypic expression of deafness-associated mitochondrial 12S rRNA mutations. A point mutation (A10S) in TRMU results in the defect in 2-thio modification in mt tRNAs, which leads to decreases of the steady-state level of mt tRNAs, subsequently causing the impairment of mitochondrial translation. Resultant biochemical defects aggravate the mitochondrial dysfunction below the threshold for normal cell function, thereby expressing the deafness phenotype. Therefore, the mutated TRMU acts in synergy with the 12S rRNA A1555G mutation, modulating the phenotypic manifestation.
Acknowledgments
This work was supported by Public Health Service grants RO1DC05230 and RO3DC04958 from the National Institute on Deafness and Other Communication Disorders and RO1NS44015 from the National Institute of Neurological Disorders and Stroke (to M.-X.G.) and grant RO1DC01402 from the National Institute on Deafness and Other Communication Disorders (to N.F.-G.) and by grants from Fundacion Ramon Areces and Programa Ramon y Cajal (to I.d.C.). We thank Drs. Gregory Grabowski, John Greinwald, and Richard Wenstrup, for control and deaf DNA samples, and Dr. Fernandez-Silva, for suggestion of mitochondrial-import experiments. We thank Professor Des Clark-Walker for critical comments of this manuscript. We are grateful to Chuck Loftice, William Gibbson, and Li Yang for skilled technical assistance.
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the complete TRMU sequences [accession number AF448221])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for deafness-associated mutations in 12S rRNA)
References
- 1.Guan MX (2004) Molecular pathogenetic mechanism of maternally inherited deafness. Ann N Y Acad Sci 1011:259–271 10.1196/annals.1293.025 [DOI] [PubMed] [Google Scholar]
- 2.Fischel-Ghodsian N (1999) Mitochondrial deafness mutations reviewed. Hum Mut 13:261–270 [DOI] [PubMed] [Google Scholar]
- 3.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N (1993) Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 4:289–294 10.1038/ng0793-289 [DOI] [PubMed] [Google Scholar]
- 4.Estivill X, Govea N, Barceló A, Perelló E, Badenas C, Romero E, Moral L, Scozzari R, D’Urbano L, Zeviani M, Torroni A (1998) Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet 62:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, Xing G, Yan M, Cao X, Liu XZ, Bu X, Guan MX (2004) Cosegregation of C-insertion at position 961 with A1555G mutation of mitochondrial 12S rRNA gene in a large Chinese family with maternally inherited hearing loss. Am J Med Genet A 124:113–117 10.1002/ajmg.a.20305 [DOI] [PubMed] [Google Scholar]
- 6.Young WY, Zhao L, Qian Y, Wang Q, Li N, Greinwald JH Jr, Guan MX (2005) Extremely low penetrance of hearing loss in four Chinese families with the mitochondrial 12S rRNA A1555G mutation. Biochem Biophys Res Commun 328:1244–1251 10.1016/j.bbrc.2005.01.085 [DOI] [PubMed] [Google Scholar]
- 7.Matthijs G, Claes S, Longo-Bbenza B, Cassiman J-J (1996) Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet 4:46–51 [DOI] [PubMed] [Google Scholar]
- 8.Zhao H, Li R, Wang Q, Yan Q, Deng J-H, Han D, Bai Y, Young W-Y, Guan M-X (2004) Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet 74:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan MX, Fischel-Ghodsian N, Attardi G (1996) Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet 5:963–997 10.1093/hmg/5.7.963 [DOI] [PubMed] [Google Scholar]
- 10.Guan MX, Fischel-Ghodsian N, Attardi G (2001) Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet 10:573–580 10.1093/hmg/10.6.573 [DOI] [PubMed] [Google Scholar]
- 11.Guan MX, Fischel-Ghodsian N, Attardi G (2000) A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet 9:1787–1793 10.1093/hmg/9.12.1787 [DOI] [PubMed] [Google Scholar]
- 12.Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, Cantor RM, Aouizerat B, Bu X, Rotter JI, Jaber L, Fischel-Ghodsian N (1998) Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet 77:421–426 [DOI] [PubMed] [Google Scholar]
- 13.Bykhovskaya Y, Estivill X, Taylor K, Hang T, Hamon M, Casano RAMS, Yang H, Rotter JI, Shohat M, Fischel-Ghodsian N (2000) Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am J Hum Genet 66:1905–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bykhovskaya Y, Mengesha E, Wang D, Yang H, Estivill, X, Shohat M, Fischel-Ghodsian N (2004) Human mitochondrial transcription factor B1 as a modifier gene for hearing loss associated with the mitochondrial A1555G mutation. Mol Genet Metab 82:27–32 10.1016/j.ymgme.2004.01.020 [DOI] [PubMed] [Google Scholar]
- 15.Li X, Guan MX (2002) A human mitochondrial GTP binding protein related to tRNA modification may modulate the phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol Cell Biol 22:7701–7711 10.1128/MCB.22.21.7701-7711.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Li R, Lin X, Guan MX (2002) Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of the deafness-associated mitochondrial 12S rRNA A1555G mutation. J Biol Chem 277:27256–27264 10.1074/jbc.M203267200 [DOI] [PubMed] [Google Scholar]
- 17.Yan Q, Li X, Faye G, Guan MX (2005) Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15S rRNA. J Biol Chem 280:29151–29157 10.1074/jbc.M504247200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decoster E, Vassal A, Faye G (1993) MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J Mol Biol 232:79–88 10.1006/jmbi.1993.1371 [DOI] [PubMed] [Google Scholar]
- 19.Colby G, Wu M, Tzagoloff A (1998) MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem 273:27945–27952 10.1074/jbc.273.43.27945 [DOI] [PubMed] [Google Scholar]
- 20.Cabedo H, Macian F, Villarroya M, Escudero JC, Martinez-Vicente M, Knecht E, Armengod ME (1999) The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J 18:7063–7076 10.1093/emboj/18.24.7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brégeon D, Colot V, Miroslav M, Radman M, Taddei F (2001) Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev 15:2295–2306 10.1101/gad.207701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kambampati R, Lauhon CT (2003) MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42:1109–1117 10.1021/bi026536+ [DOI] [PubMed] [Google Scholar]
- 23.Björk GR (1996) Stable RNA modification. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low BK, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC, pp 861–886 [Google Scholar]
- 24.Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K, Suzuki T (2005) Mitochodnria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs: implications for the molecular pathogenesis of human mitochondrial diseases. J Biol Chem 280:1613–1624 10.1074/jbc.M409306200 [DOI] [PubMed] [Google Scholar]
- 25.Sullivan MA, Cannon JF, Webb FH, Bock RM (1985) Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J Bacteriol 161:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan Q, Bykhovskaya Y, Li R, Mengesha E, Shohat M, Estivill X, Fischel-Ghodsian N, Guan MX (2006) Human TRMU encoding the mitochondrial 5-methylaminomethyl-2-thiouridylate-methyltransferase is a putative nuclear modifier gene for the phenotypic expression of the deafness-associated 12S rRNA mutations. Biochem Biophys Res Commun 342:1130–1136 10.1016/j.bbrc.2006.02.078 [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, Xiong S, Heman-Ackah S, Wu J, Choo DI, Guan MX (2005) Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet 117:9–15 10.1007/s00439-005-1276-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, Arellano B, Gallo-Teran J, Morales-Angulo C, Ramirez-Camacho R, Cruz Tapia M, Solanellas J, Martinez-Conde A, Villamar M, Moreno-Pelayo MA, Moreno F, del Castillo I (2003) Heteroplasmy for the 1555A→G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet 40:632–636 10.1136/jmg.40.8.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G (1997) The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J 16:1066–1079 10.1093/emboj/16.5.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King MP, Attardi G (1993) Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J Biol Chem 268:10228–10237 [PubMed] [Google Scholar]
- 31.Schneider A, McNally KP, Agabian N (1994) Nuclear-encoded mitochondrial tRNAs of Trypanosoma brucei have a modified cytidine in the anticodon. loop. Nucleic Acids Res 22:3699–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigi N, Suzuki T, Tamakoshi M, Oshima T, Watanabe K (2002) Conserved bases in the TΨC loop of trna are determinants for thermophile-specific 2-thiouridylation at position 54. J Biol Chem 277:39128–39135 10.1074/jbc.M207323200 [DOI] [PubMed] [Google Scholar]
- 33.Igloi GL (1988) Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial: probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27:3842–3849 10.1021/bi00410a048 [DOI] [PubMed] [Google Scholar]
- 34.Anderson S, Bankier AT, Barrell BG, deBruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Rose BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young I (1982) Comparison of the human and bovine mitochondrial genomes. In: Slonimski P, Borst P, Attardi G (eds) Mitochondrial genes. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp 5–43 [Google Scholar]
- 35.Magalhaes PJ, Andreu AL, Schon EA (1998) Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell 9:2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX (2004) Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res 32:867–877 10.1093/nar/gkh226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan MX, Enriquez JA, Fischel-Ghodsian N, Puranam R, Lin CP, Marion MA, Attardi G (1998) The Deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol Cell Biol 18:5868–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chomyn A (1996) In vivo labeling and analysis of human mitochondrial translation products. Meth Enzymol 264:197–211 [DOI] [PubMed] [Google Scholar]
- 39.Yan Q, Guan MX (2004) Identification and characterization of mouse TRMU gene encoding themitochondrial 5-methylaminomethyl-2-thiouridylate-methyltransferase. Biochim Biophys Acta 1676:119–126 [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T, Suzuki T, Wada T, Saigo K, Watanabe K (2002) Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J 21:6581–6589 10.1093/emboj/cdf656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, Hayashi N, Yokoyama S, Watanabe K (1994) A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry 33:2234–2239 10.1021/bi00174a033 [DOI] [PubMed] [Google Scholar]
- 42.Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K (2001) Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease EMBO J 20:4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann RA, Thomas CL, Wower J (1990) Structure and function of rRNA in the decoding domain and at the peptidyltransferase center. In: Hill WE, Moore PB, Dahlberg A, Schlessinger D, Garrett RA, Warner JR (eds) The ribosome: structure, function and evolution. American Society for Microbiology, Washington, DC, pp 331–347 [Google Scholar]
- 44.Neefs JM, Van de Peer Y, De Rijk P, Goris A, De Wachter R (1991) Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res Suppl 19:1987–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purohit P, Stern S (1994) Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature 370:659–662 10.1038/370659a0 [DOI] [PubMed] [Google Scholar]
- 46.Moazed D, Noller HF (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389–394 10.1038/327389a0 [DOI] [PubMed] [Google Scholar]
- 47.Hamasaki K, Rando RR (1997) Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism, which causes aminoglycoside-induced deafness. Biochemistry 36:12323–12328 10.1021/bi970962r [DOI] [PubMed] [Google Scholar]
- 48.Green R, Noller HF (1997) Ribosomes and translation. Annu Rev Biochem 66:679–716 10.1146/annurev.biochem.66.1.679 [DOI] [PubMed] [Google Scholar]
- 49.Weiss-Brummer B, Huttenhofer A (1989) The paromomycin resistance mutation (PARR454) in the 15S rRNA gene of the yeast Saccharomyces cerevisiae is involved in ribosomal frameshifting. Mol Gen Genet 217:362–369 [DOI] [PubMed] [Google Scholar]
- 50.Zhao H, Young WY, Yan Q, Li R, Cao J, Wang Q, Li X, Peters JL, Han D, Guan MX (2005) Functional characterization of the mitochondrial 12S rRNA C1494T mutation associated with aminoglycoside-induced and nonsyndromic hearing loss. Nucleic Acid Res 33:1132–1139 10.1093/nar/gki262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Stasio EA, Dahlberg AE (1990) Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16S ribosomal RNA. J Mol Biol 212:127–133 10.1016/0022-2836(90)90309-A [DOI] [PubMed] [Google Scholar]
- 52.Henley CM, Schacht J (1988) Pharmacokinetics of aminoglycoside antibiotics in blood, inner-ear fluids and tissues and their relationship to ototoxicity. Audiology 27:137–146 [DOI] [PubMed] [Google Scholar]
- 53.Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem 277:16391–16395 10.1074/jbc.M200253200 [DOI] [PubMed] [Google Scholar]